Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
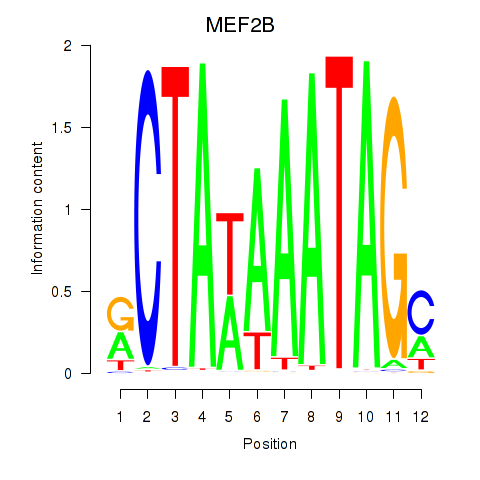
Results for MEF2B
Z-value: 0.75
Transcription factors associated with MEF2B
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2B
|
ENSG00000213999.11 | myocyte enhancer factor 2B |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_19280776_19281129 | MEF2B | 102 | 0.937982 | -0.41 | 2.7e-01 | Click! |
| chr19_19281413_19281564 | MEF2B | 390 | 0.745518 | 0.30 | 4.4e-01 | Click! |
| chr19_19281195_19281405 | MEF2B | 202 | 0.887596 | 0.24 | 5.3e-01 | Click! |
| chr19_19282584_19282735 | MEF2B | 1561 | 0.235275 | 0.14 | 7.2e-01 | Click! |
| chr19_19283135_19283286 | MEF2B | 2112 | 0.178375 | 0.11 | 7.7e-01 | Click! |
Activity of the MEF2B motif across conditions
Conditions sorted by the z-value of the MEF2B motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
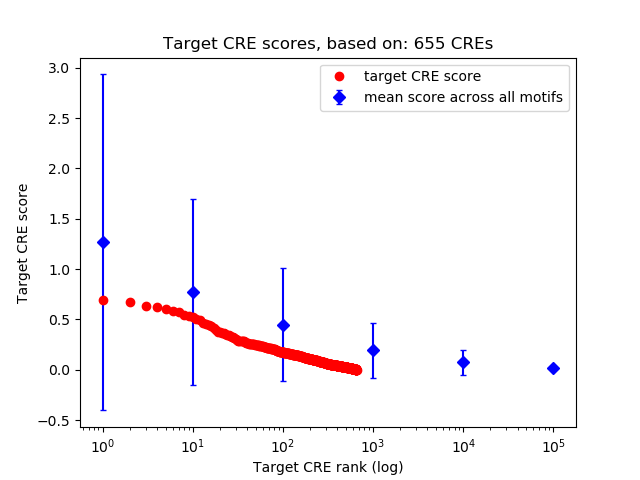
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_17699920_17700290 | 0.69 |
RAD51AP2 |
RAD51 associated protein 2 |
399 |
0.89 |
| chr1_234630809_234631231 | 0.67 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
16171 |
0.21 |
| chr20_11871763_11872529 | 0.63 |
BTBD3 |
BTB (POZ) domain containing 3 |
171 |
0.97 |
| chr6_86167890_86168219 | 0.62 |
NT5E |
5'-nucleotidase, ecto (CD73) |
8227 |
0.28 |
| chr11_94503028_94503179 | 0.61 |
AMOTL1 |
angiomotin like 1 |
1566 |
0.45 |
| chr14_89769991_89770787 | 0.59 |
RP11-356K23.1 |
|
46240 |
0.14 |
| chr11_43597772_43598095 | 0.57 |
ENSG00000199077 |
. |
5011 |
0.24 |
| chr9_124360553_124361088 | 0.55 |
RP11-524G24.2 |
|
22327 |
0.18 |
| chr2_155312257_155312697 | 0.54 |
AC009227.2 |
|
1473 |
0.49 |
| chr3_145968218_145968873 | 0.53 |
PLSCR4 |
phospholipid scramblase 4 |
109 |
0.98 |
| chr6_49293859_49294324 | 0.50 |
ENSG00000252457 |
. |
18430 |
0.26 |
| chr5_39399204_39399669 | 0.50 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
4934 |
0.29 |
| chr1_151173034_151173185 | 0.46 |
PIP5K1A |
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
2058 |
0.17 |
| chr16_88449218_88449497 | 0.45 |
ZNF469 |
zinc finger protein 469 |
44522 |
0.14 |
| chr1_93818876_93819027 | 0.44 |
ENSG00000223745 |
. |
7369 |
0.19 |
| chr9_81322437_81322726 | 0.44 |
PSAT1 |
phosphoserine aminotransferase 1 |
410522 |
0.01 |
| chr11_113649028_113649338 | 0.41 |
CLDN25 |
claudin 25 |
1286 |
0.35 |
| chr1_229762768_229763169 | 0.40 |
URB2 |
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
987 |
0.42 |
| chr8_6539493_6539644 | 0.38 |
CTD-2541M15.1 |
|
26132 |
0.18 |
| chr12_109219039_109219464 | 0.37 |
SSH1 |
slingshot protein phosphatase 1 |
811 |
0.57 |
| chr9_127999236_127999759 | 0.37 |
HSPA5 |
heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) |
4112 |
0.17 |
| chr6_106994674_106995055 | 0.37 |
AIM1 |
absent in melanoma 1 |
5835 |
0.22 |
| chr1_50575996_50576351 | 0.36 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
795 |
0.71 |
| chr2_55226424_55226816 | 0.35 |
RTN4 |
reticulon 4 |
10965 |
0.21 |
| chr13_45011783_45011934 | 0.35 |
TSC22D1 |
TSC22 domain family, member 1 |
877 |
0.72 |
| chr19_8305457_8305834 | 0.34 |
CTD-3020H12.4 |
|
1113 |
0.43 |
| chr2_98487283_98487560 | 0.33 |
TMEM131 |
transmembrane protein 131 |
36288 |
0.17 |
| chr5_14154391_14154790 | 0.32 |
TRIO |
trio Rho guanine nucleotide exchange factor |
10761 |
0.32 |
| chr20_62485470_62485890 | 0.31 |
ABHD16B |
abhydrolase domain containing 16B |
6886 |
0.08 |
| chr17_27719677_27719955 | 0.31 |
TAOK1 |
TAO kinase 1 |
1867 |
0.18 |
| chr5_88081859_88082385 | 0.30 |
MEF2C |
myocyte enhancer factor 2C |
37483 |
0.19 |
| chrX_77908364_77908810 | 0.29 |
ZCCHC5 |
zinc finger, CCHC domain containing 5 |
6238 |
0.32 |
| chr3_66139685_66140118 | 0.29 |
SLC25A26 |
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
20616 |
0.25 |
| chr1_24373488_24373760 | 0.28 |
RP11-293P20.2 |
|
6299 |
0.15 |
| chr17_65428016_65428233 | 0.28 |
ENSG00000201547 |
. |
23127 |
0.12 |
| chr22_46366779_46367595 | 0.28 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
1408 |
0.38 |
| chr3_29377379_29377667 | 0.28 |
ENSG00000216169 |
. |
33389 |
0.18 |
| chr3_99357829_99358140 | 0.28 |
COL8A1 |
collagen, type VIII, alpha 1 |
530 |
0.85 |
| chr3_48597891_48598569 | 0.27 |
PFKFB4 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
332 |
0.77 |
| chr8_56805751_56805987 | 0.27 |
RP11-318K15.2 |
|
285 |
0.88 |
| chr6_26698405_26698556 | 0.26 |
ZNF322 |
zinc finger protein 322 |
38500 |
0.16 |
| chr15_99864560_99864718 | 0.26 |
AC022819.2 |
Uncharacterized protein |
4811 |
0.23 |
| chr2_55345979_55346130 | 0.26 |
RTN4 |
reticulon 4 |
6297 |
0.2 |
| chr2_174983381_174983564 | 0.26 |
OLA1 |
Obg-like ATPase 1 |
99202 |
0.07 |
| chr9_108211383_108211534 | 0.26 |
FSD1L |
fibronectin type III and SPRY domain containing 1-like |
1054 |
0.64 |
| chr18_3579974_3580125 | 0.26 |
ENSG00000238445 |
. |
7008 |
0.16 |
| chr1_184476918_184477069 | 0.25 |
GS1-115G20.1 |
|
99522 |
0.07 |
| chr1_10168624_10168775 | 0.25 |
UBE4B |
ubiquitination factor E4B |
13401 |
0.19 |
| chr21_30504177_30504355 | 0.25 |
MAP3K7CL |
MAP3K7 C-terminal like |
688 |
0.61 |
| chr4_183167608_183167774 | 0.25 |
TENM3 |
teneurin transmembrane protein 3 |
3109 |
0.34 |
| chr18_43840452_43840603 | 0.24 |
RNF165 |
ring finger protein 165 |
66245 |
0.11 |
| chr4_41362274_41362647 | 0.24 |
LIMCH1 |
LIM and calponin homology domains 1 |
188 |
0.96 |
| chr11_85429273_85429899 | 0.24 |
SYTL2 |
synaptotagmin-like 2 |
55 |
0.97 |
| chr10_134401745_134402170 | 0.24 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
19473 |
0.23 |
| chr14_89631722_89632070 | 0.24 |
FOXN3 |
forkhead box N3 |
15192 |
0.28 |
| chr7_121012824_121012975 | 0.24 |
FAM3C |
family with sequence similarity 3, member C |
23463 |
0.22 |
| chr2_122258317_122258683 | 0.24 |
CLASP1 |
cytoplasmic linker associated protein 1 |
3064 |
0.27 |
| chr1_201819395_201819711 | 0.24 |
IPO9-AS1 |
IPO9 antisense RNA 1 |
20866 |
0.11 |
| chr5_65750356_65750571 | 0.23 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
141713 |
0.05 |
| chr2_238571219_238571370 | 0.23 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
29494 |
0.17 |
| chr14_34145464_34145720 | 0.23 |
NPAS3 |
neuronal PAS domain protein 3 |
58855 |
0.17 |
| chr17_42172843_42173098 | 0.23 |
HDAC5 |
histone deacetylase 5 |
2415 |
0.15 |
| chr1_17945358_17945612 | 0.23 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
636 |
0.82 |
| chr5_52098224_52098443 | 0.22 |
CTD-2288O8.1 |
|
14473 |
0.19 |
| chr2_145695991_145696253 | 0.22 |
ENSG00000253036 |
. |
396516 |
0.01 |
| chr3_24527242_24527393 | 0.22 |
ENSG00000228791 |
. |
8261 |
0.19 |
| chr12_50504495_50504703 | 0.22 |
COX14 |
cytochrome c oxidase assembly homolog 14 (S. cerevisiae) |
1163 |
0.28 |
| chr17_41669450_41669625 | 0.22 |
ETV4 |
ets variant 4 |
12549 |
0.16 |
| chr4_15011908_15012097 | 0.22 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
6442 |
0.31 |
| chr13_44642585_44642983 | 0.22 |
SMIM2-AS1 |
SMIM2 antisense RNA 1 |
73898 |
0.1 |
| chr21_39678468_39679041 | 0.21 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
9906 |
0.29 |
| chr5_174673965_174674120 | 0.21 |
DRD1 |
dopamine receptor D1 |
197169 |
0.03 |
| chr10_65019032_65019264 | 0.21 |
JMJD1C |
jumonji domain containing 1C |
9678 |
0.27 |
| chr7_137439335_137439486 | 0.21 |
DGKI |
diacylglycerol kinase, iota |
91872 |
0.09 |
| chr7_16751263_16751414 | 0.21 |
AC073333.8 |
|
8185 |
0.18 |
| chr12_125340893_125341100 | 0.21 |
SCARB1 |
scavenger receptor class B, member 1 |
7333 |
0.22 |
| chr20_52352609_52352999 | 0.21 |
ENSG00000238468 |
. |
67507 |
0.12 |
| chr9_98639144_98639295 | 0.20 |
ERCC6L2 |
excision repair cross-complementing rodent repair deficiency, complementation group 6-like 2 |
1236 |
0.48 |
| chr2_151338744_151338901 | 0.20 |
RND3 |
Rho family GTPase 3 |
3074 |
0.42 |
| chr15_36793558_36793709 | 0.20 |
C15orf41 |
chromosome 15 open reading frame 41 |
78179 |
0.12 |
| chr12_62115825_62115976 | 0.20 |
FAM19A2 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
105114 |
0.08 |
| chr1_31459830_31459981 | 0.20 |
PUM1 |
pumilio RNA-binding family member 1 |
8001 |
0.17 |
| chr9_80641416_80641906 | 0.19 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
3859 |
0.36 |
| chr6_94127165_94127388 | 0.19 |
EPHA7 |
EPH receptor A7 |
1968 |
0.51 |
| chr16_3999501_3999652 | 0.19 |
RP11-462G12.1 |
|
869 |
0.56 |
| chr4_140685024_140685175 | 0.19 |
ENSG00000252233 |
. |
30488 |
0.19 |
| chr2_109257559_109258085 | 0.19 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
13446 |
0.22 |
| chr21_17909068_17909746 | 0.19 |
ENSG00000207638 |
. |
2002 |
0.33 |
| chr2_33682993_33683144 | 0.19 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
45 |
0.99 |
| chr1_189407105_189407256 | 0.19 |
ENSG00000252553 |
. |
228206 |
0.02 |
| chr9_35341457_35341792 | 0.18 |
AL160274.1 |
HCG17281; PRO0038; Uncharacterized protein |
19638 |
0.2 |
| chr14_89632118_89632412 | 0.18 |
FOXN3 |
forkhead box N3 |
14823 |
0.28 |
| chr1_3087716_3087867 | 0.18 |
RP1-163G9.2 |
|
37518 |
0.18 |
| chr4_77491396_77491547 | 0.18 |
ENSG00000263445 |
. |
3250 |
0.21 |
| chr4_170585829_170585980 | 0.18 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
4691 |
0.28 |
| chr19_45931964_45932396 | 0.18 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
288 |
0.84 |
| chr7_120631344_120631706 | 0.18 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
1849 |
0.35 |
| chr7_36168506_36168695 | 0.18 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
24158 |
0.18 |
| chr13_114540399_114540680 | 0.17 |
GAS6 |
growth arrest-specific 6 |
1522 |
0.45 |
| chr2_192111446_192111930 | 0.17 |
MYO1B |
myosin IB |
679 |
0.79 |
| chr6_45413846_45414088 | 0.17 |
RUNX2 |
runt-related transcription factor 2 |
23745 |
0.2 |
| chr11_46259732_46260224 | 0.17 |
CTD-2589M5.4 |
|
36120 |
0.14 |
| chr17_53398574_53398941 | 0.17 |
RP11-515O17.2 |
|
47743 |
0.16 |
| chr12_25486836_25487071 | 0.17 |
ENSG00000201439 |
. |
70409 |
0.1 |
| chr8_108316261_108316433 | 0.17 |
ANGPT1 |
angiopoietin 1 |
32403 |
0.22 |
| chr6_74434667_74435200 | 0.17 |
RP11-553A21.3 |
|
29079 |
0.17 |
| chr12_43136229_43136380 | 0.17 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
152147 |
0.04 |
| chr3_32602518_32602669 | 0.17 |
DYNC1LI1 |
dynein, cytoplasmic 1, light intermediate chain 1 |
9152 |
0.22 |
| chr3_138066783_138067508 | 0.17 |
MRAS |
muscle RAS oncogene homolog |
169 |
0.96 |
| chr15_64391587_64391738 | 0.17 |
SNX1 |
sorting nexin 1 |
3437 |
0.18 |
| chr18_33106392_33106607 | 0.17 |
INO80C |
INO80 complex subunit C |
28544 |
0.16 |
| chr11_2834821_2835116 | 0.16 |
KCNQ1-AS1 |
KCNQ1 antisense RNA 1 |
47830 |
0.11 |
| chr2_227555497_227555648 | 0.16 |
ENSG00000263363 |
. |
32063 |
0.22 |
| chr4_86747653_86747804 | 0.16 |
ARHGAP24 |
Rho GTPase activating protein 24 |
1170 |
0.61 |
| chr22_46082903_46083054 | 0.16 |
ATXN10 |
ataxin 10 |
15299 |
0.21 |
| chr7_75341548_75341699 | 0.16 |
HIP1 |
huntingtin interacting protein 1 |
26636 |
0.18 |
| chr12_131200528_131200853 | 0.16 |
RIMBP2 |
RIMS binding protein 2 |
136 |
0.98 |
| chr1_208393211_208393379 | 0.16 |
PLXNA2 |
plexin A2 |
24370 |
0.28 |
| chr3_188108011_188108162 | 0.16 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
150434 |
0.04 |
| chr13_51252397_51252695 | 0.16 |
DLEU7-AS1 |
DLEU7 antisense RNA 1 |
129446 |
0.05 |
| chr10_125608306_125608596 | 0.16 |
CPXM2 |
carboxypeptidase X (M14 family), member 2 |
42879 |
0.15 |
| chr20_7566360_7566511 | 0.16 |
ENSG00000240584 |
. |
48063 |
0.19 |
| chr9_89738562_89738713 | 0.16 |
C9orf170 |
chromosome 9 open reading frame 170 |
24922 |
0.26 |
| chr14_76327643_76327914 | 0.16 |
RP11-270M14.4 |
|
13737 |
0.25 |
| chr7_98028694_98029033 | 0.16 |
BAIAP2L1 |
BAI1-associated protein 2-like 1 |
1517 |
0.48 |
| chr13_94996950_94997101 | 0.16 |
ENSG00000212057 |
. |
120619 |
0.06 |
| chr19_3818575_3818945 | 0.16 |
MATK |
megakaryocyte-associated tyrosine kinase |
16633 |
0.11 |
| chr14_63951066_63951267 | 0.15 |
ENSG00000252800 |
. |
5176 |
0.16 |
| chr6_35687700_35688402 | 0.15 |
FKBP5 |
FK506 binding protein 5 |
8309 |
0.12 |
| chr10_119132010_119132432 | 0.15 |
PDZD8 |
PDZ domain containing 8 |
2757 |
0.3 |
| chr8_107281700_107282399 | 0.15 |
OXR1 |
oxidation resistance 1 |
424 |
0.92 |
| chr6_152784305_152784456 | 0.15 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
9385 |
0.22 |
| chr19_17790996_17791451 | 0.15 |
UNC13A |
unc-13 homolog A (C. elegans) |
4245 |
0.17 |
| chr4_146601616_146601767 | 0.15 |
C4orf51 |
chromosome 4 open reading frame 51 |
214 |
0.94 |
| chr6_16722645_16722796 | 0.15 |
RP1-151F17.1 |
|
38649 |
0.19 |
| chr8_81934177_81934401 | 0.15 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
90014 |
0.09 |
| chr6_14639094_14639727 | 0.15 |
ENSG00000206960 |
. |
7356 |
0.34 |
| chr17_67056911_67057062 | 0.15 |
ABCA9 |
ATP-binding cassette, sub-family A (ABC1), member 9 |
61 |
0.98 |
| chr8_145598687_145598886 | 0.15 |
ADCK5 |
aarF domain containing kinase 5 |
1036 |
0.26 |
| chr21_17584209_17584483 | 0.15 |
ENSG00000201025 |
. |
72743 |
0.13 |
| chr3_52039748_52040001 | 0.15 |
RPL29 |
ribosomal protein L29 |
9916 |
0.09 |
| chr16_84328151_84329018 | 0.15 |
WFDC1 |
WAP four-disulfide core domain 1 |
132 |
0.94 |
| chr4_186488570_186488721 | 0.14 |
RP11-301L8.2 |
|
20418 |
0.16 |
| chr1_186834383_186834534 | 0.14 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
36336 |
0.23 |
| chr16_68281342_68281493 | 0.14 |
PLA2G15 |
phospholipase A2, group XV |
2122 |
0.13 |
| chr2_179728640_179728888 | 0.14 |
CCDC141 |
coiled-coil domain containing 141 |
20914 |
0.17 |
| chr3_137834054_137834699 | 0.14 |
DZIP1L |
DAZ interacting zinc finger protein 1-like |
75 |
0.97 |
| chr5_167831410_167831561 | 0.14 |
WWC1 |
WW and C2 domain containing 1 |
1799 |
0.45 |
| chr5_36692416_36692567 | 0.14 |
CTD-2353F22.1 |
|
32784 |
0.22 |
| chr11_46360410_46360982 | 0.14 |
DGKZ |
diacylglycerol kinase, zeta |
548 |
0.72 |
| chr6_25179503_25179654 | 0.14 |
ENSG00000222373 |
. |
13063 |
0.19 |
| chr16_19016973_19017124 | 0.14 |
ENSG00000207167 |
. |
6431 |
0.16 |
| chr19_58521610_58521765 | 0.14 |
ZNF606 |
zinc finger protein 606 |
6970 |
0.11 |
| chr1_145524889_145525225 | 0.14 |
ITGA10 |
integrin, alpha 10 |
22 |
0.95 |
| chr1_217308938_217309289 | 0.14 |
ESRRG |
estrogen-related receptor gamma |
1983 |
0.51 |
| chr14_92864159_92864342 | 0.14 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
10473 |
0.28 |
| chr5_154138190_154138341 | 0.14 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
1531 |
0.39 |
| chr14_76620436_76620587 | 0.13 |
GPATCH2L |
G patch domain containing 2-like |
522 |
0.85 |
| chr13_45009518_45010564 | 0.13 |
TSC22D1 |
TSC22 domain family, member 1 |
940 |
0.69 |
| chr5_159208675_159208826 | 0.13 |
ADRA1B |
adrenoceptor alpha 1B |
135040 |
0.05 |
| chr17_13502555_13502706 | 0.13 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
2614 |
0.37 |
| chr9_100264063_100264279 | 0.13 |
TMOD1 |
tropomodulin 1 |
250 |
0.92 |
| chr5_125031921_125032072 | 0.13 |
ENSG00000222107 |
. |
345172 |
0.01 |
| chr4_57974250_57974401 | 0.13 |
IGFBP7-AS1 |
IGFBP7 antisense RNA 1 |
1603 |
0.28 |
| chr1_82048437_82048588 | 0.13 |
RP5-837I24.2 |
|
25125 |
0.21 |
| chr4_143766947_143768160 | 0.13 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
99 |
0.99 |
| chr2_25599883_25600214 | 0.13 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
34589 |
0.16 |
| chr10_56993124_56993275 | 0.13 |
PCDH15 |
protocadherin-related 15 |
86175 |
0.11 |
| chr2_227665986_227666395 | 0.13 |
IRS1 |
insulin receptor substrate 1 |
1715 |
0.35 |
| chr7_115670129_115670280 | 0.12 |
TFEC |
transcription factor EC |
591 |
0.85 |
| chr12_1626749_1626900 | 0.12 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
12233 |
0.24 |
| chr2_54195795_54196058 | 0.12 |
ACYP2 |
acylphosphatase 2, muscle type |
2049 |
0.28 |
| chr6_153414041_153414192 | 0.12 |
RGS17 |
regulator of G-protein signaling 17 |
38268 |
0.14 |
| chr4_159731146_159731380 | 0.12 |
FNIP2 |
folliculin interacting protein 2 |
3863 |
0.25 |
| chrX_105431716_105431867 | 0.12 |
MUM1L1 |
melanoma associated antigen (mutated) 1-like 1 |
13938 |
0.29 |
| chr8_38205081_38205318 | 0.12 |
RP11-513D5.2 |
|
11700 |
0.13 |
| chr2_134894818_134895366 | 0.12 |
ENSG00000263813 |
. |
10396 |
0.24 |
| chr12_12686379_12686530 | 0.12 |
DUSP16 |
dual specificity phosphatase 16 |
12395 |
0.24 |
| chr7_157148744_157148916 | 0.12 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
16315 |
0.23 |
| chr5_44943313_44943551 | 0.12 |
MRPS30 |
mitochondrial ribosomal protein S30 |
134405 |
0.05 |
| chr2_203159927_203160078 | 0.12 |
ENSG00000238317 |
. |
2230 |
0.18 |
| chr1_192803524_192803675 | 0.12 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
25428 |
0.21 |
| chr20_1356402_1356657 | 0.12 |
RP11-314N13.3 |
|
14263 |
0.13 |
| chr13_38113810_38113961 | 0.12 |
POSTN |
periostin, osteoblast specific factor |
58978 |
0.16 |
| chr2_197852125_197852688 | 0.12 |
ANKRD44 |
ankyrin repeat domain 44 |
12820 |
0.25 |
| chr10_94039164_94039387 | 0.11 |
ENSG00000199881 |
. |
7268 |
0.17 |
| chr10_6234346_6234592 | 0.11 |
RP11-414H17.5 |
|
10187 |
0.15 |
| chr1_115753443_115753594 | 0.11 |
RP4-663N10.1 |
|
72137 |
0.11 |
| chr10_126753112_126753370 | 0.11 |
ENSG00000264572 |
. |
31802 |
0.18 |
| chrX_119751985_119752136 | 0.11 |
C1GALT1C1 |
C1GALT1-specific chaperone 1 |
11790 |
0.18 |
| chr8_144512945_144513817 | 0.11 |
MAFA |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A |
805 |
0.48 |
| chr1_230272340_230272635 | 0.11 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
69469 |
0.11 |
| chr7_125333680_125333831 | 0.11 |
ENSG00000221418 |
. |
346528 |
0.01 |
| chr20_19991582_19991882 | 0.11 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
6028 |
0.22 |
| chr5_66935802_66935953 | 0.11 |
ENSG00000222939 |
. |
175441 |
0.03 |
| chr7_90085000_90085151 | 0.11 |
CDK14 |
cyclin-dependent kinase 14 |
10663 |
0.23 |
| chr18_57549496_57550010 | 0.11 |
PMAIP1 |
phorbol-12-myristate-13-acetate-induced protein 1 |
17427 |
0.24 |
| chr5_148796790_148796941 | 0.11 |
ENSG00000208035 |
. |
11616 |
0.13 |
| chr6_129822270_129822588 | 0.11 |
RP1-69D17.3 |
|
19874 |
0.26 |
| chr12_90484641_90484914 | 0.11 |
ENSG00000252823 |
. |
336941 |
0.01 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.2 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.2 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.1 | GO:0072205 | metanephric collecting duct development(GO:0072205) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0090273 | regulation of glucagon secretion(GO:0070092) regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:0032497 | detection of lipopolysaccharide(GO:0032497) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.0 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0051818 | disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:1901978 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.0 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0009200 | deoxyribonucleoside triphosphate metabolic process(GO:0009200) |
| 0.0 | 0.1 | GO:1903670 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.1 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.3 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.1 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |