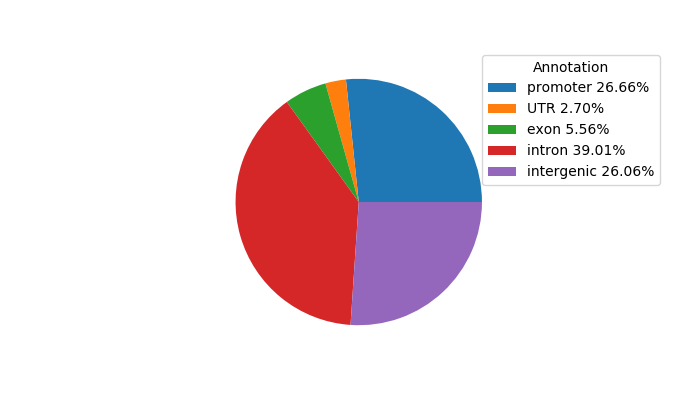
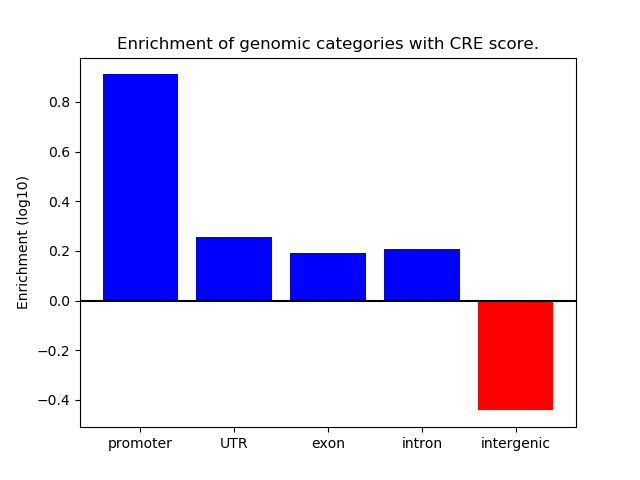
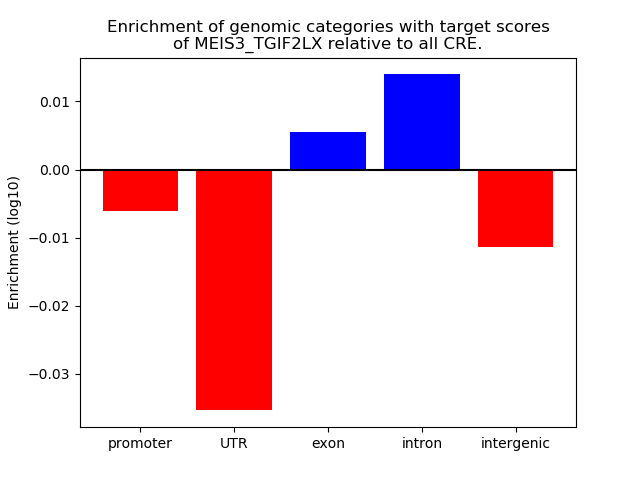
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
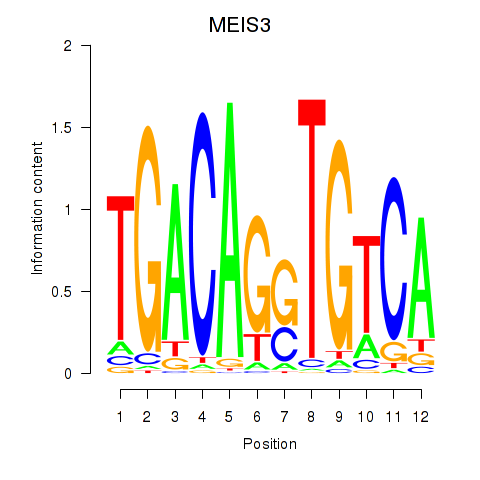
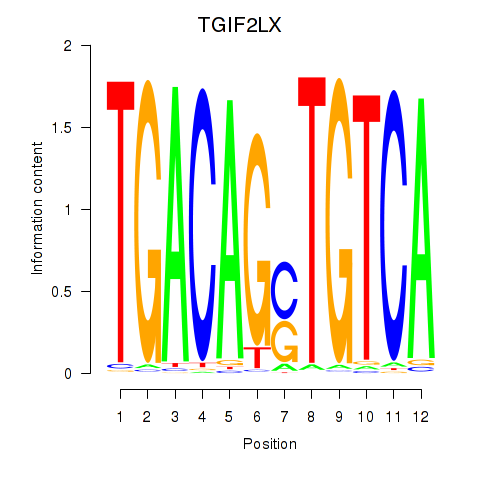
Results for MEIS3_TGIF2LX
Z-value: 0.96
Transcription factors associated with MEIS3_TGIF2LX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEIS3
|
ENSG00000105419.13 | Meis homeobox 3 |
|
TGIF2LX
|
ENSG00000153779.8 | TGFB induced factor homeobox 2 like X-linked |
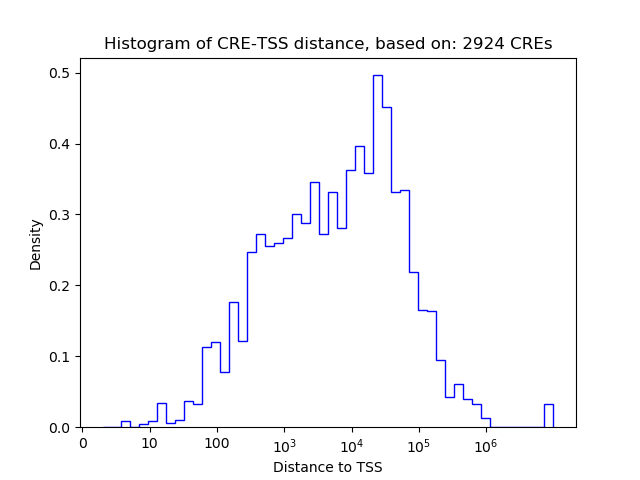
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_47920152_47920303 | MEIS3 | 245 | 0.910513 | 0.72 | 3.0e-02 | Click! |
| chr19_47885629_47885780 | MEIS3 | 26705 | 0.122103 | 0.67 | 5.0e-02 | Click! |
| chr19_47894051_47894202 | MEIS3 | 18283 | 0.140572 | 0.59 | 9.6e-02 | Click! |
| chr19_47921712_47921897 | MEIS3 | 575 | 0.715024 | 0.57 | 1.1e-01 | Click! |
| chr19_47922582_47922733 | MEIS3 | 85 | 0.965214 | 0.54 | 1.4e-01 | Click! |
Activity of the MEIS3_TGIF2LX motif across conditions
Conditions sorted by the z-value of the MEIS3_TGIF2LX motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
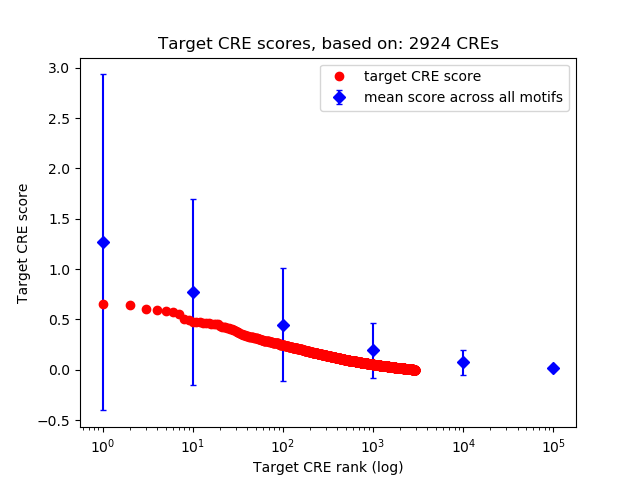
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_119597594_119597745 | 0.65 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
1594 |
0.37 |
| chr7_27191813_27192140 | 0.64 |
HOXA-AS3 |
HOXA cluster antisense RNA 3 |
2305 |
0.1 |
| chr2_65547750_65548155 | 0.61 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
13955 |
0.23 |
| chr20_58512577_58512765 | 0.59 |
PPP1R3D |
protein phosphatase 1, regulatory subunit 3D |
2681 |
0.21 |
| chr3_132349108_132349259 | 0.58 |
UBA5 |
ubiquitin-like modifier activating enzyme 5 |
24107 |
0.16 |
| chr11_120110982_120111133 | 0.57 |
POU2F3 |
POU class 2 homeobox 3 |
154 |
0.95 |
| chr7_40611540_40611691 | 0.55 |
AC004988.1 |
|
25088 |
0.27 |
| chr20_48989399_48989550 | 0.50 |
ENSG00000244376 |
. |
56546 |
0.13 |
| chr14_37570606_37570757 | 0.50 |
RP11-537P22.2 |
|
2913 |
0.31 |
| chr10_99442034_99442185 | 0.48 |
AVPI1 |
arginine vasopressin-induced 1 |
4971 |
0.16 |
| chr1_205462241_205462485 | 0.48 |
CDK18 |
cyclin-dependent kinase 18 |
11360 |
0.17 |
| chr4_181055224_181055668 | 0.47 |
NA |
NA |
> 106 |
NA |
| chr16_85673169_85673320 | 0.47 |
GSE1 |
Gse1 coiled-coil protein |
14794 |
0.16 |
| chr20_22384501_22385014 | 0.47 |
FOXA2 |
forkhead box A2 |
180344 |
0.03 |
| chr9_18689231_18689382 | 0.47 |
ENSG00000252960 |
. |
38161 |
0.2 |
| chr6_86114788_86115158 | 0.46 |
NT5E |
5'-nucleotidase, ecto (CD73) |
44836 |
0.19 |
| chr3_16308473_16308624 | 0.46 |
OXNAD1 |
oxidoreductase NAD-binding domain containing 1 |
1575 |
0.33 |
| chr2_9441510_9441661 | 0.46 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
94691 |
0.07 |
| chr22_33725446_33725597 | 0.45 |
SC22CB-1D7.1 |
|
8670 |
0.27 |
| chr6_106961305_106961653 | 0.43 |
AIM1 |
absent in melanoma 1 |
1749 |
0.38 |
| chr17_62195939_62196108 | 0.43 |
ERN1 |
endoplasmic reticulum to nucleus signaling 1 |
11462 |
0.16 |
| chr12_53264228_53264379 | 0.43 |
KRT78 |
keratin 78 |
21427 |
0.12 |
| chr4_125065453_125065672 | 0.43 |
ANKRD50 |
ankyrin repeat domain 50 |
568325 |
0.0 |
| chr5_112824477_112824726 | 0.42 |
MCC |
mutated in colorectal cancers |
74 |
0.97 |
| chr1_159111648_159111897 | 0.42 |
AIM2 |
absent in melanoma 2 |
1474 |
0.37 |
| chr4_31171719_31171870 | 0.41 |
RP11-619J20.1 |
|
376202 |
0.01 |
| chr17_50235287_50235438 | 0.40 |
CA10 |
carbonic anhydrase X |
61 |
0.99 |
| chr2_165696044_165696353 | 0.40 |
AC019181.2 |
|
1061 |
0.42 |
| chr2_166131422_166131573 | 0.40 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
19044 |
0.25 |
| chr5_2747693_2748131 | 0.39 |
IRX2 |
iroquois homeobox 2 |
3857 |
0.28 |
| chr4_2066511_2066662 | 0.38 |
NAT8L |
N-acetyltransferase 8-like (GCN5-related, putative) |
5047 |
0.17 |
| chr12_132915168_132915319 | 0.38 |
RP13-895J2.7 |
|
8807 |
0.19 |
| chr16_30078571_30078900 | 0.36 |
ALDOA |
aldolase A, fructose-bisphosphate |
89 |
0.92 |
| chr15_67311354_67311505 | 0.36 |
SMAD3 |
SMAD family member 3 |
44672 |
0.19 |
| chr20_10286915_10287750 | 0.35 |
ENSG00000211588 |
. |
55546 |
0.13 |
| chr11_17411398_17411576 | 0.35 |
KCNJ11 |
potassium inwardly-rectifying channel, subfamily J, member 11 |
609 |
0.69 |
| chrX_7053368_7053519 | 0.35 |
ENSG00000264268 |
. |
12458 |
0.23 |
| chr1_24706252_24706403 | 0.34 |
STPG1 |
sperm-tail PG-rich repeat containing 1 |
14 |
0.98 |
| chr1_177274527_177274678 | 0.34 |
ENSG00000222749 |
. |
119014 |
0.06 |
| chr14_45559377_45559745 | 0.33 |
ENSG00000212615 |
. |
2115 |
0.2 |
| chr22_30624979_30625130 | 0.33 |
RP1-102K2.6 |
|
10129 |
0.11 |
| chr1_209923190_209923341 | 0.33 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
6112 |
0.15 |
| chr5_4258108_4258399 | 0.33 |
CTD-2012M11.3 |
|
657951 |
0.0 |
| chr8_28208694_28208845 | 0.32 |
ZNF395 |
zinc finger protein 395 |
314 |
0.87 |
| chr2_238804511_238804662 | 0.32 |
ENSG00000263723 |
. |
26039 |
0.17 |
| chr10_36555876_36556027 | 0.32 |
ENSG00000237002 |
. |
171638 |
0.04 |
| chr19_6239308_6239459 | 0.32 |
CTC-503J8.4 |
|
19785 |
0.13 |
| chr20_16514306_16514457 | 0.32 |
KIF16B |
kinesin family member 16B |
39697 |
0.2 |
| chr2_54557164_54557870 | 0.32 |
C2orf73 |
chromosome 2 open reading frame 73 |
346 |
0.92 |
| chr20_19915434_19915955 | 0.31 |
RIN2 |
Ras and Rab interactor 2 |
45484 |
0.16 |
| chr5_119723718_119723919 | 0.31 |
ENSG00000251975 |
. |
50470 |
0.18 |
| chr3_58012091_58012242 | 0.31 |
FLNB |
filamin B, beta |
18039 |
0.25 |
| chr1_27928830_27929490 | 0.31 |
AHDC1 |
AT hook, DNA binding motif, containing 1 |
942 |
0.51 |
| chr3_126248153_126248304 | 0.31 |
CHST13 |
carbohydrate (chondroitin 4) sulfotransferase 13 |
5102 |
0.16 |
| chr19_42712082_42712233 | 0.31 |
DEDD2 |
death effector domain containing 2 |
9664 |
0.09 |
| chr7_29234619_29234840 | 0.30 |
CPVL |
carboxypeptidase, vitellogenic-like |
73 |
0.64 |
| chr7_35792332_35792483 | 0.30 |
SEPT7 |
septin 7 |
48135 |
0.14 |
| chr15_38543239_38543468 | 0.29 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
1174 |
0.65 |
| chr19_31830357_31830584 | 0.29 |
AC007796.1 |
|
9317 |
0.25 |
| chr17_3814774_3814925 | 0.29 |
P2RX1 |
purinergic receptor P2X, ligand-gated ion channel, 1 |
4945 |
0.17 |
| chr2_46524867_46525365 | 0.29 |
EPAS1 |
endothelial PAS domain protein 1 |
575 |
0.84 |
| chr22_40078600_40078751 | 0.29 |
CACNA1I |
calcium channel, voltage-dependent, T type, alpha 1I subunit |
111917 |
0.05 |
| chr6_130010303_130010454 | 0.29 |
ARHGAP18 |
Rho GTPase activating protein 18 |
20992 |
0.24 |
| chr6_45751303_45751454 | 0.29 |
ENSG00000252738 |
. |
137537 |
0.05 |
| chrX_281313_281835 | 0.29 |
LINC00685 |
long intergenic non-protein coding RNA 685 |
151 |
0.97 |
| chr7_113985866_113986017 | 0.29 |
FOXP2 |
forkhead box P2 |
68388 |
0.12 |
| chrX_42836312_42836609 | 0.29 |
MAOA |
monoamine oxidase A |
679007 |
0.0 |
| chr6_53659860_53660726 | 0.28 |
LRRC1 |
leucine rich repeat containing 1 |
405 |
0.79 |
| chr17_10703635_10703786 | 0.28 |
PIRT |
phosphoinositide-interacting regulator of transient receptor potential channels |
38052 |
0.13 |
| chr20_33585796_33585947 | 0.28 |
MYH7B |
myosin, heavy chain 7B, cardiac muscle, beta |
3177 |
0.19 |
| chr19_42636826_42637026 | 0.28 |
CTC-378H22.1 |
|
219 |
0.52 |
| chr16_70562877_70563028 | 0.28 |
ENSG00000221514 |
. |
460 |
0.7 |
| chr13_80454610_80454761 | 0.28 |
NDFIP2 |
Nedd4 family interacting protein 2 |
399097 |
0.01 |
| chrX_22153788_22153939 | 0.28 |
PHEX-AS1 |
PHEX antisense RNA 1 |
37237 |
0.17 |
| chr1_19278015_19278216 | 0.27 |
IFFO2 |
intermediate filament family orphan 2 |
4077 |
0.23 |
| chr1_11619306_11619457 | 0.27 |
PTCHD2 |
patched domain containing 2 |
25648 |
0.18 |
| chr17_40274740_40274891 | 0.27 |
HSPB9 |
heat shock protein, alpha-crystallin-related, B9 |
59 |
0.94 |
| chr11_60939398_60939549 | 0.27 |
VPS37C |
vacuolar protein sorting 37 homolog C (S. cerevisiae) |
10384 |
0.15 |
| chr7_99474179_99474330 | 0.27 |
OR2AE1 |
olfactory receptor, family 2, subfamily AE, member 1 |
426 |
0.76 |
| chr1_13910795_13911459 | 0.27 |
PDPN |
podoplanin |
362 |
0.87 |
| chr7_104908860_104909472 | 0.27 |
SRPK2 |
SRSF protein kinase 2 |
296 |
0.92 |
| chr13_53765255_53765406 | 0.27 |
ENSG00000241613 |
. |
66882 |
0.14 |
| chr8_99961674_99961862 | 0.26 |
OSR2 |
odd-skipped related transciption factor 2 |
1148 |
0.46 |
| chr6_157097564_157098248 | 0.26 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
1157 |
0.47 |
| chrY_231310_231809 | 0.26 |
NA |
NA |
> 106 |
NA |
| chr12_618388_618539 | 0.26 |
B4GALNT3 |
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
33836 |
0.15 |
| chr8_97602904_97603273 | 0.26 |
SDC2 |
syndecan 2 |
4473 |
0.31 |
| chr15_69868625_69868776 | 0.26 |
ENSG00000207119 |
. |
118212 |
0.05 |
| chr8_83687250_83687401 | 0.26 |
SNX16 |
sorting nexin 16 |
932224 |
0.0 |
| chr1_185529229_185529380 | 0.26 |
ENSG00000207108 |
. |
70356 |
0.11 |
| chr5_135347341_135347492 | 0.25 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
17168 |
0.2 |
| chr2_178032482_178032633 | 0.25 |
ENSG00000201102 |
. |
27154 |
0.12 |
| chr6_1960344_1960495 | 0.25 |
GMDS |
GDP-mannose 4,6-dehydratase |
215806 |
0.02 |
| chr3_112964540_112964768 | 0.25 |
BOC |
BOC cell adhesion associated, oncogene regulated |
29456 |
0.19 |
| chr2_113593759_113594336 | 0.25 |
IL1B |
interleukin 1, beta |
36 |
0.97 |
| chr4_74485025_74485176 | 0.25 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
1029 |
0.65 |
| chr19_4866504_4867667 | 0.25 |
PLIN3 |
perilipin 3 |
39 |
0.96 |
| chr4_77561140_77561386 | 0.25 |
AC107072.2 |
|
2487 |
0.32 |
| chr1_235132676_235132959 | 0.24 |
ENSG00000239690 |
. |
92884 |
0.08 |
| chr18_9611313_9611464 | 0.24 |
PPP4R1 |
protein phosphatase 4, regulatory subunit 1 |
2897 |
0.23 |
| chr10_6101040_6101191 | 0.24 |
IL2RA |
interleukin 2 receptor, alpha |
3138 |
0.2 |
| chrX_71288852_71289006 | 0.24 |
RGAG4 |
retrotransposon gag domain containing 4 |
62749 |
0.1 |
| chr12_131199797_131200212 | 0.24 |
RIMBP2 |
RIMS binding protein 2 |
822 |
0.73 |
| chr6_91424436_91424686 | 0.24 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
127797 |
0.06 |
| chr11_128142089_128142381 | 0.24 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
233054 |
0.02 |
| chr2_113875856_113876007 | 0.24 |
IL1RN |
interleukin 1 receptor antagonist |
457 |
0.77 |
| chr6_128580481_128581088 | 0.24 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
62542 |
0.14 |
| chr1_37195790_37195941 | 0.24 |
RP4-614N24.1 |
|
42876 |
0.19 |
| chr18_18823015_18823269 | 0.24 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
823 |
0.71 |
| chr12_106531293_106531501 | 0.24 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
2414 |
0.35 |
| chr21_44614471_44614622 | 0.23 |
CRYAA |
crystallin, alpha A |
24280 |
0.16 |
| chr13_45008185_45008336 | 0.23 |
TSC22D1 |
TSC22 domain family, member 1 |
2721 |
0.38 |
| chr5_114505235_114505578 | 0.23 |
TRIM36 |
tripartite motif containing 36 |
200 |
0.94 |
| chr2_109934979_109935130 | 0.23 |
ENSG00000265965 |
. |
4973 |
0.34 |
| chr2_236569141_236569292 | 0.23 |
AGAP1 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1 |
9032 |
0.28 |
| chr21_32537085_32537236 | 0.23 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
34621 |
0.22 |
| chr14_102050713_102051023 | 0.23 |
DIO3 |
deiodinase, iodothyronine, type III |
23180 |
0.17 |
| chr6_44083954_44084715 | 0.23 |
TMEM63B |
transmembrane protein 63B |
10317 |
0.15 |
| chr20_6749993_6750220 | 0.23 |
BMP2 |
bone morphogenetic protein 2 |
1795 |
0.51 |
| chr8_63967849_63968000 | 0.23 |
GGH |
gamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) |
16194 |
0.18 |
| chr12_95509986_95510137 | 0.23 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
688 |
0.74 |
| chr16_70749372_70749620 | 0.22 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
15348 |
0.13 |
| chr17_38178315_38178466 | 0.22 |
MED24 |
mediator complex subunit 24 |
1023 |
0.38 |
| chr3_113464275_113464538 | 0.22 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
235 |
0.9 |
| chr17_3715887_3716038 | 0.22 |
C17orf85 |
chromosome 17 open reading frame 85 |
582 |
0.7 |
| chr1_185528978_185529129 | 0.22 |
ENSG00000207108 |
. |
70607 |
0.11 |
| chr4_55650955_55651106 | 0.22 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
126945 |
0.06 |
| chr12_25206187_25206540 | 0.22 |
LRMP |
lymphoid-restricted membrane protein |
689 |
0.71 |
| chr1_203648008_203648159 | 0.22 |
ATP2B4 |
ATPase, Ca++ transporting, plasma membrane 4 |
3854 |
0.27 |
| chr20_31472115_31472266 | 0.22 |
EFCAB8 |
EF-hand calcium binding domain 8 |
25461 |
0.15 |
| chr16_51671642_51672070 | 0.22 |
ENSG00000223168 |
. |
33618 |
0.25 |
| chr2_192709104_192709385 | 0.22 |
AC098617.1 |
|
2021 |
0.33 |
| chr4_29610349_29610500 | 0.22 |
ENSG00000252303 |
. |
98167 |
0.09 |
| chr17_39821847_39822265 | 0.22 |
EIF1 |
eukaryotic translation initiation factor 1 |
23083 |
0.09 |
| chr22_27441616_27441767 | 0.22 |
ENSG00000200443 |
. |
9841 |
0.32 |
| chr17_28552731_28552882 | 0.22 |
SLC6A4 |
solute carrier family 6 (neurotransmitter transporter), member 4 |
9910 |
0.11 |
| chr10_73532392_73533248 | 0.22 |
C10orf54 |
chromosome 10 open reading frame 54 |
435 |
0.83 |
| chr2_74220808_74220959 | 0.21 |
TET3 |
tet methylcytosine dioxygenase 3 |
8957 |
0.16 |
| chr4_145621271_145621422 | 0.21 |
HHIP-AS1 |
HHIP antisense RNA 1 |
38837 |
0.19 |
| chr19_18670319_18670560 | 0.21 |
KXD1 |
KxDL motif containing 1 |
517 |
0.55 |
| chr1_210465594_210466382 | 0.21 |
HHAT |
hedgehog acyltransferase |
35608 |
0.17 |
| chr9_34990286_34990475 | 0.21 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
117 |
0.95 |
| chr3_99349930_99350081 | 0.21 |
COL8A1 |
collagen, type VIII, alpha 1 |
7314 |
0.29 |
| chr1_75811389_75811540 | 0.21 |
SLC44A5 |
solute carrier family 44, member 5 |
50857 |
0.17 |
| chr18_67712930_67713081 | 0.21 |
RTTN |
rotatin |
25051 |
0.25 |
| chr8_845837_845988 | 0.21 |
ERICH1-AS1 |
ERICH1 antisense RNA 1 |
12584 |
0.3 |
| chr3_30544040_30544191 | 0.21 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
103879 |
0.08 |
| chr7_76140575_76140726 | 0.21 |
UPK3B |
uroplakin 3B |
718 |
0.6 |
| chr16_89381066_89381217 | 0.21 |
AC137932.6 |
|
6400 |
0.14 |
| chr15_78112406_78112810 | 0.21 |
LINGO1 |
leucine rich repeat and Ig domain containing 1 |
66 |
0.97 |
| chr6_90119364_90119517 | 0.21 |
RRAGD |
Ras-related GTP binding D |
2349 |
0.29 |
| chr22_48027568_48027719 | 0.21 |
LL22NC03-75H12.2 |
Novel protein; Uncharacterized protein |
144783 |
0.05 |
| chr19_39154900_39155424 | 0.21 |
ACTN4 |
actinin, alpha 4 |
16784 |
0.11 |
| chr3_10050371_10050522 | 0.21 |
EMC3 |
ER membrane protein complex subunit 3 |
2354 |
0.15 |
| chr9_110495286_110496024 | 0.20 |
AL162389.1 |
Uncharacterized protein |
44764 |
0.15 |
| chr9_100870862_100871013 | 0.20 |
TRIM14 |
tripartite motif containing 14 |
10543 |
0.19 |
| chr2_57825783_57825934 | 0.20 |
ENSG00000212168 |
. |
54188 |
0.18 |
| chr17_8368616_8368767 | 0.20 |
NDEL1 |
nudE neurodevelopment protein 1-like 1 |
14592 |
0.14 |
| chr1_39624513_39624771 | 0.20 |
ENSG00000222378 |
. |
4674 |
0.21 |
| chr8_846023_846174 | 0.20 |
ERICH1-AS1 |
ERICH1 antisense RNA 1 |
12770 |
0.3 |
| chr5_36151340_36151953 | 0.20 |
LMBRD2 |
LMBR1 domain containing 2 |
417 |
0.46 |
| chr9_11751779_11751930 | 0.20 |
ENSG00000222581 |
. |
548597 |
0.0 |
| chr12_104657438_104657589 | 0.20 |
RP11-818F20.5 |
|
16987 |
0.18 |
| chr20_62586715_62586914 | 0.20 |
UCKL1 |
uridine-cytidine kinase 1-like 1 |
954 |
0.28 |
| chr11_128773455_128773606 | 0.20 |
C11orf45 |
chromosome 11 open reading frame 45 |
2062 |
0.28 |
| chr4_184427310_184427800 | 0.20 |
ING2 |
inhibitor of growth family, member 2 |
195 |
0.93 |
| chr3_99363600_99363751 | 0.20 |
COL8A1 |
collagen, type VIII, alpha 1 |
6221 |
0.28 |
| chr7_148473091_148473297 | 0.20 |
ENSG00000251712 |
. |
44910 |
0.12 |
| chr2_106886104_106886958 | 0.19 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
75736 |
0.1 |
| chr22_45624284_45624435 | 0.19 |
KIAA0930 |
KIAA0930 |
1575 |
0.36 |
| chr8_96128376_96128527 | 0.19 |
PLEKHF2 |
pleckstrin homology domain containing, family F (with FYVE domain) member 2 |
17581 |
0.16 |
| chr2_60525179_60525372 | 0.19 |
ENSG00000200807 |
. |
86465 |
0.09 |
| chrX_17342148_17342299 | 0.19 |
ENSG00000238764 |
. |
33830 |
0.19 |
| chr12_110720958_110721647 | 0.19 |
ATP2A2 |
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
1816 |
0.41 |
| chr15_74873270_74873421 | 0.19 |
CLK3 |
CDC-like kinase 3 |
27368 |
0.12 |
| chr15_72529055_72529849 | 0.19 |
PKM |
pyruvate kinase, muscle |
5288 |
0.17 |
| chr11_13300075_13300637 | 0.19 |
ARNTL |
aryl hydrocarbon receptor nuclear translocator-like |
924 |
0.7 |
| chr19_2503722_2503936 | 0.19 |
ENSG00000252962 |
. |
690 |
0.64 |
| chr12_123356373_123356852 | 0.19 |
HIP1R |
huntingtin interacting protein 1 related |
11900 |
0.16 |
| chr18_29028288_29028439 | 0.19 |
DSG3 |
desmoglein 3 |
605 |
0.71 |
| chr20_11137905_11138056 | 0.19 |
C20orf187 |
chromosome 20 open reading frame 187 |
129169 |
0.06 |
| chr6_27858007_27858288 | 0.19 |
HIST1H3J |
histone cluster 1, H3j |
423 |
0.61 |
| chr12_110440425_110440804 | 0.19 |
ANKRD13A |
ankyrin repeat domain 13A |
3286 |
0.23 |
| chr4_30725920_30726277 | 0.19 |
PCDH7 |
protocadherin 7 |
2121 |
0.46 |
| chr2_105320423_105320574 | 0.18 |
ENSG00000207249 |
. |
29194 |
0.21 |
| chr5_54455785_54456014 | 0.18 |
GPX8 |
glutathione peroxidase 8 (putative) |
47 |
0.96 |
| chr8_97864758_97864909 | 0.18 |
CPQ |
carboxypeptidase Q |
91365 |
0.1 |
| chr13_76079768_76079919 | 0.18 |
TBC1D4 |
TBC1 domain family, member 4 |
23593 |
0.17 |
| chr6_139433656_139433807 | 0.18 |
HECA |
headcase homolog (Drosophila) |
22518 |
0.22 |
| chr8_32412534_32412685 | 0.18 |
NRG1 |
neuregulin 1 |
6364 |
0.32 |
| chr5_139721787_139722024 | 0.18 |
HBEGF |
heparin-binding EGF-like growth factor |
4283 |
0.14 |
| chr15_62599631_62599782 | 0.18 |
RP11-299H22.5 |
|
36827 |
0.17 |
| chr2_175197056_175197207 | 0.18 |
SP9 |
Sp9 transcription factor |
2543 |
0.24 |
| chr5_107285614_107285882 | 0.18 |
ENSG00000251732 |
. |
139418 |
0.05 |
| chrX_128789131_128789414 | 0.18 |
APLN |
apelin |
339 |
0.92 |
| chr20_62670859_62671010 | 0.18 |
ZNF512B |
zinc finger protein 512B |
996 |
0.34 |
| chr15_68143074_68143225 | 0.18 |
ENSG00000206625 |
. |
10766 |
0.24 |
| chr11_110166292_110166969 | 0.18 |
RDX |
radixin |
707 |
0.81 |
| chr4_185713844_185713995 | 0.18 |
ACSL1 |
acyl-CoA synthetase long-chain family member 1 |
13023 |
0.18 |
| chr11_36170786_36170937 | 0.18 |
ENSG00000263389 |
. |
139213 |
0.04 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.1 | 0.3 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.1 | 0.2 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.2 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.1 | 0.2 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.2 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0003171 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:2000696 | nephron tubule epithelial cell differentiation(GO:0072160) regulation of nephron tubule epithelial cell differentiation(GO:0072182) regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 0.1 | GO:0003179 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.2 | GO:0036303 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 | 0.1 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.2 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0009448 | gamma-aminobutyric acid metabolic process(GO:0009448) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.3 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.2 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.5 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.1 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.1 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.0 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.0 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.1 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.1 | GO:0031274 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0010662 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0071501 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:2000095 | regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.0 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0006837 | serotonin transport(GO:0006837) |
| 0.0 | 0.0 | GO:1902305 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.0 | GO:0060157 | urinary bladder development(GO:0060157) |
| 0.0 | 0.0 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.0 | GO:2000189 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0048821 | erythrocyte development(GO:0048821) myeloid cell development(GO:0061515) |
| 0.0 | 0.0 | GO:0060742 | epithelial cell differentiation involved in prostate gland development(GO:0060742) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.0 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0006893 | Golgi to plasma membrane transport(GO:0006893) vesicle-mediated transport to the plasma membrane(GO:0098876) |
| 0.0 | 0.1 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0002070 | epithelial cell maturation(GO:0002070) |
| 0.0 | 0.0 | GO:0003283 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.0 | GO:0043921 | modulation by host of viral transcription(GO:0043921) modulation by host of symbiont transcription(GO:0052472) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.0 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.0 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0052805 | histidine catabolic process(GO:0006548) imidazole-containing compound catabolic process(GO:0052805) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.1 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.2 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.2 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0019907 | cyclin-dependent protein kinase activating kinase holoenzyme complex(GO:0019907) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.0 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.2 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.2 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.3 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.0 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0071855 | neuropeptide receptor binding(GO:0071855) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.0 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.0 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | PID CXCR4 PATHWAY | CXCR4-mediated signaling events |
| 0.0 | 0.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.0 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |