Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
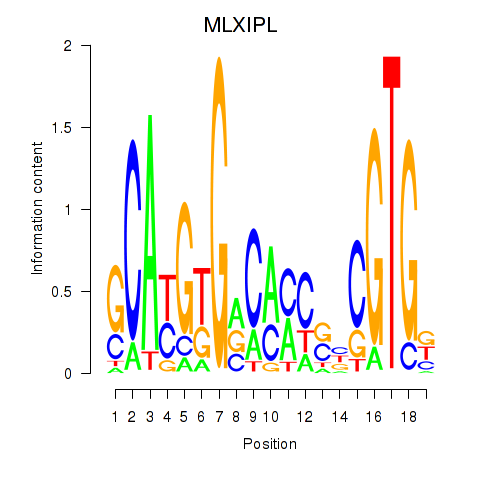
Results for MLXIPL
Z-value: 0.78
Transcription factors associated with MLXIPL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MLXIPL
|
ENSG00000009950.11 | MLX interacting protein like |
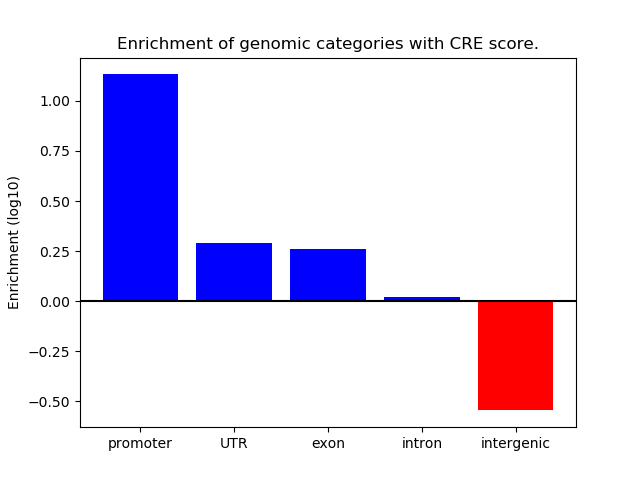
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_73028930_73029081 | MLXIPL | 1486 | 0.325724 | 0.24 | 5.4e-01 | Click! |
| chr7_73021530_73021732 | MLXIPL | 8860 | 0.141963 | 0.23 | 5.6e-01 | Click! |
| chr7_73038945_73039096 | MLXIPL | 147 | 0.941543 | 0.18 | 6.4e-01 | Click! |
| chr7_73037398_73037611 | MLXIPL | 1318 | 0.358771 | -0.13 | 7.5e-01 | Click! |
| chr7_73038556_73038914 | MLXIPL | 87 | 0.959310 | 0.08 | 8.4e-01 | Click! |
Activity of the MLXIPL motif across conditions
Conditions sorted by the z-value of the MLXIPL motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
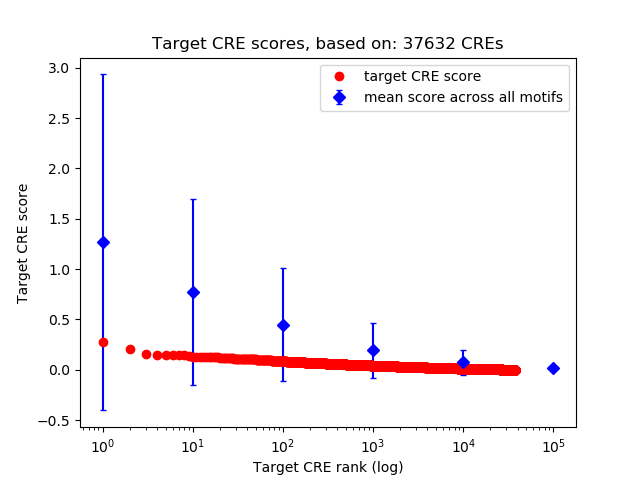
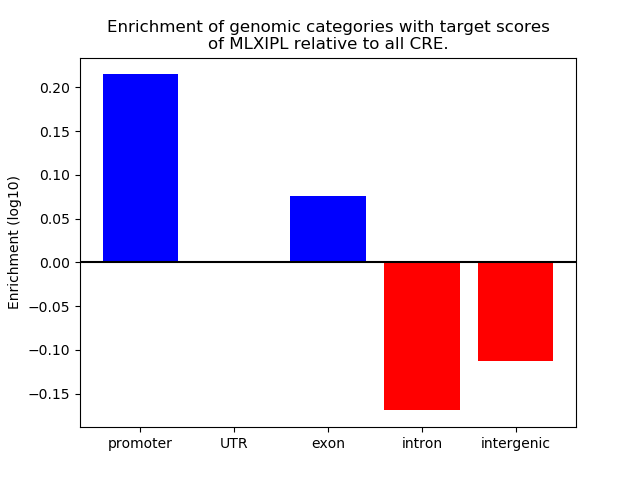
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_114965108_114965301 | 0.28 |
CDC16 |
cell division cycle 16 |
35158 |
0.14 |
| chr22_46988879_46989444 | 0.21 |
GRAMD4 |
GRAM domain containing 4 |
16135 |
0.21 |
| chr16_89042817_89042968 | 0.16 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
509 |
0.75 |
| chr16_1429359_1429577 | 0.15 |
ENSG00000265529 |
. |
44 |
0.66 |
| chr19_38423084_38423604 | 0.15 |
SIPA1L3 |
signal-induced proliferation-associated 1 like 3 |
25476 |
0.15 |
| chr8_13423939_13424235 | 0.15 |
C8orf48 |
chromosome 8 open reading frame 48 |
265 |
0.93 |
| chr10_20106092_20106642 | 0.14 |
PLXDC2 |
plexin domain containing 2 |
1199 |
0.49 |
| chr4_156589035_156589493 | 0.14 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
449 |
0.88 |
| chr17_3819186_3819649 | 0.13 |
P2RX1 |
purinergic receptor P2X, ligand-gated ion channel, 1 |
377 |
0.84 |
| chr15_39871853_39872515 | 0.13 |
THBS1 |
thrombospondin 1 |
1096 |
0.52 |
| chr7_37325864_37326015 | 0.13 |
ELMO1 |
engulfment and cell motility 1 |
56428 |
0.13 |
| chr15_33010722_33010990 | 0.13 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
651 |
0.73 |
| chr11_47399612_47399968 | 0.13 |
SPI1 |
spleen focus forming virus (SFFV) proviral integration oncogene |
152 |
0.91 |
| chr18_12253921_12254616 | 0.13 |
CIDEA |
cell death-inducing DFFA-like effector a |
50 |
0.98 |
| chr2_160918347_160919586 | 0.13 |
PLA2R1 |
phospholipase A2 receptor 1, 180kDa |
155 |
0.98 |
| chr16_3057439_3057643 | 0.13 |
LA16c-380H5.2 |
|
2439 |
0.09 |
| chr9_101014697_101014848 | 0.13 |
TBC1D2 |
TBC1 domain family, member 2 |
3090 |
0.33 |
| chr18_25755743_25755974 | 0.12 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
1552 |
0.58 |
| chr7_138348970_138349258 | 0.12 |
SVOPL |
SVOP-like |
145 |
0.97 |
| chr11_1328206_1328468 | 0.12 |
TOLLIP |
toll interacting protein |
2512 |
0.21 |
| chr20_3766593_3766776 | 0.12 |
CENPB |
centromere protein B, 80kDa |
653 |
0.45 |
| chr17_55939321_55939589 | 0.12 |
CUEDC1 |
CUE domain containing 1 |
5321 |
0.19 |
| chr6_144357186_144358268 | 0.12 |
PLAGL1 |
pleiomorphic adenoma gene-like 1 |
28008 |
0.21 |
| chr19_33685813_33686340 | 0.11 |
LRP3 |
low density lipoprotein receptor-related protein 3 |
586 |
0.65 |
| chr6_125684588_125685144 | 0.11 |
RP11-735G4.1 |
|
10604 |
0.27 |
| chr10_97049818_97050125 | 0.11 |
PDLIM1 |
PDZ and LIM domain 1 |
810 |
0.7 |
| chr7_27190086_27190291 | 0.11 |
HOXA-AS3 |
HOXA cluster antisense RNA 3 |
517 |
0.49 |
| chr11_68082046_68082433 | 0.11 |
LRP5 |
low density lipoprotein receptor-related protein 5 |
2162 |
0.35 |
| chr17_34599013_34599342 | 0.11 |
TBC1D3C |
TBC1 domain family, member 3C |
7145 |
0.15 |
| chr2_238768253_238768964 | 0.11 |
RAMP1 |
receptor (G protein-coupled) activity modifying protein 1 |
11 |
0.98 |
| chr12_113860437_113860657 | 0.11 |
SDSL |
serine dehydratase-like |
289 |
0.88 |
| chr5_121647834_121648295 | 0.11 |
SNCAIP |
synuclein, alpha interacting protein |
132 |
0.9 |
| chr2_208365436_208365587 | 0.11 |
CREB1 |
cAMP responsive element binding protein 1 |
28950 |
0.18 |
| chr2_45395914_45396553 | 0.11 |
SIX2 |
SIX homeobox 2 |
159664 |
0.04 |
| chr6_150464730_150464955 | 0.11 |
PPP1R14C |
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
630 |
0.74 |
| chr16_70719186_70719575 | 0.11 |
MTSS1L |
metastasis suppressor 1-like |
589 |
0.67 |
| chr1_178063691_178064117 | 0.11 |
RASAL2 |
RAS protein activator like 2 |
628 |
0.84 |
| chr6_72129322_72130506 | 0.11 |
ENSG00000207827 |
. |
16590 |
0.22 |
| chr2_109211293_109211679 | 0.11 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
6564 |
0.25 |
| chr17_34510955_34511411 | 0.11 |
AC131056.3 |
|
342 |
0.79 |
| chr3_43811358_43811652 | 0.11 |
ABHD5 |
abhydrolase domain containing 5 |
58306 |
0.14 |
| chr7_158996146_158996297 | 0.11 |
VIPR2 |
vasoactive intestinal peptide receptor 2 |
58572 |
0.16 |
| chr6_54711000_54712080 | 0.11 |
FAM83B |
family with sequence similarity 83, member B |
29 |
0.98 |
| chr12_562387_562595 | 0.11 |
B4GALNT3 |
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
7039 |
0.21 |
| chr19_8559121_8559517 | 0.11 |
PRAM1 |
PML-RARA regulated adaptor molecule 1 |
8176 |
0.12 |
| chr3_149094469_149095370 | 0.10 |
TM4SF1-AS1 |
TM4SF1 antisense RNA 1 |
646 |
0.47 |
| chr1_231163919_231164070 | 0.10 |
ENSG00000221290 |
. |
8324 |
0.15 |
| chr22_50608207_50608432 | 0.10 |
PANX2 |
pannexin 2 |
841 |
0.41 |
| chr10_29010987_29011704 | 0.10 |
ENSG00000201001 |
. |
38387 |
0.14 |
| chr11_13031575_13031768 | 0.10 |
ENSG00000266625 |
. |
46484 |
0.16 |
| chr6_39281750_39281998 | 0.10 |
KCNK17 |
potassium channel, subfamily K, member 17 |
363 |
0.89 |
| chr8_11057742_11057983 | 0.10 |
XKR6 |
XK, Kell blood group complex subunit-related family, member 6 |
986 |
0.53 |
| chr3_35680671_35680927 | 0.10 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
282 |
0.93 |
| chr20_50720613_50721041 | 0.10 |
ZFP64 |
ZFP64 zinc finger protein |
562 |
0.86 |
| chr14_103058041_103058305 | 0.10 |
RCOR1 |
REST corepressor 1 |
825 |
0.6 |
| chr6_116719287_116719943 | 0.10 |
DSE |
dermatan sulfate epimerase |
27505 |
0.14 |
| chr4_134071254_134071890 | 0.10 |
PCDH10 |
protocadherin 10 |
1102 |
0.7 |
| chr13_21634768_21635319 | 0.10 |
LATS2 |
large tumor suppressor kinase 2 |
614 |
0.69 |
| chr20_25213879_25214247 | 0.10 |
AL035252.1 |
HCG2018895; Uncharacterized protein |
6693 |
0.16 |
| chr1_213090021_213090397 | 0.10 |
VASH2 |
vasohibin 2 |
33653 |
0.16 |
| chr18_74534859_74535018 | 0.10 |
ZNF236 |
zinc finger protein 236 |
375 |
0.68 |
| chr10_28968795_28969240 | 0.10 |
BAMBI |
BMP and activin membrane-bound inhibitor |
2746 |
0.26 |
| chr16_89630259_89630469 | 0.10 |
RPL13 |
ribosomal protein L13 |
1586 |
0.22 |
| chr1_181455162_181455370 | 0.10 |
CACNA1E |
calcium channel, voltage-dependent, R type, alpha 1E subunit |
2385 |
0.45 |
| chr11_94501626_94502525 | 0.10 |
AMOTL1 |
angiomotin like 1 |
538 |
0.82 |
| chr12_16501800_16502026 | 0.10 |
MGST1 |
microsomal glutathione S-transferase 1 |
1182 |
0.59 |
| chr12_120662282_120662577 | 0.09 |
PXN |
paxillin |
124 |
0.94 |
| chr1_65613306_65613822 | 0.09 |
AK4 |
adenylate kinase 4 |
51 |
0.98 |
| chr2_91635039_91635306 | 0.09 |
IGKV1OR2-118 |
immunoglobulin kappa variable 1/OR2-118 (pseudogene) |
43915 |
0.21 |
| chrX_9754895_9755291 | 0.09 |
SHROOM2 |
shroom family member 2 |
597 |
0.54 |
| chr4_157890038_157890662 | 0.09 |
PDGFC |
platelet derived growth factor C |
1705 |
0.41 |
| chr16_81466874_81467025 | 0.09 |
CMIP |
c-Maf inducing protein |
11826 |
0.23 |
| chr16_89043094_89043288 | 0.09 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
210 |
0.92 |
| chr18_52626293_52626952 | 0.09 |
CCDC68 |
coiled-coil domain containing 68 |
117 |
0.97 |
| chr17_47077080_47077320 | 0.09 |
IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
2092 |
0.16 |
| chr8_125739093_125740062 | 0.09 |
MTSS1 |
metastasis suppressor 1 |
280 |
0.94 |
| chr18_52495652_52496658 | 0.09 |
RAB27B |
RAB27B, member RAS oncogene family |
725 |
0.77 |
| chr5_146832482_146833250 | 0.09 |
DPYSL3 |
dihydropyrimidinase-like 3 |
385 |
0.91 |
| chr11_61448104_61448359 | 0.09 |
DAGLA |
diacylglycerol lipase, alpha |
326 |
0.89 |
| chr11_3181150_3181578 | 0.09 |
OSBPL5 |
oxysterol binding protein-like 5 |
1259 |
0.4 |
| chr3_157820963_157821448 | 0.09 |
SHOX2 |
short stature homeobox 2 |
1878 |
0.36 |
| chr6_43044172_43044516 | 0.09 |
PTK7 |
protein tyrosine kinase 7 |
208 |
0.85 |
| chrX_151141627_151141778 | 0.09 |
GABRE |
gamma-aminobutyric acid (GABA) A receptor, epsilon |
1441 |
0.33 |
| chr2_62798336_62798579 | 0.09 |
TMEM17 |
transmembrane protein 17 |
64981 |
0.11 |
| chr15_83875054_83875562 | 0.09 |
HDGFRP3 |
Hepatoma-derived growth factor-related protein 3 |
1462 |
0.4 |
| chr5_156886857_156887334 | 0.09 |
CTB-109A12.1 |
|
9 |
0.54 |
| chr16_70729499_70729650 | 0.09 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
78 |
0.96 |
| chr4_185268825_185269197 | 0.09 |
ENSG00000244512 |
. |
68922 |
0.09 |
| chr16_23912477_23912952 | 0.09 |
PRKCB |
protein kinase C, beta |
64170 |
0.12 |
| chr12_63328180_63328595 | 0.09 |
PPM1H |
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
430 |
0.76 |
| chr12_88973452_88974158 | 0.09 |
KITLG |
KIT ligand |
433 |
0.82 |
| chr14_106376117_106376268 | 0.09 |
ENSG00000228131 |
. |
94 |
0.69 |
| chr4_91049201_91049889 | 0.09 |
CCSER1 |
coiled-coil serine-rich protein 1 |
809 |
0.78 |
| chr4_41215795_41216596 | 0.09 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
280 |
0.92 |
| chr21_46573548_46573916 | 0.09 |
ADARB1 |
adenosine deaminase, RNA-specific, B1 |
17825 |
0.17 |
| chr11_747907_748726 | 0.09 |
TALDO1 |
transaldolase 1 |
888 |
0.33 |
| chr16_31276973_31277152 | 0.09 |
ENSG00000252876 |
. |
1750 |
0.22 |
| chr10_72431780_72432013 | 0.09 |
ADAMTS14 |
ADAM metallopeptidase with thrombospondin type 1 motif, 14 |
663 |
0.78 |
| chr9_139929883_139930034 | 0.09 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
2496 |
0.09 |
| chr4_57976289_57976477 | 0.08 |
IGFBP7 |
insulin-like growth factor binding protein 7 |
168 |
0.72 |
| chr16_85603204_85603491 | 0.08 |
GSE1 |
Gse1 coiled-coil protein |
41668 |
0.16 |
| chr21_44831868_44832019 | 0.08 |
SIK1 |
salt-inducible kinase 1 |
15065 |
0.27 |
| chrX_10587548_10588615 | 0.08 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
378 |
0.91 |
| chr10_18429523_18430148 | 0.08 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
52 |
0.99 |
| chr19_1017181_1017478 | 0.08 |
TMEM259 |
transmembrane protein 259 |
93 |
0.91 |
| chr18_31157392_31158104 | 0.08 |
ASXL3 |
additional sex combs like 3 (Drosophila) |
848 |
0.76 |
| chr11_11642193_11642674 | 0.08 |
GALNT18 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 18 |
1119 |
0.56 |
| chr11_3043840_3044115 | 0.08 |
CARS-AS1 |
CARS antisense RNA 1 |
6647 |
0.13 |
| chrX_51637827_51638413 | 0.08 |
MAGED1 |
melanoma antigen family D, 1 |
1385 |
0.5 |
| chr7_70159815_70160908 | 0.08 |
AUTS2 |
autism susceptibility candidate 2 |
33764 |
0.25 |
| chr11_67210518_67210749 | 0.08 |
CORO1B |
coronin, actin binding protein, 1B |
310 |
0.72 |
| chr15_38545953_38546360 | 0.08 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
774 |
0.79 |
| chr5_150021079_150021262 | 0.08 |
SYNPO |
synaptopodin |
930 |
0.5 |
| chr15_99558654_99558941 | 0.08 |
PGPEP1L |
pyroglutamyl-peptidase I-like |
7773 |
0.22 |
| chr19_16556537_16556749 | 0.08 |
CTD-2013N17.4 |
|
4886 |
0.15 |
| chr20_36433171_36433322 | 0.08 |
CTNNBL1 |
catenin, beta like 1 |
27571 |
0.22 |
| chr15_80270114_80270265 | 0.08 |
BCL2A1 |
BCL2-related protein A1 |
6401 |
0.2 |
| chr16_57723286_57723437 | 0.08 |
CCDC135 |
coiled-coil domain containing 135 |
5344 |
0.14 |
| chr14_53301814_53302501 | 0.08 |
FERMT2 |
fermitin family member 2 |
29082 |
0.15 |
| chr14_100204275_100204562 | 0.08 |
EML1 |
echinoderm microtubule associated protein like 1 |
332 |
0.9 |
| chr14_72050667_72050818 | 0.08 |
SIPA1L1 |
signal-induced proliferation-associated 1 like 1 |
2256 |
0.45 |
| chr9_137393023_137393337 | 0.08 |
RXRA |
retinoid X receptor, alpha |
94752 |
0.07 |
| chr3_159482295_159482916 | 0.08 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
287 |
0.9 |
| chr16_88218272_88218475 | 0.08 |
BANP |
BTG3 associated nuclear protein |
214749 |
0.02 |
| chr7_116311713_116312022 | 0.08 |
MET |
met proto-oncogene |
577 |
0.82 |
| chr14_38053784_38053935 | 0.08 |
FOXA1 |
forkhead box A1 |
10380 |
0.23 |
| chr3_123602332_123603013 | 0.08 |
MYLK |
myosin light chain kinase |
477 |
0.85 |
| chr6_78172796_78173983 | 0.08 |
HTR1B |
5-hydroxytryptamine (serotonin) receptor 1B, G protein-coupled |
101 |
0.99 |
| chr17_56348208_56348359 | 0.08 |
MPO |
myeloperoxidase |
2570 |
0.2 |
| chr18_74178798_74178949 | 0.08 |
ZNF516 |
zinc finger protein 516 |
23862 |
0.16 |
| chr8_127837007_127837644 | 0.08 |
ENSG00000212451 |
. |
153558 |
0.04 |
| chr9_140345898_140346049 | 0.08 |
NSMF |
NMDA receptor synaptonuclear signaling and neuronal migration factor |
2971 |
0.16 |
| chr9_130308551_130308785 | 0.08 |
FAM129B |
family with sequence similarity 129, member B |
22699 |
0.15 |
| chrX_76703737_76703926 | 0.08 |
FGF16 |
fibroblast growth factor 16 |
5817 |
0.34 |
| chr1_158087708_158088406 | 0.08 |
KIRREL |
kin of IRRE like (Drosophila) |
31587 |
0.18 |
| chr12_66218806_66219120 | 0.08 |
HMGA2 |
high mobility group AT-hook 2 |
60 |
0.98 |
| chr10_54074428_54074776 | 0.08 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
546 |
0.57 |
| chr3_72396838_72397278 | 0.08 |
ENSG00000212070 |
. |
85479 |
0.1 |
| chr20_19193596_19194206 | 0.08 |
RP11-97N19.2 |
|
220 |
0.81 |
| chr5_159739674_159740114 | 0.08 |
CCNJL |
cyclin J-like |
287 |
0.9 |
| chr1_15484794_15484945 | 0.08 |
TMEM51 |
transmembrane protein 51 |
4584 |
0.24 |
| chr1_196577911_196578299 | 0.08 |
KCNT2 |
potassium channel, subfamily T, member 2 |
250 |
0.94 |
| chr13_95619763_95620412 | 0.08 |
ENSG00000252335 |
. |
51402 |
0.18 |
| chrX_130962862_130963372 | 0.08 |
ENSG00000200587 |
. |
109867 |
0.07 |
| chr11_17756507_17757113 | 0.08 |
KCNC1 |
potassium voltage-gated channel, Shaw-related subfamily, member 1 |
451 |
0.83 |
| chr14_106383409_106383560 | 0.08 |
ENSG00000211931 |
. |
769 |
0.15 |
| chr6_14453382_14453777 | 0.08 |
ENSG00000206960 |
. |
193187 |
0.03 |
| chr17_75278220_75278512 | 0.08 |
SEPT9 |
septin 9 |
833 |
0.68 |
| chr10_71812550_71812868 | 0.08 |
H2AFY2 |
H2A histone family, member Y2 |
73 |
0.98 |
| chr2_24307248_24307399 | 0.08 |
TP53I3 |
tumor protein p53 inducible protein 3 |
127 |
0.94 |
| chr2_220435062_220435638 | 0.07 |
OBSL1 |
obscurin-like 1 |
613 |
0.46 |
| chr11_24518160_24518510 | 0.07 |
LUZP2 |
leucine zipper protein 2 |
389 |
0.91 |
| chr1_204098492_204098674 | 0.07 |
RP11-74C13.4 |
|
1607 |
0.3 |
| chr19_41104181_41104507 | 0.07 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
1203 |
0.38 |
| chr15_82338737_82339392 | 0.07 |
MEX3B |
mex-3 RNA binding family member B |
582 |
0.73 |
| chr18_61498851_61499100 | 0.07 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
39951 |
0.14 |
| chr17_80175288_80175536 | 0.07 |
RP13-516M14.2 |
|
3309 |
0.13 |
| chr19_14886701_14887039 | 0.07 |
EMR2 |
egf-like module containing, mucin-like, hormone receptor-like 2 |
698 |
0.62 |
| chr19_45121599_45121750 | 0.07 |
IGSF23 |
immunoglobulin superfamily, member 23 |
4434 |
0.17 |
| chr17_68165729_68165994 | 0.07 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
185 |
0.66 |
| chr4_188916560_188916711 | 0.07 |
ZFP42 |
ZFP42 zinc finger protein |
290 |
0.94 |
| chr20_61467867_61468113 | 0.07 |
COL9A3 |
collagen, type IX, alpha 3 |
18827 |
0.11 |
| chr20_23065943_23066176 | 0.07 |
CD93 |
CD93 molecule |
918 |
0.56 |
| chr2_62887339_62887490 | 0.07 |
AC092155.4 |
|
2349 |
0.33 |
| chr12_124990844_124991534 | 0.07 |
NCOR2 |
nuclear receptor corepressor 2 |
11391 |
0.3 |
| chr10_122216781_122217284 | 0.07 |
PPAPDC1A |
phosphatidic acid phosphatase type 2 domain containing 1A |
273 |
0.93 |
| chr7_119913514_119913995 | 0.07 |
KCND2 |
potassium voltage-gated channel, Shal-related subfamily, member 2 |
32 |
0.99 |
| chr15_99557644_99557868 | 0.07 |
PGPEP1L |
pyroglutamyl-peptidase I-like |
6732 |
0.23 |
| chr7_1963923_1964074 | 0.07 |
MAD1L1 |
MAD1 mitotic arrest deficient-like 1 (yeast) |
16187 |
0.23 |
| chr19_680149_680337 | 0.07 |
FSTL3 |
follistatin-like 3 (secreted glycoprotein) |
2301 |
0.16 |
| chr14_93552815_93553084 | 0.07 |
ITPK1-AS1 |
ITPK1 antisense RNA 1 |
19152 |
0.18 |
| chr5_77943226_77943942 | 0.07 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
1064 |
0.69 |
| chr3_44063258_44064101 | 0.07 |
ENSG00000252980 |
. |
48900 |
0.17 |
| chr2_159825200_159826543 | 0.07 |
TANC1 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
688 |
0.73 |
| chr7_5467738_5468320 | 0.07 |
TNRC18 |
trinucleotide repeat containing 18 |
2984 |
0.19 |
| chr5_141293636_141294105 | 0.07 |
KIAA0141 |
KIAA0141 |
9503 |
0.15 |
| chr4_151504492_151504761 | 0.07 |
MAB21L2 |
mab-21-like 2 (C. elegans) |
1549 |
0.43 |
| chr16_3156089_3156516 | 0.07 |
RP11-473M20.11 |
|
463 |
0.57 |
| chr8_37555995_37556200 | 0.07 |
ZNF703 |
zinc finger protein 703 |
2828 |
0.18 |
| chr16_81790383_81790667 | 0.07 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
17823 |
0.27 |
| chr14_59104798_59105385 | 0.07 |
DACT1 |
dishevelled-binding antagonist of beta-catenin 1 |
194 |
0.97 |
| chr19_48928010_48928416 | 0.07 |
GRWD1 |
glutamate-rich WD repeat containing 1 |
20817 |
0.09 |
| chr14_70170115_70170348 | 0.07 |
SRSF5 |
serine/arginine-rich splicing factor 5 |
23386 |
0.21 |
| chr7_116312458_116312799 | 0.07 |
MET |
met proto-oncogene |
169 |
0.97 |
| chr1_198901485_198902338 | 0.07 |
ENSG00000207759 |
. |
73629 |
0.11 |
| chr1_156426947_156427327 | 0.07 |
MEF2D |
myocyte enhancer factor 2D |
26085 |
0.1 |
| chr7_5466302_5466453 | 0.07 |
TNRC18 |
trinucleotide repeat containing 18 |
1332 |
0.35 |
| chr4_157997002_157997188 | 0.07 |
GLRB |
glycine receptor, beta |
114 |
0.97 |
| chr9_96717847_96718052 | 0.07 |
BARX1 |
BARX homeobox 1 |
295 |
0.94 |
| chr11_64099501_64099713 | 0.07 |
AP003774.1 |
|
2631 |
0.11 |
| chr2_106008139_106008408 | 0.07 |
FHL2 |
four and a half LIM domains 2 |
4969 |
0.21 |
| chr7_95546876_95547027 | 0.07 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
113355 |
0.07 |
| chr16_57576641_57576837 | 0.07 |
GPR114 |
G protein-coupled receptor 114 |
138 |
0.95 |
| chr9_137809478_137809629 | 0.07 |
FCN1 |
ficolin (collagen/fibrinogen domain containing) 1 |
256 |
0.88 |
| chr15_25123043_25123575 | 0.07 |
ENSG00000238615 |
. |
15585 |
0.12 |
| chr6_27205685_27206711 | 0.07 |
PRSS16 |
protease, serine, 16 (thymus) |
9282 |
0.19 |
| chr2_85632386_85633119 | 0.07 |
CAPG |
capping protein (actin filament), gelsolin-like |
3186 |
0.11 |
| chr1_208137959_208138110 | 0.07 |
CD34 |
CD34 molecule |
53287 |
0.17 |
| chr19_8567028_8567223 | 0.07 |
PRAM1 |
PML-RARA regulated adaptor molecule 1 |
370 |
0.79 |
| chr16_54407600_54408115 | 0.07 |
IRX3 |
iroquois homeobox 3 |
87182 |
0.09 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.1 | 0.1 | GO:0060462 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.1 | 0.2 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.1 | GO:0061298 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.2 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.0 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0021938 | smoothened signaling pathway involved in regulation of cerebellar granule cell precursor cell proliferation(GO:0021938) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0001820 | serotonin secretion(GO:0001820) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0060912 | negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cardioblast cell fate specification(GO:0009997) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.1 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0010890 | positive regulation of sequestering of triglyceride(GO:0010890) |
| 0.0 | 0.0 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.0 | 0.1 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0061318 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0048342 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.1 | GO:0050961 | detection of temperature stimulus involved in sensory perception(GO:0050961) detection of temperature stimulus involved in sensory perception of pain(GO:0050965) |
| 0.0 | 0.1 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.0 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.3 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.0 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.0 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.0 | GO:0060033 | anatomical structure regression(GO:0060033) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0048262 | determination of dorsal/ventral asymmetry(GO:0048262) determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.0 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.0 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0051873 | disruption by host of symbiont cells(GO:0051852) killing by host of symbiont cells(GO:0051873) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.0 | GO:0003416 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 | 0.0 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0070977 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.0 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.1 | GO:0006991 | response to sterol depletion(GO:0006991) |
| 0.0 | 0.0 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.0 | GO:0030540 | female genitalia development(GO:0030540) |
| 0.0 | 0.1 | GO:0033603 | positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.1 | GO:0030259 | lipid glycosylation(GO:0030259) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0060071 | Wnt signaling pathway, planar cell polarity pathway(GO:0060071) regulation of establishment of planar polarity(GO:0090175) |
| 0.0 | 0.1 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.4 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.1 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0060900 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.1 | GO:0070373 | negative regulation of ERK1 and ERK2 cascade(GO:0070373) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.2 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0048875 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0042441 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0060536 | trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0045835 | negative regulation of meiotic nuclear division(GO:0045835) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0071545 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.0 | GO:0060662 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.0 | GO:0051549 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.2 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0016102 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.0 | GO:0045990 | carbon catabolite regulation of transcription(GO:0045990) regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.0 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.0 | GO:0003321 | positive regulation of blood pressure by epinephrine-norepinephrine(GO:0003321) |
| 0.0 | 0.2 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.0 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.0 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.0 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.0 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.0 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.0 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.0 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.0 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0016717 | oxidoreductase activity, acting on paired donors, with oxidation of a pair of donors resulting in the reduction of molecular oxygen to two molecules of water(GO:0016717) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0016934 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.2 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.2 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.2 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.0 | GO:0003909 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.0 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.0 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.0 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.0 | GO:0001846 | opsonin binding(GO:0001846) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0050733 | RS domain binding(GO:0050733) |
| 0.0 | 0.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0070717 | poly(A) binding(GO:0008143) poly-purine tract binding(GO:0070717) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.0 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.5 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.0 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | Genes involved in CDT1 association with the CDC6:ORC:origin complex |
| 0.0 | 0.0 | REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | Genes involved in Platelet activation, signaling and aggregation |
| 0.0 | 0.0 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.0 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.0 | REACTOME HEMOSTASIS | Genes involved in Hemostasis |
| 0.0 | 0.5 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.0 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.1 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.0 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.0 | REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | Genes involved in E2F mediated regulation of DNA replication |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |