Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for MEF2D_MEF2A
Z-value: 1.20
Transcription factors associated with MEF2D_MEF2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
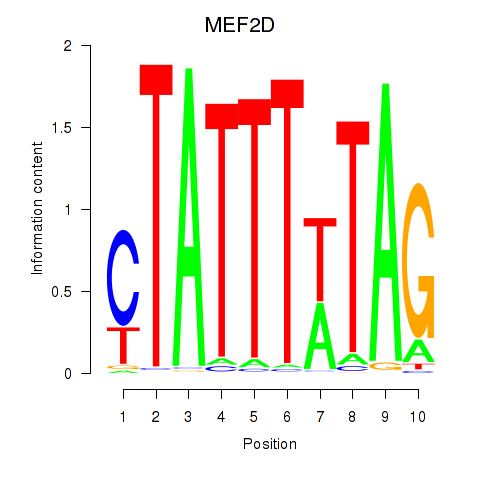
MEF2D
|
ENSG00000116604.13 | myocyte enhancer factor 2D |
|
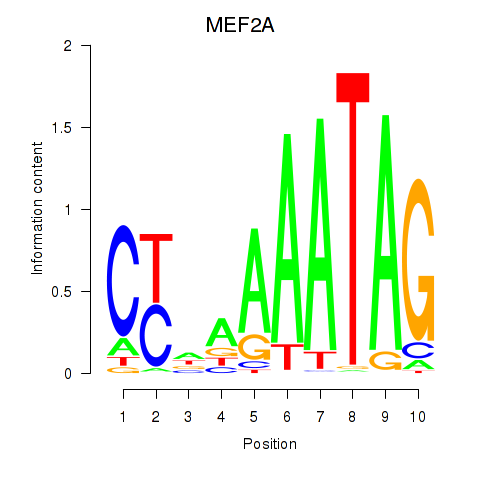
MEF2A
|
ENSG00000068305.13 | myocyte enhancer factor 2A |
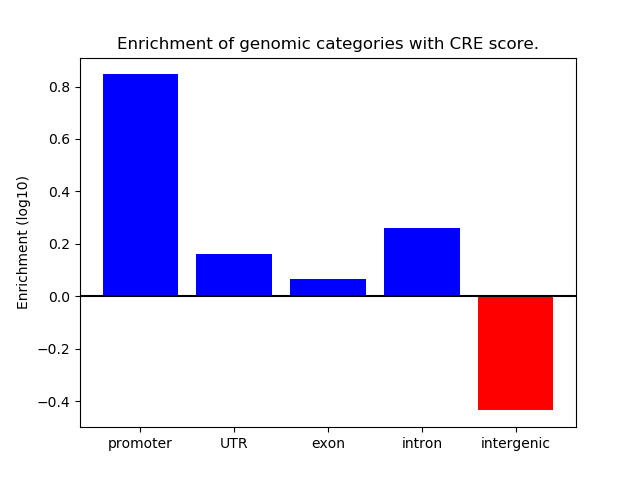
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr15_100027229_100027393 | MEF2A | 9941 | 0.204639 | 0.79 | 1.1e-02 | Click! |
| chr15_99991178_99991413 | MEF2A | 26075 | 0.174136 | -0.69 | 3.8e-02 | Click! |
| chr15_100026519_100026720 | MEF2A | 9249 | 0.206838 | 0.66 | 5.1e-02 | Click! |
| chr15_99982440_99982591 | MEF2A | 34855 | 0.151225 | 0.61 | 7.8e-02 | Click! |
| chr15_100108821_100109055 | MEF2A | 2250 | 0.363158 | 0.59 | 9.3e-02 | Click! |
| chr1_156466320_156466471 | MEF2D | 4134 | 0.143467 | 0.82 | 6.6e-03 | Click! |
| chr1_156480380_156480531 | MEF2D | 9835 | 0.115540 | 0.71 | 3.3e-02 | Click! |
| chr1_156458657_156458808 | MEF2D | 1659 | 0.256349 | -0.56 | 1.1e-01 | Click! |
| chr1_156466052_156466203 | MEF2D | 4402 | 0.140380 | 0.55 | 1.2e-01 | Click! |
| chr1_156471272_156471435 | MEF2D | 733 | 0.543760 | -0.54 | 1.3e-01 | Click! |
Activity of the MEF2D_MEF2A motif across conditions
Conditions sorted by the z-value of the MEF2D_MEF2A motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
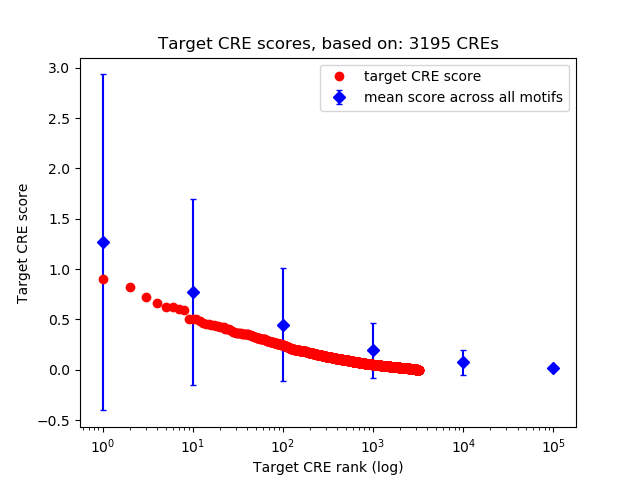
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_88081859_88082385 | 0.90 |
MEF2C |
myocyte enhancer factor 2C |
37483 |
0.19 |
| chr3_112353843_112353994 | 0.83 |
CCDC80 |
coiled-coil domain containing 80 |
3026 |
0.32 |
| chr3_66139685_66140118 | 0.72 |
SLC25A26 |
solute carrier family 25 (S-adenosylmethionine carrier), member 26 |
20616 |
0.25 |
| chr7_120631344_120631706 | 0.67 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
1849 |
0.35 |
| chr16_86775280_86775446 | 0.63 |
FOXL1 |
forkhead box L1 |
163248 |
0.03 |
| chr2_155312257_155312697 | 0.63 |
AC009227.2 |
|
1473 |
0.49 |
| chr11_43597772_43598095 | 0.60 |
ENSG00000199077 |
. |
5011 |
0.24 |
| chr1_151173034_151173185 | 0.59 |
PIP5K1A |
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
2058 |
0.17 |
| chr5_39399204_39399669 | 0.51 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
4934 |
0.29 |
| chr1_17945358_17945612 | 0.51 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
636 |
0.82 |
| chr2_17699920_17700290 | 0.50 |
RAD51AP2 |
RAD51 associated protein 2 |
399 |
0.89 |
| chr13_94996950_94997101 | 0.49 |
ENSG00000212057 |
. |
120619 |
0.06 |
| chr12_109219039_109219464 | 0.47 |
SSH1 |
slingshot protein phosphatase 1 |
811 |
0.57 |
| chr17_13502555_13502706 | 0.45 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
2614 |
0.37 |
| chr3_48597891_48598569 | 0.45 |
PFKFB4 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
332 |
0.77 |
| chr1_234630809_234631231 | 0.45 |
TARBP1 |
TAR (HIV-1) RNA binding protein 1 |
16171 |
0.21 |
| chr11_85429273_85429899 | 0.44 |
SYTL2 |
synaptotagmin-like 2 |
55 |
0.97 |
| chr7_112124877_112125132 | 0.44 |
LSMEM1 |
leucine-rich single-pass membrane protein 1 |
3958 |
0.27 |
| chr14_89769991_89770787 | 0.43 |
RP11-356K23.1 |
|
46240 |
0.14 |
| chr3_99357829_99358140 | 0.43 |
COL8A1 |
collagen, type VIII, alpha 1 |
530 |
0.85 |
| chr19_11305569_11305933 | 0.42 |
KANK2 |
KN motif and ankyrin repeat domains 2 |
117 |
0.94 |
| chr1_170632786_170633084 | 0.42 |
PRRX1 |
paired related homeobox 1 |
112 |
0.98 |
| chr1_157962818_157962980 | 0.41 |
KIRREL |
kin of IRRE like (Drosophila) |
164 |
0.96 |
| chr17_57564845_57564996 | 0.41 |
ENSG00000200889 |
. |
49940 |
0.12 |
| chr16_88449218_88449497 | 0.41 |
ZNF469 |
zinc finger protein 469 |
44522 |
0.14 |
| chr1_24373488_24373760 | 0.39 |
RP11-293P20.2 |
|
6299 |
0.15 |
| chr9_130615735_130616098 | 0.38 |
ENG |
endoglin |
999 |
0.31 |
| chr5_65750356_65750571 | 0.38 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
141713 |
0.05 |
| chr9_81322437_81322726 | 0.37 |
PSAT1 |
phosphoserine aminotransferase 1 |
410522 |
0.01 |
| chr3_145968218_145968873 | 0.37 |
PLSCR4 |
phospholipid scramblase 4 |
109 |
0.98 |
| chr10_134401745_134402170 | 0.37 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
19473 |
0.23 |
| chr4_41362274_41362647 | 0.37 |
LIMCH1 |
LIM and calponin homology domains 1 |
188 |
0.96 |
| chr2_55226424_55226816 | 0.37 |
RTN4 |
reticulon 4 |
10965 |
0.21 |
| chr14_34145464_34145720 | 0.37 |
NPAS3 |
neuronal PAS domain protein 3 |
58855 |
0.17 |
| chr20_32979012_32979163 | 0.36 |
ENSG00000201498 |
. |
7016 |
0.16 |
| chr6_121760422_121760623 | 0.36 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
3684 |
0.22 |
| chr11_109842804_109842955 | 0.36 |
ZC3H12C |
zinc finger CCCH-type containing 12C |
121208 |
0.07 |
| chr9_98639144_98639295 | 0.36 |
ERCC6L2 |
excision repair cross-complementing rodent repair deficiency, complementation group 6-like 2 |
1236 |
0.48 |
| chr1_229762768_229763169 | 0.35 |
URB2 |
URB2 ribosome biogenesis 2 homolog (S. cerevisiae) |
987 |
0.42 |
| chr1_224805949_224806359 | 0.35 |
CNIH3 |
cornichon family AMPA receptor auxiliary protein 3 |
2159 |
0.29 |
| chr5_132385127_132385278 | 0.35 |
HSPA4 |
heat shock 70kDa protein 4 |
2452 |
0.27 |
| chr13_45011783_45011934 | 0.35 |
TSC22D1 |
TSC22 domain family, member 1 |
877 |
0.72 |
| chr6_46458168_46458592 | 0.34 |
RCAN2 |
regulator of calcineurin 2 |
719 |
0.62 |
| chr2_85389562_85389713 | 0.34 |
TCF7L1-IT1 |
TCF7L1 intronic transcript 1 (non-protein coding) |
23895 |
0.15 |
| chr1_93818876_93819027 | 0.34 |
ENSG00000223745 |
. |
7369 |
0.19 |
| chr12_9268515_9268666 | 0.34 |
A2M |
alpha-2-macroglobulin |
163 |
0.95 |
| chr5_15499822_15500142 | 0.34 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
323 |
0.94 |
| chr21_30504177_30504355 | 0.33 |
MAP3K7CL |
MAP3K7 C-terminal like |
688 |
0.61 |
| chr1_50575996_50576351 | 0.33 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
795 |
0.71 |
| chrX_69700453_69700680 | 0.33 |
DLG3-AS1 |
DLG3 antisense RNA 1 |
24722 |
0.14 |
| chr5_174673965_174674120 | 0.32 |
DRD1 |
dopamine receptor D1 |
197169 |
0.03 |
| chr2_179728640_179728888 | 0.32 |
CCDC141 |
coiled-coil domain containing 141 |
20914 |
0.17 |
| chr7_90085000_90085151 | 0.32 |
CDK14 |
cyclin-dependent kinase 14 |
10663 |
0.23 |
| chr18_3579974_3580125 | 0.31 |
ENSG00000238445 |
. |
7008 |
0.16 |
| chr5_14146856_14147086 | 0.31 |
TRIO |
trio Rho guanine nucleotide exchange factor |
3142 |
0.4 |
| chr4_55095719_55096018 | 0.31 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
142 |
0.98 |
| chr2_97018640_97018821 | 0.31 |
NCAPH |
non-SMC condensin I complex, subunit H |
11488 |
0.15 |
| chr2_71356365_71356651 | 0.31 |
MCEE |
methylmalonyl CoA epimerase |
836 |
0.37 |
| chr3_137834054_137834699 | 0.31 |
DZIP1L |
DAZ interacting zinc finger protein 1-like |
75 |
0.97 |
| chr2_197852125_197852688 | 0.31 |
ANKRD44 |
ankyrin repeat domain 44 |
12820 |
0.25 |
| chr4_15011908_15012097 | 0.30 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
6442 |
0.31 |
| chr20_19991582_19991882 | 0.30 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
6028 |
0.22 |
| chr4_86747653_86747804 | 0.30 |
ARHGAP24 |
Rho GTPase activating protein 24 |
1170 |
0.61 |
| chr2_55345979_55346130 | 0.30 |
RTN4 |
reticulon 4 |
6297 |
0.2 |
| chr21_17584209_17584483 | 0.30 |
ENSG00000201025 |
. |
72743 |
0.13 |
| chr19_11314227_11314401 | 0.29 |
CTC-510F12.2 |
|
10 |
0.95 |
| chr5_52098224_52098443 | 0.29 |
CTD-2288O8.1 |
|
14473 |
0.19 |
| chr10_63724125_63724283 | 0.29 |
ENSG00000221272 |
. |
37427 |
0.2 |
| chr6_49293859_49294324 | 0.29 |
ENSG00000252457 |
. |
18430 |
0.26 |
| chr4_77491396_77491547 | 0.28 |
ENSG00000263445 |
. |
3250 |
0.21 |
| chr11_1930082_1930233 | 0.28 |
TNNT3 |
troponin T type 3 (skeletal, fast) |
10635 |
0.1 |
| chr20_50180013_50180188 | 0.28 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
730 |
0.78 |
| chr1_184476918_184477069 | 0.28 |
GS1-115G20.1 |
|
99522 |
0.07 |
| chr20_50417404_50418104 | 0.28 |
SALL4 |
spalt-like transcription factor 4 |
1193 |
0.55 |
| chr15_38547811_38548278 | 0.28 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
2662 |
0.42 |
| chr5_36692416_36692567 | 0.28 |
CTD-2353F22.1 |
|
32784 |
0.22 |
| chr6_25179503_25179654 | 0.27 |
ENSG00000222373 |
. |
13063 |
0.19 |
| chr7_143581670_143581927 | 0.27 |
FAM115A |
family with sequence similarity 115, member A |
665 |
0.7 |
| chr15_99864560_99864718 | 0.27 |
AC022819.2 |
Uncharacterized protein |
4811 |
0.23 |
| chr9_80641416_80641906 | 0.27 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
3859 |
0.36 |
| chr2_198539715_198539970 | 0.26 |
RFTN2 |
raftlin family member 2 |
877 |
0.6 |
| chr5_88129679_88129830 | 0.26 |
MEF2C |
myocyte enhancer factor 2C |
6779 |
0.28 |
| chr13_42269392_42269543 | 0.26 |
ENSG00000241406 |
. |
112346 |
0.06 |
| chr18_59249295_59249706 | 0.26 |
CDH20 |
cadherin 20, type 2 |
32401 |
0.25 |
| chr1_211306419_211307099 | 0.26 |
KCNH1-IT1 |
KCNH1 intronic transcript 1 (non-protein coding) |
43 |
0.92 |
| chr20_62485470_62485890 | 0.26 |
ABHD16B |
abhydrolase domain containing 16B |
6886 |
0.08 |
| chr6_154800458_154800845 | 0.26 |
CNKSR3 |
CNKSR family member 3 |
1026 |
0.69 |
| chr3_46033980_46034131 | 0.26 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
3252 |
0.24 |
| chr10_16870751_16871209 | 0.26 |
RSU1 |
Ras suppressor protein 1 |
11453 |
0.28 |
| chr4_54230912_54231063 | 0.26 |
SCFD2 |
sec1 family domain containing 2 |
1241 |
0.52 |
| chr16_31022230_31022574 | 0.26 |
STX1B |
syntaxin 1B |
453 |
0.64 |
| chr20_11871763_11872529 | 0.26 |
BTBD3 |
BTB (POZ) domain containing 3 |
171 |
0.97 |
| chr15_36793558_36793709 | 0.25 |
C15orf41 |
chromosome 15 open reading frame 41 |
78179 |
0.12 |
| chr19_37096291_37096844 | 0.25 |
ZNF382 |
zinc finger protein 382 |
330 |
0.52 |
| chr1_8713025_8713176 | 0.25 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
3330 |
0.34 |
| chr17_36571464_36571657 | 0.25 |
ENSG00000266103 |
. |
5518 |
0.17 |
| chr2_164589458_164589609 | 0.25 |
FIGN |
fidgetin |
2984 |
0.42 |
| chr5_148796790_148796941 | 0.25 |
ENSG00000208035 |
. |
11616 |
0.13 |
| chr8_81934177_81934401 | 0.25 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
90014 |
0.09 |
| chr13_45242299_45242450 | 0.25 |
ENSG00000238932 |
. |
39565 |
0.2 |
| chr4_186488570_186488721 | 0.24 |
RP11-301L8.2 |
|
20418 |
0.16 |
| chr5_141826116_141826340 | 0.24 |
AC005592.2 |
|
42445 |
0.18 |
| chr2_25017882_25018033 | 0.24 |
CENPO |
centromere protein O |
1624 |
0.28 |
| chr13_45059417_45059568 | 0.24 |
TSC22D1 |
TSC22 domain family, member 1 |
11106 |
0.29 |
| chr2_25599883_25600214 | 0.23 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
34589 |
0.16 |
| chr2_192111446_192111930 | 0.23 |
MYO1B |
myosin IB |
679 |
0.79 |
| chr7_16751263_16751414 | 0.23 |
AC073333.8 |
|
8185 |
0.18 |
| chr11_12714851_12715219 | 0.23 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
17985 |
0.26 |
| chr6_152784305_152784456 | 0.23 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
9385 |
0.22 |
| chr1_217308938_217309289 | 0.23 |
ESRRG |
estrogen-related receptor gamma |
1983 |
0.51 |
| chr2_201021102_201021310 | 0.23 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
149398 |
0.04 |
| chr3_52741792_52741943 | 0.23 |
SPCS1 |
signal peptidase complex subunit 1 homolog (S. cerevisiae) |
1773 |
0.15 |
| chr6_72205767_72206133 | 0.22 |
ENSG00000212099 |
. |
42729 |
0.17 |
| chr9_100264063_100264279 | 0.22 |
TMOD1 |
tropomodulin 1 |
250 |
0.92 |
| chr10_56993124_56993275 | 0.22 |
PCDH15 |
protocadherin-related 15 |
86175 |
0.11 |
| chr14_76327643_76327914 | 0.22 |
RP11-270M14.4 |
|
13737 |
0.25 |
| chr7_132737872_132738088 | 0.22 |
CHCHD3 |
coiled-coil-helix-coiled-coil-helix domain containing 3 |
28820 |
0.2 |
| chr3_24527242_24527393 | 0.22 |
ENSG00000228791 |
. |
8261 |
0.19 |
| chr14_56047342_56047590 | 0.22 |
KTN1 |
kinectin 1 (kinesin receptor) |
424 |
0.62 |
| chr5_111071431_111071582 | 0.21 |
STARD4-AS1 |
STARD4 antisense RNA 1 |
5420 |
0.22 |
| chr4_14864468_14865022 | 0.21 |
CPEB2 |
cytoplasmic polyadenylation element binding protein 2 |
139553 |
0.05 |
| chr2_109257559_109258085 | 0.21 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
13446 |
0.22 |
| chr4_141347804_141348226 | 0.21 |
CLGN |
calmegin |
529 |
0.78 |
| chr19_17790996_17791451 | 0.21 |
UNC13A |
unc-13 homolog A (C. elegans) |
4245 |
0.17 |
| chr15_31528566_31528785 | 0.21 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
75199 |
0.1 |
| chr5_82372177_82372328 | 0.21 |
TMEM167A |
transmembrane protein 167A |
1063 |
0.38 |
| chr9_124320076_124320249 | 0.21 |
DAB2IP |
DAB2 interacting protein |
9174 |
0.18 |
| chr11_113649028_113649338 | 0.21 |
CLDN25 |
claudin 25 |
1286 |
0.35 |
| chr10_62335914_62336175 | 0.21 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
3623 |
0.38 |
| chr13_23489541_23489915 | 0.20 |
ENSG00000240341 |
. |
36390 |
0.2 |
| chr9_5518324_5518475 | 0.20 |
PDCD1LG2 |
programmed cell death 1 ligand 2 |
7829 |
0.25 |
| chr1_145524889_145525225 | 0.20 |
ITGA10 |
integrin, alpha 10 |
22 |
0.95 |
| chr4_114897314_114897610 | 0.20 |
ARSJ |
arylsulfatase family, member J |
2690 |
0.37 |
| chr14_92864159_92864342 | 0.20 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
10473 |
0.28 |
| chr19_45931964_45932396 | 0.20 |
ERCC1 |
excision repair cross-complementing rodent repair deficiency, complementation group 1 (includes overlapping antisense sequence) |
288 |
0.84 |
| chr2_3286362_3286843 | 0.20 |
TSSC1-IT1 |
TSSC1 intronic transcript 1 (non-protein coding) |
18634 |
0.24 |
| chr10_33251129_33251359 | 0.20 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
967 |
0.68 |
| chr21_32707517_32707668 | 0.20 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
9002 |
0.31 |
| chr8_49014264_49014415 | 0.20 |
UBE2V2 |
ubiquitin-conjugating enzyme E2 variant 2 |
60032 |
0.14 |
| chr15_70994869_70995020 | 0.20 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
297 |
0.94 |
| chr7_18545603_18545779 | 0.20 |
HDAC9 |
histone deacetylase 9 |
3205 |
0.37 |
| chr1_31459830_31459981 | 0.20 |
PUM1 |
pumilio RNA-binding family member 1 |
8001 |
0.17 |
| chr21_17386302_17386519 | 0.20 |
ENSG00000252273 |
. |
21419 |
0.28 |
| chr1_21348527_21348859 | 0.20 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
28702 |
0.19 |
| chr18_32290997_32291170 | 0.20 |
DTNA |
dystrobrevin, alpha |
822 |
0.73 |
| chr11_124595320_124595471 | 0.19 |
NRGN |
neurogranin (protein kinase C substrate, RC3) |
14347 |
0.1 |
| chr9_124360553_124361088 | 0.19 |
RP11-524G24.2 |
|
22327 |
0.18 |
| chr1_39882885_39883036 | 0.19 |
MACF1 |
microtubule-actin crosslinking factor 1 |
2888 |
0.25 |
| chr1_225879081_225879232 | 0.19 |
RP11-145A3.1 |
|
9150 |
0.18 |
| chr5_154138190_154138341 | 0.19 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
1531 |
0.39 |
| chr7_50889730_50889881 | 0.19 |
GRB10 |
growth factor receptor-bound protein 10 |
28646 |
0.21 |
| chr5_167831410_167831561 | 0.19 |
WWC1 |
WW and C2 domain containing 1 |
1799 |
0.45 |
| chr2_44742897_44743048 | 0.19 |
CAMKMT |
calmodulin-lysine N-methyltransferase |
143091 |
0.04 |
| chrX_39504439_39504590 | 0.19 |
ENSG00000263730 |
. |
15956 |
0.29 |
| chr4_86853777_86853998 | 0.19 |
ARHGAP24 |
Rho GTPase activating protein 24 |
2461 |
0.32 |
| chr16_3999501_3999652 | 0.19 |
RP11-462G12.1 |
|
869 |
0.56 |
| chr15_68406642_68406961 | 0.19 |
PIAS1 |
protein inhibitor of activated STAT, 1 |
59887 |
0.12 |
| chr9_97781118_97781407 | 0.19 |
C9orf3 |
chromosome 9 open reading frame 3 |
13984 |
0.18 |
| chr13_51252397_51252695 | 0.19 |
DLEU7-AS1 |
DLEU7 antisense RNA 1 |
129446 |
0.05 |
| chr12_25486836_25487071 | 0.19 |
ENSG00000201439 |
. |
70409 |
0.1 |
| chr13_24901797_24901969 | 0.19 |
C1QTNF9-AS1 |
C1QTNF9 antisense RNA 1 |
6147 |
0.17 |
| chr2_145780897_145781240 | 0.19 |
ENSG00000253036 |
. |
311570 |
0.01 |
| chr13_91032921_91033108 | 0.19 |
ENSG00000207858 |
. |
149578 |
0.05 |
| chr5_76012905_76013056 | 0.19 |
CTD-2384B11.2 |
|
940 |
0.39 |
| chr2_196978205_196978455 | 0.19 |
RP11-347P5.1 |
|
37657 |
0.15 |
| chr7_18810006_18810178 | 0.19 |
ENSG00000222164 |
. |
37810 |
0.22 |
| chr8_128902590_128902741 | 0.19 |
TMEM75 |
transmembrane protein 75 |
57926 |
0.12 |
| chr12_1697602_1697753 | 0.19 |
FBXL14 |
F-box and leucine-rich repeat protein 14 |
5654 |
0.22 |
| chr16_56658952_56660008 | 0.19 |
MT1E |
metallothionein 1E |
93 |
0.91 |
| chr20_56283633_56283861 | 0.19 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
1069 |
0.63 |
| chr10_3087908_3088170 | 0.19 |
PFKP |
phosphofructokinase, platelet |
20486 |
0.26 |
| chr6_121869662_121869813 | 0.18 |
ENSG00000201379 |
. |
5965 |
0.25 |
| chrX_43831538_43831824 | 0.18 |
NDP |
Norrie disease (pseudoglioma) |
1069 |
0.61 |
| chr7_137439335_137439486 | 0.18 |
DGKI |
diacylglycerol kinase, iota |
91872 |
0.09 |
| chr7_36168506_36168695 | 0.18 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
24158 |
0.18 |
| chr5_6722325_6722630 | 0.18 |
PAPD7 |
PAP associated domain containing 7 |
7732 |
0.28 |
| chr14_85739826_85739977 | 0.18 |
ENSG00000252529 |
. |
1625 |
0.56 |
| chr7_4763069_4763220 | 0.18 |
FOXK1 |
forkhead box K1 |
17346 |
0.17 |
| chr11_33398887_33399038 | 0.18 |
ENSG00000223134 |
. |
22951 |
0.23 |
| chr14_39735421_39735572 | 0.18 |
RP11-407N17.3 |
cTAGE family member 5 isoform 4 |
6 |
0.81 |
| chr9_106853866_106854017 | 0.18 |
SMC2 |
structural maintenance of chromosomes 2 |
2600 |
0.44 |
| chr8_105373378_105373573 | 0.18 |
DCSTAMP |
dendrocyte expressed seven transmembrane protein |
12706 |
0.2 |
| chr2_227835215_227835366 | 0.18 |
ENSG00000212391 |
. |
1452 |
0.52 |
| chr21_46550403_46550801 | 0.18 |
ADARB1 |
adenosine deaminase, RNA-specific, B1 |
9787 |
0.16 |
| chr5_54776796_54776996 | 0.18 |
ENSG00000265135 |
. |
27858 |
0.18 |
| chr12_59227648_59227799 | 0.18 |
RP11-362K2.2 |
Protein LOC100506869 |
33569 |
0.19 |
| chr2_174361615_174361790 | 0.18 |
CDCA7 |
cell division cycle associated 7 |
142102 |
0.05 |
| chr21_17909068_17909746 | 0.17 |
ENSG00000207638 |
. |
2002 |
0.33 |
| chr3_178787181_178787457 | 0.17 |
ZMAT3 |
zinc finger, matrin-type 3 |
2035 |
0.36 |
| chr11_19599028_19599179 | 0.17 |
ENSG00000265210 |
. |
2246 |
0.31 |
| chr8_96443387_96443671 | 0.17 |
C8orf37 |
chromosome 8 open reading frame 37 |
162100 |
0.04 |
| chr1_109401335_109401486 | 0.17 |
SPATA42 |
spermatogenesis associated 42 (non-protein coding) |
1544 |
0.29 |
| chr8_38030618_38031135 | 0.17 |
BAG4 |
BCL2-associated athanogene 4 |
3230 |
0.13 |
| chr15_55652527_55652678 | 0.17 |
CCPG1 |
cell cycle progression 1 |
197 |
0.93 |
| chr2_18157972_18158481 | 0.17 |
ENSG00000212455 |
. |
63773 |
0.13 |
| chr2_33682993_33683144 | 0.17 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
45 |
0.99 |
| chr17_74736392_74736543 | 0.17 |
MFSD11 |
major facilitator superfamily domain containing 11 |
2314 |
0.13 |
| chr1_212841495_212841646 | 0.17 |
ENSG00000207491 |
. |
24036 |
0.13 |
| chr8_32084910_32085127 | 0.17 |
NRG1-IT2 |
NRG1 intronic transcript 2 (non-protein coding) |
6574 |
0.22 |
| chr10_94039164_94039387 | 0.17 |
ENSG00000199881 |
. |
7268 |
0.17 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.1 | 0.3 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.3 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.1 | 0.5 | GO:0001553 | luteinization(GO:0001553) |
| 0.1 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.1 | 0.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.1 | 0.2 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.1 | 0.2 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.2 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.5 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:2000051 | regulation of non-canonical Wnt signaling pathway(GO:2000050) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.3 | GO:1901532 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.2 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0015868 | purine ribonucleotide transport(GO:0015868) |
| 0.0 | 0.1 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.6 | GO:0006904 | vesicle docking involved in exocytosis(GO:0006904) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.7 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.2 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.2 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.1 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.1 | GO:0071688 | skeletal muscle myosin thick filament assembly(GO:0030241) myosin filament assembly(GO:0031034) striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:1904376 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0045040 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.0 | GO:2001279 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.0 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0051103 | DNA ligation involved in DNA repair(GO:0051103) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0051883 | disruption of cells of other organism involved in symbiotic interaction(GO:0051818) killing of cells in other organism involved in symbiotic interaction(GO:0051883) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.2 | GO:0009147 | pyrimidine nucleoside triphosphate metabolic process(GO:0009147) |
| 0.0 | 0.0 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) positive regulation of thymocyte apoptotic process(GO:0070245) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.0 | GO:1903312 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0060900 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.2 | GO:0015936 | coenzyme A metabolic process(GO:0015936) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.0 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0043001 | Golgi to plasma membrane protein transport(GO:0043001) |
| 0.0 | 0.0 | GO:0060536 | trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.0 | GO:0032277 | regulation of gonadotropin secretion(GO:0032276) negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.2 | GO:0030239 | myofibril assembly(GO:0030239) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.0 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.4 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.5 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.0 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.1 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.3 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.5 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.2 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.0 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.0 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.4 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.0 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.0 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |