Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
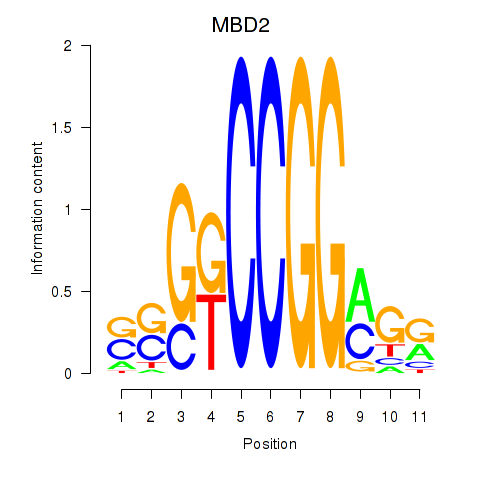
Results for MBD2
Z-value: 0.57
Transcription factors associated with MBD2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MBD2
|
ENSG00000134046.7 | methyl-CpG binding domain protein 2 |
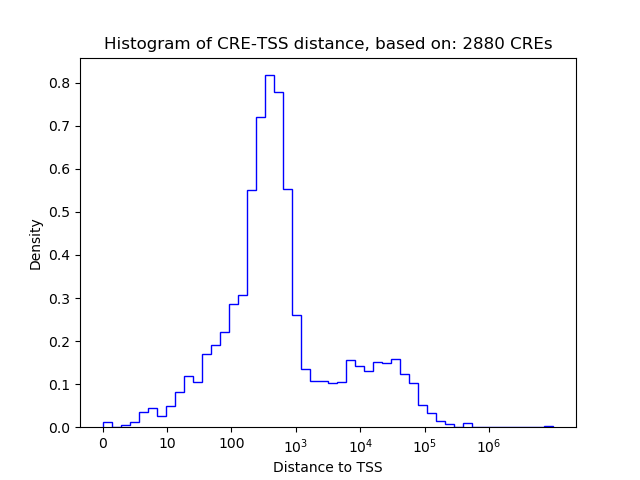
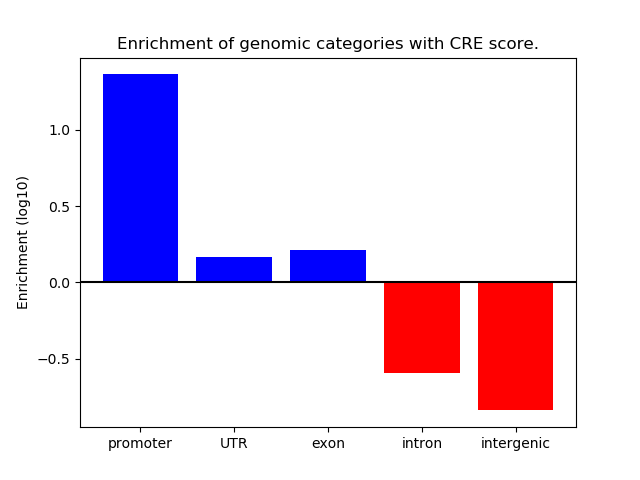
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_51751599_51751750 | MBD2 | 516 | 0.785603 | -0.85 | 3.8e-03 | Click! |
| chr18_51766045_51766196 | MBD2 | 14962 | 0.186217 | 0.38 | 3.1e-01 | Click! |
| chr18_51749801_51749975 | MBD2 | 1099 | 0.380751 | -0.32 | 4.0e-01 | Click! |
| chr18_51750360_51750511 | MBD2 | 552 | 0.724562 | -0.13 | 7.3e-01 | Click! |
| chr18_51751166_51751526 | MBD2 | 188 | 0.944061 | -0.13 | 7.3e-01 | Click! |
Activity of the MBD2 motif across conditions
Conditions sorted by the z-value of the MBD2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
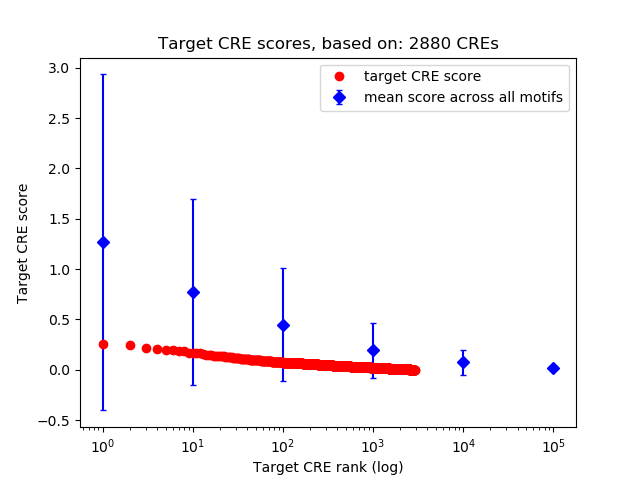
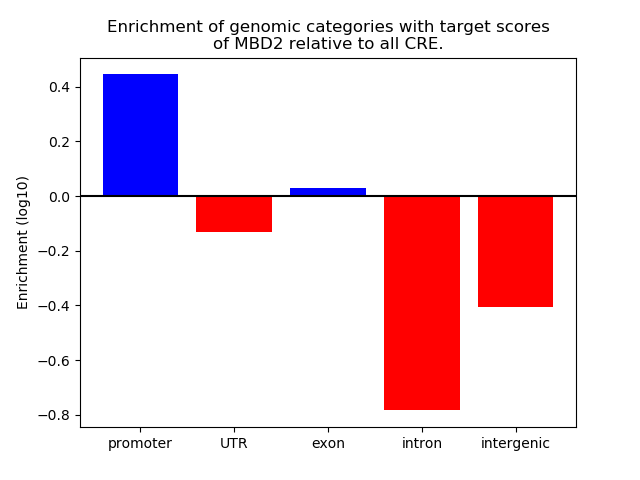
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_80189068_80189889 | 0.26 |
SLC16A3 |
solute carrier family 16 (monocarboxylate transporter), member 3 |
630 |
0.57 |
| chr6_166721057_166722024 | 0.25 |
PRR18 |
proline rich 18 |
331 |
0.9 |
| chr4_74809740_74810515 | 0.22 |
PF4 |
platelet factor 4 |
37714 |
0.11 |
| chr3_36985452_36986747 | 0.20 |
TRANK1 |
tetratricopeptide repeat and ankyrin repeat containing 1 |
449 |
0.83 |
| chr12_3308931_3310293 | 0.19 |
TSPAN9 |
tetraspanin 9 |
729 |
0.75 |
| chr3_142681892_142682427 | 0.19 |
PAQR9 |
progestin and adipoQ receptor family member IX |
19 |
0.84 |
| chr20_43729394_43729952 | 0.19 |
KCNS1 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 |
77 |
0.96 |
| chr21_44496501_44497065 | 0.18 |
CBS |
cystathionine-beta-synthase |
270 |
0.91 |
| chrX_48748285_48749195 | 0.17 |
TIMM17B |
translocase of inner mitochondrial membrane 17 homolog B (yeast) |
6290 |
0.1 |
| chr2_27485345_27486489 | 0.17 |
SLC30A3 |
solute carrier family 30 (zinc transporter), member 3 |
178 |
0.89 |
| chr3_196594834_196595961 | 0.17 |
SENP5 |
SUMO1/sentrin specific peptidase 5 |
581 |
0.72 |
| chr10_94449633_94450257 | 0.16 |
HHEX |
hematopoietically expressed homeobox |
898 |
0.6 |
| chr3_192126971_192127690 | 0.15 |
FGF12 |
fibroblast growth factor 12 |
492 |
0.87 |
| chr18_70210992_70211675 | 0.15 |
CBLN2 |
cerebellin 2 precursor |
441 |
0.91 |
| chr7_128171824_128173030 | 0.15 |
RP11-274B21.1 |
|
1279 |
0.43 |
| chr5_149864820_149865733 | 0.14 |
NDST1 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 |
105 |
0.97 |
| chr10_98592747_98593792 | 0.14 |
LCOR |
ligand dependent nuclear receptor corepressor |
532 |
0.78 |
| chr3_39194337_39194962 | 0.14 |
CSRNP1 |
cysteine-serine-rich nuclear protein 1 |
417 |
0.84 |
| chr15_56035451_56035609 | 0.14 |
PRTG |
protogenin |
242 |
0.94 |
| chr17_78234751_78235551 | 0.14 |
RNF213 |
ring finger protein 213 |
482 |
0.76 |
| chr4_71570546_71572213 | 0.13 |
RUFY3 |
RUN and FYVE domain containing 3 |
861 |
0.36 |
| chr5_74907170_74908023 | 0.13 |
ANKDD1B |
ankyrin repeat and death domain containing 1B |
312 |
0.9 |
| chr2_185463143_185463634 | 0.13 |
ZNF804A |
zinc finger protein 804A |
295 |
0.95 |
| chr13_29068702_29069490 | 0.13 |
FLT1 |
fms-related tyrosine kinase 1 |
136 |
0.98 |
| chr17_73584494_73585142 | 0.13 |
MYO15B |
myosin XVB pseudogene |
2040 |
0.21 |
| chrX_39964334_39965624 | 0.13 |
BCOR |
BCL6 corepressor |
8323 |
0.32 |
| chr1_180881914_180882730 | 0.12 |
KIAA1614 |
KIAA1614 |
3 |
0.98 |
| chr3_195384928_195385957 | 0.12 |
SDHAP2 |
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 2 |
475 |
0.81 |
| chr15_99192776_99194432 | 0.12 |
IGF1R |
insulin-like growth factor 1 receptor |
1331 |
0.47 |
| chr17_20491916_20492779 | 0.12 |
CDRT15L2 |
CMT1A duplicated region transcript 15-like 2 |
9310 |
0.21 |
| chr16_691284_692401 | 0.12 |
FAM195A |
family with sequence similarity 195, member A |
29 |
0.92 |
| chr2_160653969_160654659 | 0.11 |
CD302 |
CD302 molecule |
439 |
0.87 |
| chr22_20003723_20004755 | 0.11 |
ARVCF |
armadillo repeat gene deleted in velocardiofacial syndrome |
92 |
0.75 |
| chr21_37529152_37530122 | 0.11 |
DOPEY2 |
dopey family member 2 |
557 |
0.57 |
| chr14_100706520_100706671 | 0.11 |
YY1 |
YY1 transcription factor |
499 |
0.64 |
| chr22_29601932_29602692 | 0.11 |
EMID1 |
EMI domain containing 1 |
400 |
0.8 |
| chr8_144512945_144513817 | 0.11 |
MAFA |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A |
805 |
0.48 |
| chr5_134825154_134825603 | 0.11 |
TIFAB |
TRAF-interacting protein with forkhead-associated domain, family member B |
37289 |
0.11 |
| chr18_77558508_77558774 | 0.10 |
KCNG2 |
potassium voltage-gated channel, subfamily G, member 2 |
65027 |
0.12 |
| chr18_11149170_11149978 | 0.10 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
987 |
0.72 |
| chr7_2394785_2395825 | 0.10 |
EIF3B |
eukaryotic translation initiation factor 3, subunit B |
788 |
0.53 |
| chr10_102278895_102279852 | 0.10 |
SEC31B |
SEC31 homolog B (S. cerevisiae) |
218 |
0.92 |
| chr11_62455448_62455813 | 0.10 |
LRRN4CL |
LRRN4 C-terminal like |
1741 |
0.12 |
| chr11_119019412_119020504 | 0.10 |
ABCG4 |
ATP-binding cassette, sub-family G (WHITE), member 4 |
5 |
0.94 |
| chr3_141379125_141379497 | 0.10 |
RNF7 |
ring finger protein 7 |
77735 |
0.09 |
| chr13_21634768_21635319 | 0.10 |
LATS2 |
large tumor suppressor kinase 2 |
614 |
0.69 |
| chr19_10982465_10983821 | 0.10 |
CARM1 |
coactivator-associated arginine methyltransferase 1 |
764 |
0.53 |
| chr3_197463352_197463883 | 0.10 |
KIAA0226 |
KIAA0226 |
27 |
0.88 |
| chr2_8818889_8820001 | 0.10 |
AC011747.7 |
|
259 |
0.7 |
| chrX_36975622_36976198 | 0.10 |
FAM47C |
family with sequence similarity 47, member C |
50522 |
0.19 |
| chrX_53349448_53350467 | 0.10 |
IQSEC2 |
IQ motif and Sec7 domain 2 |
565 |
0.73 |
| chr15_69113585_69114214 | 0.10 |
ANP32A |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member A |
665 |
0.75 |
| chr11_63535609_63536120 | 0.09 |
ENSG00000264519 |
. |
29679 |
0.13 |
| chr20_3073179_3074190 | 0.09 |
ENSG00000263905 |
. |
1177 |
0.34 |
| chr1_159750747_159751482 | 0.09 |
DUSP23 |
dual specificity phosphatase 23 |
321 |
0.85 |
| chr18_74843262_74844680 | 0.09 |
MBP |
myelin basic protein |
331 |
0.94 |
| chrX_119384747_119385998 | 0.09 |
ZBTB33 |
zinc finger and BTB domain containing 33 |
762 |
0.69 |
| chr12_6937518_6938631 | 0.09 |
LEPREL2 |
leprecan-like 2 |
502 |
0.55 |
| chr2_103235205_103235939 | 0.09 |
SLC9A2 |
solute carrier family 9, subfamily A (NHE2, cation proton antiporter 2), member 2 |
594 |
0.82 |
| chr12_14720104_14720509 | 0.09 |
RP11-695J4.2 |
|
378 |
0.73 |
| chr16_66638822_66640143 | 0.09 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
58 |
0.95 |
| chr5_42944152_42944647 | 0.09 |
SEPP1 |
selenoprotein P, plasma, 1 |
56905 |
0.11 |
| chr5_76506770_76507546 | 0.09 |
PDE8B |
phosphodiesterase 8B |
407 |
0.88 |
| chr1_17446030_17446253 | 0.09 |
PADI2 |
peptidyl arginine deiminase, type II |
193 |
0.94 |
| chr16_70221940_70222878 | 0.09 |
CLEC18C |
C-type lectin domain family 18, member C |
14349 |
0.1 |
| chr4_128543868_128544388 | 0.09 |
INTU |
inturned planar cell polarity protein |
298 |
0.94 |
| chr1_45008521_45008672 | 0.09 |
ENSG00000263381 |
. |
2569 |
0.31 |
| chr7_18125753_18126403 | 0.08 |
HDAC9 |
histone deacetylase 9 |
494 |
0.84 |
| chr1_36923329_36923682 | 0.08 |
MRPS15 |
mitochondrial ribosomal protein S15 |
6533 |
0.14 |
| chr6_85472920_85473300 | 0.08 |
TBX18 |
T-box 18 |
37 |
0.99 |
| chr20_49347136_49348386 | 0.08 |
PARD6B |
par-6 family cell polarity regulator beta |
320 |
0.89 |
| chr1_103573929_103574109 | 0.08 |
COL11A1 |
collagen, type XI, alpha 1 |
16 |
0.99 |
| chr20_32273512_32274295 | 0.08 |
E2F1 |
E2F transcription factor 1 |
307 |
0.81 |
| chr14_101034010_101035263 | 0.08 |
BEGAIN |
brain-enriched guanylate kinase-associated |
192 |
0.93 |
| chr8_142138821_142140144 | 0.08 |
RP11-809O17.1 |
|
578 |
0.46 |
| chr5_133861629_133862874 | 0.08 |
JADE2 |
jade family PHD finger 2 |
76 |
0.97 |
| chr9_77501796_77502826 | 0.08 |
TRPM6 |
transient receptor potential cation channel, subfamily M, member 6 |
48 |
0.97 |
| chr13_114822852_114823008 | 0.08 |
RASA3 |
RAS p21 protein activator 3 |
20508 |
0.23 |
| chr18_499765_500605 | 0.08 |
COLEC12 |
collectin sub-family member 12 |
537 |
0.79 |
| chr20_31351193_31351609 | 0.08 |
DNMT3B |
DNA (cytosine-5-)-methyltransferase 3 beta |
953 |
0.56 |
| chr11_68780491_68780695 | 0.08 |
MRGPRF |
MAS-related GPR, member F |
121 |
0.88 |
| chr20_39657787_39658990 | 0.08 |
TOP1 |
topoisomerase (DNA) I |
930 |
0.61 |
| chr13_27131903_27132772 | 0.08 |
WASF3 |
WAS protein family, member 3 |
450 |
0.89 |
| chr10_27547544_27548544 | 0.08 |
ARMC4P1 |
armadillo repeat containing 4 pseudogene 1 |
3842 |
0.17 |
| chr3_195635375_195636127 | 0.08 |
TNK2 |
tyrosine kinase, non-receptor, 2 |
204 |
0.83 |
| chr10_6186918_6187892 | 0.08 |
PFKFB3 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
508 |
0.72 |
| chr8_67624599_67625774 | 0.08 |
SGK3 |
serum/glucocorticoid regulated kinase family, member 3 |
533 |
0.76 |
| chr17_79268296_79269061 | 0.08 |
SLC38A10 |
solute carrier family 38, member 10 |
389 |
0.8 |
| chr7_92218956_92219793 | 0.08 |
FAM133B |
family with sequence similarity 133, member B |
10 |
0.98 |
| chr13_99227754_99228977 | 0.08 |
STK24 |
serine/threonine kinase 24 |
752 |
0.54 |
| chr1_53685385_53686250 | 0.08 |
C1orf123 |
chromosome 1 open reading frame 123 |
472 |
0.43 |
| chr17_38498589_38499681 | 0.08 |
CTD-2267D19.2 |
|
253 |
0.69 |
| chr16_85793355_85793506 | 0.08 |
C16orf74 |
chromosome 16 open reading frame 74 |
8695 |
0.11 |
| chr4_682374_683342 | 0.07 |
MFSD7 |
major facilitator superfamily domain containing 7 |
74 |
0.94 |
| chr18_12376105_12377115 | 0.07 |
AFG3L2 |
AFG3-like AAA ATPase 2 |
391 |
0.83 |
| chr5_1110808_1112337 | 0.07 |
SLC12A7 |
solute carrier family 12 (potassium/chloride transporter), member 7 |
578 |
0.75 |
| chr10_27148230_27149879 | 0.07 |
ABI1 |
abl-interactor 1 |
738 |
0.67 |
| chr17_2296252_2297077 | 0.07 |
MNT |
MAX network transcriptional repressor |
7734 |
0.11 |
| chr6_7051751_7052548 | 0.07 |
ENSG00000251762 |
. |
11370 |
0.24 |
| chr7_127671114_127671650 | 0.07 |
LRRC4 |
leucine rich repeat containing 4 |
259 |
0.94 |
| chr8_136470073_136470726 | 0.07 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
69 |
0.96 |
| chr1_40942691_40943841 | 0.07 |
ZFP69 |
ZFP69 zinc finger protein |
36 |
0.96 |
| chr17_73521710_73521861 | 0.07 |
LLGL2 |
lethal giant larvae homolog 2 (Drosophila) |
22 |
0.96 |
| chr17_1962412_1962588 | 0.07 |
HIC1 |
hypermethylated in cancer 1 |
2896 |
0.12 |
| chr2_243030793_243031816 | 0.07 |
AC093642.5 |
|
460 |
0.62 |
| chr17_79848117_79849118 | 0.07 |
ANAPC11 |
anaphase promoting complex subunit 11 |
49 |
0.89 |
| chr10_15138786_15140121 | 0.07 |
RPP38 |
ribonuclease P/MRP 38kDa subunit |
34 |
0.62 |
| chr4_1341026_1341962 | 0.07 |
UVSSA |
UV-stimulated scaffold protein A |
0 |
0.97 |
| chr5_32522838_32523035 | 0.07 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
8803 |
0.26 |
| chr1_146714369_146715035 | 0.07 |
CHD1L |
chromodomain helicase DNA binding protein 1-like |
346 |
0.88 |
| chr1_246952376_246952607 | 0.07 |
ENSG00000227953 |
. |
1435 |
0.37 |
| chrX_103357104_103358067 | 0.07 |
ZCCHC18 |
zinc finger, CCHC domain containing 18 |
383 |
0.86 |
| chr15_90294233_90294789 | 0.07 |
MESP1 |
mesoderm posterior 1 homolog (mouse) |
30 |
0.96 |
| chr11_48002643_48003787 | 0.07 |
PTPRJ |
protein tyrosine phosphatase, receptor type, J |
936 |
0.61 |
| chr9_35489468_35490886 | 0.07 |
RUSC2 |
RUN and SH3 domain containing 2 |
53 |
0.97 |
| chr5_176432878_176433695 | 0.07 |
UIMC1 |
ubiquitin interaction motif containing 1 |
123 |
0.96 |
| chr1_1207989_1209115 | 0.07 |
UBE2J2 |
ubiquitin-conjugating enzyme E2, J2 |
299 |
0.76 |
| chr18_3262315_3263346 | 0.07 |
MYL12B |
myosin, light chain 12B, regulatory |
124 |
0.92 |
| chr8_145747877_145748779 | 0.07 |
LRRC24 |
leucine rich repeat containing 24 |
4074 |
0.07 |
| chr17_37856067_37857084 | 0.07 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
242 |
0.86 |
| chr5_138089111_138089974 | 0.07 |
CTNNA1 |
catenin (cadherin-associated protein), alpha 1, 102kDa |
192 |
0.8 |
| chr17_34611752_34612896 | 0.07 |
CCL3L1 |
chemokine (C-C motif) ligand 3-like 1 |
13395 |
0.14 |
| chr17_235901_236198 | 0.07 |
RPH3AL |
rabphilin 3A-like (without C2 domains) |
4 |
0.97 |
| chr3_8809480_8810320 | 0.07 |
OXTR |
oxytocin receptor |
463 |
0.76 |
| chr19_36545772_36546481 | 0.07 |
WDR62 |
WD repeat domain 62 |
234 |
0.67 |
| chr17_36508115_36509219 | 0.07 |
SOCS7 |
suppressor of cytokine signaling 7 |
159 |
0.94 |
| chr9_91926364_91927170 | 0.07 |
ENSG00000265112 |
. |
373 |
0.66 |
| chr3_181413371_181413945 | 0.07 |
SOX2-OT |
SOX2 overlapping transcript (non-protein coding) |
3623 |
0.29 |
| chrX_9432622_9433173 | 0.07 |
TBL1X |
transducin (beta)-like 1X-linked |
151 |
0.98 |
| chr17_29814991_29815971 | 0.07 |
RAB11FIP4 |
RAB11 family interacting protein 4 (class II) |
355 |
0.83 |
| chrX_131622714_131623644 | 0.07 |
MBNL3 |
muscleblind-like splicing regulator 3 |
136 |
0.98 |
| chr15_95388341_95388759 | 0.07 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
489347 |
0.01 |
| chr3_75707498_75708269 | 0.07 |
FRG2C |
FSHD region gene 2 family, member C |
5598 |
0.22 |
| chr2_121102314_121103173 | 0.07 |
INHBB |
inhibin, beta B |
976 |
0.66 |
| chr15_62360405_62361178 | 0.07 |
C2CD4A |
C2 calcium-dependent domain containing 4A |
1615 |
0.36 |
| chr7_29605545_29606286 | 0.07 |
PRR15 |
proline rich 15 |
849 |
0.6 |
| chr14_106347518_106347669 | 0.07 |
ENSG00000225825 |
. |
179 |
0.62 |
| chr9_140500205_140501463 | 0.07 |
ARRDC1 |
arrestin domain containing 1 |
679 |
0.56 |
| chr3_128210659_128211025 | 0.07 |
GATA2 |
GATA binding protein 2 |
1174 |
0.41 |
| chr7_26437839_26438861 | 0.07 |
AC004540.4 |
|
22029 |
0.21 |
| chr4_8632079_8632376 | 0.07 |
CPZ |
carboxypeptidase Z |
28530 |
0.2 |
| chr12_117036919_117037510 | 0.07 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
23558 |
0.24 |
| chr13_20766378_20767168 | 0.06 |
GJB2 |
gap junction protein, beta 2, 26kDa |
264 |
0.91 |
| chr8_27491561_27492240 | 0.06 |
SCARA3 |
scavenger receptor class A, member 3 |
202 |
0.93 |
| chr11_118305386_118306890 | 0.06 |
RP11-770J1.4 |
|
217 |
0.83 |
| chr9_133813639_133813991 | 0.06 |
FIBCD1 |
fibrinogen C domain containing 1 |
422 |
0.82 |
| chr4_77342002_77342745 | 0.06 |
SHROOM3 |
shroom family member 3 |
13880 |
0.15 |
| chr17_40332753_40333403 | 0.06 |
KCNH4 |
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
21 |
0.95 |
| chr10_49732228_49732379 | 0.06 |
ARHGAP22 |
Rho GTPase activating protein 22 |
22 |
0.98 |
| chr19_15489473_15490549 | 0.06 |
ENSG00000268189 |
. |
112 |
0.87 |
| chr22_17601691_17602165 | 0.06 |
CECR6 |
cat eye syndrome chromosome region, candidate 6 |
215 |
0.77 |
| chrX_109246139_109246998 | 0.06 |
TMEM164 |
transmembrane protein 164 |
225 |
0.95 |
| chr12_95043269_95044318 | 0.06 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
545 |
0.85 |
| chr7_150783520_150783695 | 0.06 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
222 |
0.85 |
| chr7_75986838_75988224 | 0.06 |
YWHAG |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma |
817 |
0.59 |
| chr11_75479321_75480435 | 0.06 |
DGAT2 |
diacylglycerol O-acyltransferase 2 |
21 |
0.69 |
| chr12_52263267_52263773 | 0.06 |
ANKRD33 |
ankyrin repeat domain 33 |
18224 |
0.17 |
| chr11_73019949_73020282 | 0.06 |
RP11-800A3.7 |
|
291 |
0.76 |
| chr4_118954790_118955212 | 0.06 |
NDST3 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 3 |
499 |
0.89 |
| chr1_206831789_206831940 | 0.06 |
DYRK3 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 3 |
22690 |
0.12 |
| chr5_76475983_76476746 | 0.06 |
PDE8B |
phosphodiesterase 8B |
29910 |
0.15 |
| chr20_32030739_32031661 | 0.06 |
SNTA1 |
syntrophin, alpha 1 |
498 |
0.77 |
| chr11_61595087_61595308 | 0.06 |
FADS2 |
fatty acid desaturase 2 |
36 |
0.95 |
| chr1_247694177_247694328 | 0.06 |
OR2C3 |
olfactory receptor, family 2, subfamily C, member 3 |
2889 |
0.17 |
| chr1_45278722_45278873 | 0.06 |
BTBD19 |
BTB (POZ) domain containing 19 |
4602 |
0.08 |
| chr1_223743422_223744240 | 0.06 |
CAPN8 |
calpain 8 |
19451 |
0.25 |
| chr11_62367822_62369222 | 0.06 |
MTA2 |
metastasis associated 1 family, member 2 |
225 |
0.8 |
| chr12_111051969_111052303 | 0.06 |
TCTN1 |
tectonic family member 1 |
167 |
0.94 |
| chr1_244816518_244817656 | 0.06 |
DESI2 |
desumoylating isopeptidase 2 |
34 |
0.98 |
| chr7_155601099_155601405 | 0.06 |
SHH |
sonic hedgehog |
514 |
0.83 |
| chr12_26348638_26349346 | 0.06 |
SSPN |
sarcospan |
386 |
0.88 |
| chr12_124155530_124156430 | 0.06 |
TCTN2 |
tectonic family member 2 |
320 |
0.85 |
| chr15_41851700_41852372 | 0.06 |
TYRO3 |
TYRO3 protein tyrosine kinase |
804 |
0.57 |
| chr1_6672501_6673393 | 0.06 |
PHF13 |
PHD finger protein 13 |
798 |
0.46 |
| chr18_77547944_77548277 | 0.06 |
KCNG2 |
potassium voltage-gated channel, subfamily G, member 2 |
75558 |
0.1 |
| chrX_153639895_153640843 | 0.06 |
DNASE1L1 |
deoxyribonuclease I-like 1 |
51 |
0.66 |
| chrX_12156590_12157176 | 0.06 |
FRMPD4 |
FERM and PDZ domain containing 4 |
298 |
0.9 |
| chr15_56034170_56035376 | 0.06 |
PRTG |
protogenin |
41 |
0.98 |
| chr19_57828794_57829125 | 0.06 |
ZNF543 |
zinc finger protein 543 |
2918 |
0.14 |
| chr8_28269075_28269620 | 0.06 |
ENSG00000207361 |
. |
4618 |
0.15 |
| chr11_14665333_14666638 | 0.06 |
PDE3B |
phosphodiesterase 3B, cGMP-inhibited |
608 |
0.56 |
| chr12_133263459_133264038 | 0.06 |
POLE |
polymerase (DNA directed), epsilon, catalytic subunit |
166 |
0.73 |
| chr6_18277489_18278102 | 0.06 |
ENSG00000199715 |
. |
4298 |
0.21 |
| chr9_138987258_138988057 | 0.06 |
NACC2 |
NACC family member 2, BEN and BTB (POZ) domain containing |
526 |
0.8 |
| chr10_88280283_88281468 | 0.06 |
WAPAL |
wings apart-like homolog (Drosophila) |
697 |
0.48 |
| chr12_106640524_106641264 | 0.06 |
RP11-651L5.2 |
|
160 |
0.9 |
| chr6_41702621_41703238 | 0.06 |
TFEB |
transcription factor EB |
131 |
0.93 |
| chr9_123837276_123837840 | 0.06 |
CNTRL |
centriolin |
268 |
0.93 |
| chr10_116634887_116635113 | 0.06 |
FAM160B1 |
family with sequence similarity 160, member B1 |
14450 |
0.25 |
| chr7_145813891_145814290 | 0.06 |
CNTNAP2 |
contactin associated protein-like 2 |
637 |
0.8 |
| chr1_2005099_2005545 | 0.06 |
PRKCZ |
protein kinase C, zeta |
103 |
0.96 |
| chr16_89766857_89768141 | 0.06 |
SPATA2L |
spermatogenesis associated 2-like |
549 |
0.59 |
| chr7_99595147_99595680 | 0.06 |
AZGP1P1 |
alpha-2-glycoprotein 1, zinc-binding pseudogene 1 |
17024 |
0.08 |
| chr9_127240198_127240462 | 0.06 |
NR5A1 |
nuclear receptor subfamily 5, group A, member 1 |
22935 |
0.17 |
| chr2_120189452_120190154 | 0.06 |
TMEM37 |
transmembrane protein 37 |
358 |
0.88 |
| chr1_246948051_246948632 | 0.06 |
ENSG00000227953 |
. |
5585 |
0.17 |
| chr11_117185897_117186711 | 0.06 |
BACE1 |
beta-site APP-cleaving enzyme 1 |
210 |
0.87 |
| chr3_126076169_126076320 | 0.06 |
KLF15 |
Kruppel-like factor 15 |
41 |
0.98 |
| chr12_54589848_54590188 | 0.06 |
SMUG1 |
single-strand-selective monofunctional uracil-DNA glycosylase 1 |
7240 |
0.12 |
| chr4_47916858_47917055 | 0.06 |
NFXL1 |
nuclear transcription factor, X-box binding-like 1 |
323 |
0.9 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0003161 | cardiac conduction system development(GO:0003161) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.1 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.1 | GO:0050910 | detection of mechanical stimulus involved in sensory perception of sound(GO:0050910) |
| 0.0 | 0.0 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.0 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.0 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0044321 | cellular response to leptin stimulus(GO:0044320) response to leptin(GO:0044321) |
| 0.0 | 0.1 | GO:0010507 | negative regulation of autophagy(GO:0010507) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.0 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.1 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0051136 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.1 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0060199 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.0 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.0 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0043140 | ATP-dependent 3'-5' DNA helicase activity(GO:0043140) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.0 | REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | Genes involved in Neurotransmitter Receptor Binding And Downstream Transmission In The Postsynaptic Cell |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.1 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |