Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
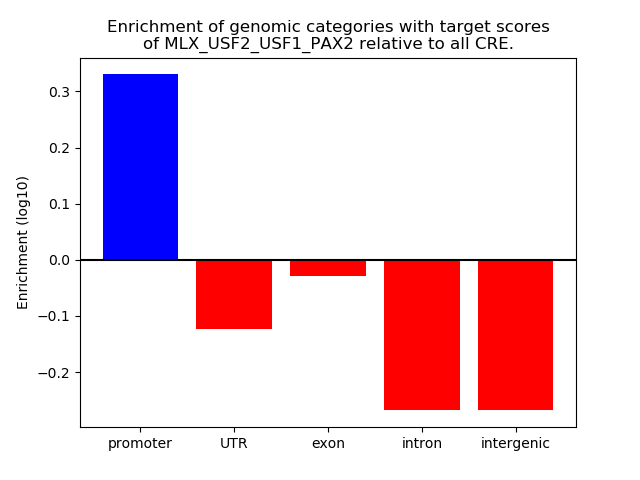
Results for MLX_USF2_USF1_PAX2
Z-value: 1.23
Transcription factors associated with MLX_USF2_USF1_PAX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
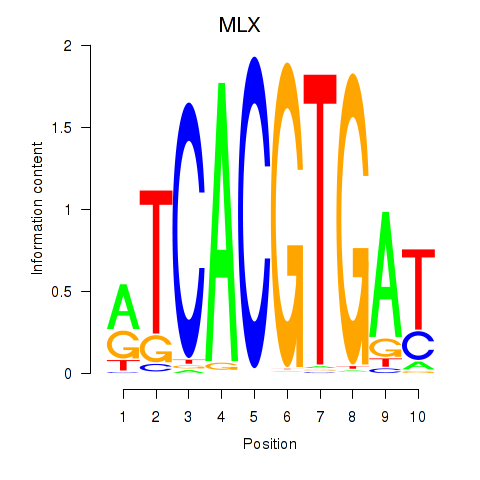
MLX
|
ENSG00000108788.7 | MAX dimerization protein MLX |
|
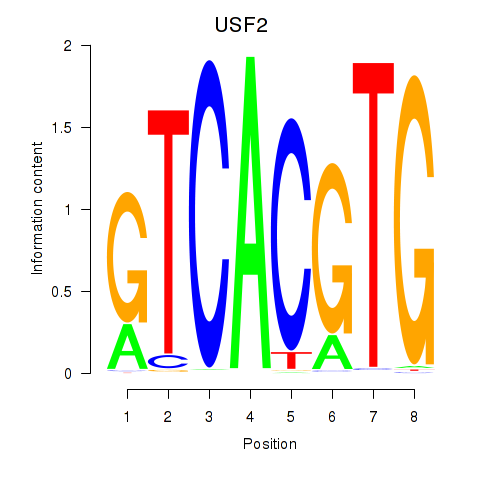
USF2
|
ENSG00000105698.11 | upstream transcription factor 2, c-fos interacting |
|
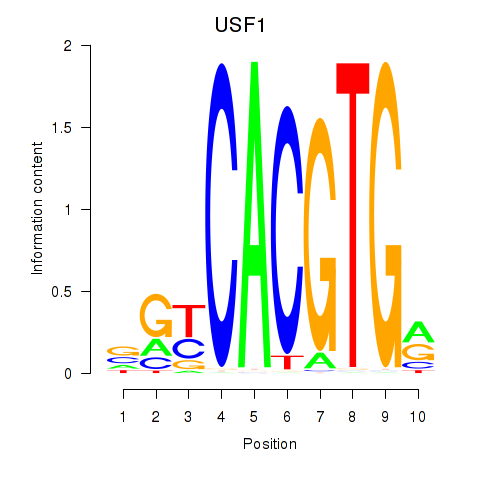
USF1
|
ENSG00000158773.10 | upstream transcription factor 1 |
|
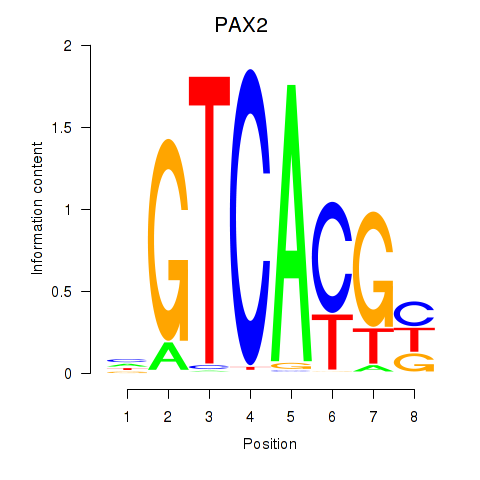
PAX2
|
ENSG00000075891.17 | paired box 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_40719513_40720031 | MLX | 680 | 0.450555 | 0.78 | 1.3e-02 | Click! |
| chr17_40719217_40719503 | MLX | 268 | 0.794788 | 0.78 | 1.3e-02 | Click! |
| chr17_40718813_40719216 | MLX | 72 | 0.928407 | 0.65 | 5.8e-02 | Click! |
| chr10_102490645_102490857 | PAX2 | 14717 | 0.235970 | 0.71 | 3.1e-02 | Click! |
| chr10_102497346_102497574 | PAX2 | 8008 | 0.254579 | 0.69 | 3.9e-02 | Click! |
| chr10_102414163_102414356 | PAX2 | 91209 | 0.068494 | 0.61 | 8.2e-02 | Click! |
| chr10_102414840_102414991 | PAX2 | 90553 | 0.069282 | 0.51 | 1.6e-01 | Click! |
| chr10_102490453_102490604 | PAX2 | 14940 | 0.235454 | 0.50 | 1.7e-01 | Click! |
| chr1_161015245_161015686 | USF1 | 198 | 0.869665 | 0.96 | 2.9e-05 | Click! |
| chr1_161015832_161016049 | USF1 | 173 | 0.886637 | 0.88 | 1.6e-03 | Click! |
| chr1_161014942_161015228 | USF1 | 354 | 0.739730 | 0.86 | 3.2e-03 | Click! |
| chr1_161014456_161014882 | USF1 | 62 | 0.939571 | 0.81 | 8.3e-03 | Click! |
| chr1_161016115_161016266 | USF1 | 423 | 0.677451 | 0.64 | 6.3e-02 | Click! |
| chr19_35758325_35758961 | USF2 | 1238 | 0.254951 | 0.85 | 4.0e-03 | Click! |
| chr19_35759494_35759654 | USF2 | 307 | 0.777066 | 0.61 | 8.3e-02 | Click! |
| chr19_35760287_35760594 | USF2 | 422 | 0.674240 | 0.53 | 1.4e-01 | Click! |
| chr19_35759189_35759340 | USF2 | 617 | 0.517168 | 0.50 | 1.7e-01 | Click! |
| chr19_35760748_35760986 | USF2 | 299 | 0.784561 | 0.41 | 2.8e-01 | Click! |
Activity of the MLX_USF2_USF1_PAX2 motif across conditions
Conditions sorted by the z-value of the MLX_USF2_USF1_PAX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
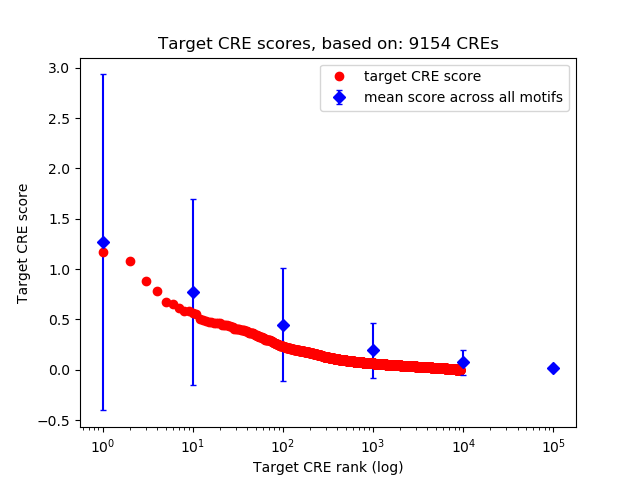
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_202896403_202896578 | 1.17 |
KLHL12 |
kelch-like family member 12 |
100 |
0.95 |
| chr20_4795670_4795862 | 1.09 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
3 |
0.98 |
| chr19_4402510_4402661 | 0.89 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
74 |
0.93 |
| chr17_6915641_6915886 | 0.78 |
RNASEK |
ribonuclease, RNase K |
27 |
0.55 |
| chr3_19988451_19988692 | 0.67 |
RAB5A |
RAB5A, member RAS oncogene family |
0 |
0.54 |
| chr10_70480705_70480856 | 0.65 |
CCAR1 |
cell division cycle and apoptosis regulator 1 |
11 |
0.97 |
| chr12_90103095_90103246 | 0.61 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
93 |
0.97 |
| chr1_154193354_154193505 | 0.58 |
UBAP2L |
ubiquitin associated protein 2-like |
6 |
0.81 |
| chr14_21852235_21852445 | 0.58 |
SUPT16H |
suppressor of Ty 16 homolog (S. cerevisiae) |
85 |
0.52 |
| chr17_17140687_17141182 | 0.56 |
FLCN |
folliculin |
438 |
0.74 |
| chr3_13460524_13460714 | 0.55 |
NUP210 |
nucleoporin 210kDa |
1190 |
0.56 |
| chr5_171542027_171542178 | 0.50 |
ENSG00000266671 |
. |
34122 |
0.17 |
| chr13_49684124_49684410 | 0.50 |
FNDC3A |
fibronectin type III domain containing 3A |
182 |
0.96 |
| chr19_10764777_10764928 | 0.48 |
ILF3 |
interleukin enhancer binding factor 3, 90kDa |
85 |
0.95 |
| chr19_49457811_49458033 | 0.48 |
BAX |
BCL2-associated X protein |
195 |
0.85 |
| chr16_84401582_84401733 | 0.47 |
ATP2C2 |
ATPase, Ca++ transporting, type 2C, member 2 |
476 |
0.82 |
| chr17_47841796_47842381 | 0.46 |
FAM117A |
family with sequence similarity 117, member A |
559 |
0.72 |
| chrY_1604612_1604993 | 0.46 |
NA |
NA |
> 106 |
NA |
| chr3_184031996_184032281 | 0.46 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
145 |
0.91 |
| chr5_139782206_139782432 | 0.46 |
ANKHD1 |
ankyrin repeat and KH domain containing 1 |
813 |
0.3 |
| chr7_100271099_100271250 | 0.45 |
GNB2 |
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
16 |
0.95 |
| chr11_126081501_126081652 | 0.45 |
RPUSD4 |
RNA pseudouridylate synthase domain containing 4 |
11 |
0.56 |
| chrX_1654663_1654996 | 0.45 |
P2RY8 |
purinergic receptor P2Y, G-protein coupled, 8 |
1171 |
0.53 |
| chr12_123849561_123849862 | 0.44 |
SBNO1 |
strawberry notch homolog 1 (Drosophila) |
321 |
0.85 |
| chr15_68132393_68132701 | 0.44 |
ENSG00000206625 |
. |
164 |
0.97 |
| chr1_222885954_222886552 | 0.43 |
AIDA |
axin interactor, dorsalization associated |
273 |
0.56 |
| chr11_118094529_118095128 | 0.43 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
26 |
0.97 |
| chr1_42921657_42922194 | 0.42 |
ZMYND12 |
zinc finger, MYND-type containing 12 |
13 |
0.64 |
| chr16_15737211_15737772 | 0.41 |
ENSG00000272213 |
. |
340 |
0.38 |
| chr3_148709208_148709359 | 0.41 |
GYG1 |
glycogenin 1 |
16 |
0.98 |
| chr19_15575302_15575574 | 0.41 |
RASAL3 |
RAS protein activator like 3 |
56 |
0.96 |
| chr9_131084429_131084680 | 0.41 |
TRUB2 |
TruB pseudouridine (psi) synthase family member 2 |
57 |
0.74 |
| chr16_745521_745735 | 0.40 |
FBXL16 |
F-box and leucine-rich repeat protein 16 |
397 |
0.57 |
| chr10_3822916_3823697 | 0.40 |
KLF6 |
Kruppel-like factor 6 |
4079 |
0.25 |
| chr12_46777195_46777346 | 0.39 |
SLC38A2 |
solute carrier family 38, member 2 |
10620 |
0.24 |
| chr9_96793015_96793166 | 0.39 |
PTPDC1 |
protein tyrosine phosphatase domain containing 1 |
14 |
0.99 |
| chr9_125667781_125668094 | 0.39 |
RC3H2 |
ring finger and CCCH-type domains 2 |
317 |
0.82 |
| chr15_74392593_74392993 | 0.38 |
GOLGA6A |
golgin A6 family, member A |
17902 |
0.12 |
| chr2_6868388_6868539 | 0.38 |
AC017076.5 |
|
105199 |
0.07 |
| chr6_106439645_106440509 | 0.38 |
ENSG00000200198 |
. |
87829 |
0.09 |
| chr9_131450959_131451258 | 0.38 |
SET |
SET nuclear oncogene |
397 |
0.73 |
| chr10_46090174_46090398 | 0.37 |
MARCH8 |
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
68 |
0.98 |
| chr19_11546030_11546251 | 0.37 |
PRKCSH |
protein kinase C substrate 80K-H |
31 |
0.69 |
| chr19_3990141_3990406 | 0.37 |
EEF2 |
eukaryotic translation elongation factor 2 |
4806 |
0.11 |
| chr17_75451832_75452118 | 0.37 |
SEPT9 |
septin 9 |
473 |
0.77 |
| chr16_28306637_28306788 | 0.37 |
SBK1 |
SH3 domain binding kinase 1 |
2872 |
0.28 |
| chr14_91879963_91880132 | 0.36 |
CCDC88C |
coiled-coil domain containing 88C |
3643 |
0.29 |
| chrX_100872700_100873033 | 0.35 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
125 |
0.93 |
| chr20_18118508_18118767 | 0.35 |
PET117 |
PET117 homolog (S. cerevisiae) |
120 |
0.5 |
| chr8_142427875_142428034 | 0.34 |
PTP4A3 |
protein tyrosine phosphatase type IVA, member 3 |
4053 |
0.16 |
| chr7_150065813_150066064 | 0.34 |
REPIN1 |
replication initiator 1 |
22 |
0.52 |
| chr5_1800677_1801081 | 0.34 |
MRPL36 |
mitochondrial ribosomal protein L36 |
601 |
0.45 |
| chr17_48450293_48450521 | 0.33 |
MRPL27 |
mitochondrial ribosomal protein L27 |
86 |
0.61 |
| chrX_154012790_154012941 | 0.33 |
ENSG00000206693 |
. |
9592 |
0.11 |
| chr9_132388509_132388893 | 0.33 |
NTMT1 |
N-terminal Xaa-Pro-Lys N-methyltransferase 1 |
22 |
0.96 |
| chrX_71002679_71002935 | 0.32 |
ENSG00000221684 |
. |
23502 |
0.23 |
| chr11_117857636_117858098 | 0.32 |
IL10RA |
interleukin 10 receptor, alpha |
758 |
0.66 |
| chr17_66201588_66201894 | 0.32 |
AMZ2 |
archaelysin family metallopeptidase 2 |
41974 |
0.12 |
| chr15_34610151_34611295 | 0.32 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
257 |
0.87 |
| chr3_177040689_177040840 | 0.32 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
125503 |
0.06 |
| chr13_52377985_52378296 | 0.31 |
DHRS12 |
dehydrogenase/reductase (SDR family) member 12 |
94 |
0.76 |
| chr10_129706310_129706549 | 0.31 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
1013 |
0.58 |
| chr2_27805637_27805914 | 0.30 |
ZNF512 |
zinc finger protein 512 |
122 |
0.9 |
| chr10_74114795_74115085 | 0.30 |
DNAJB12 |
DnaJ (Hsp40) homolog, subfamily B, member 12 |
33 |
0.98 |
| chr3_31573575_31573846 | 0.30 |
STT3B |
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
572 |
0.85 |
| chrY_2709555_2710183 | 0.30 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
92 |
0.98 |
| chr1_145611013_145611189 | 0.29 |
RNF115 |
ring finger protein 115 |
65 |
0.63 |
| chr21_27443178_27443674 | 0.29 |
APP |
amyloid beta (A4) precursor protein |
18951 |
0.24 |
| chr1_40283034_40283198 | 0.29 |
RP1-118J21.25 |
|
28468 |
0.11 |
| chr10_22609853_22610108 | 0.29 |
BMI1 |
BMI1 polycomb ring finger oncogene |
160 |
0.94 |
| chr19_15236279_15236535 | 0.29 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
63 |
0.94 |
| chr2_174129320_174129481 | 0.29 |
MLK7-AS1 |
MLK7 antisense RNA 1 |
6948 |
0.3 |
| chr1_101360427_101360745 | 0.29 |
RP4-549L20.3 |
|
102 |
0.63 |
| chr1_154908770_154909108 | 0.29 |
PMVK |
phosphomevalonate kinase |
528 |
0.6 |
| chr17_65821514_65821703 | 0.29 |
BPTF |
bromodomain PHD finger transcription factor |
32 |
0.97 |
| chr6_170580859_170581356 | 0.28 |
RP5-1086L22.1 |
|
9450 |
0.18 |
| chr11_67211190_67211466 | 0.28 |
CORO1B |
coronin, actin binding protein, 1B |
65 |
0.92 |
| chrX_72783084_72783245 | 0.28 |
CHIC1 |
cysteine-rich hydrophobic domain 1 |
120 |
0.98 |
| chr8_28259515_28259714 | 0.27 |
ZNF395 |
zinc finger protein 395 |
604 |
0.66 |
| chrX_10066246_10067411 | 0.27 |
WWC3 |
WWC family member 3 |
35329 |
0.15 |
| chrX_62780920_62781228 | 0.26 |
ENSG00000235437 |
. |
123 |
0.98 |
| chr17_25659717_25660000 | 0.26 |
WSB1 |
WD repeat and SOCS box containing 1 |
38486 |
0.16 |
| chr7_116593207_116593413 | 0.26 |
ST7 |
suppression of tumorigenicity 7 |
18 |
0.9 |
| chrX_102941872_102942023 | 0.26 |
MORF4L2 |
mortality factor 4 like 2 |
201 |
0.57 |
| chr1_192128557_192128890 | 0.26 |
RGS18 |
regulator of G-protein signaling 18 |
1136 |
0.67 |
| chr2_190305327_190305991 | 0.26 |
WDR75 |
WD repeat domain 75 |
500 |
0.87 |
| chr16_25118548_25118859 | 0.26 |
LCMT1 |
leucine carboxyl methyltransferase 1 |
4347 |
0.25 |
| chr8_38240572_38240933 | 0.26 |
WHSC1L1 |
Wolf-Hirschhorn syndrome candidate 1-like 1 |
962 |
0.42 |
| chr10_3871001_3871747 | 0.25 |
KLF6 |
Kruppel-like factor 6 |
43901 |
0.17 |
| chr19_35939103_35939363 | 0.25 |
FFAR2 |
free fatty acid receptor 2 |
30 |
0.96 |
| chr17_80406652_80406803 | 0.24 |
C17orf62 |
chromosome 17 open reading frame 62 |
754 |
0.43 |
| chr22_40406174_40406325 | 0.24 |
FAM83F |
family with sequence similarity 83, member F |
557 |
0.75 |
| chr7_50727728_50727879 | 0.24 |
GRB10 |
growth factor receptor-bound protein 10 |
33176 |
0.21 |
| chr19_12886036_12886246 | 0.24 |
HOOK2 |
hook microtubule-tethering protein 2 |
196 |
0.83 |
| chr17_21156722_21156881 | 0.24 |
C17orf103 |
chromosome 17 open reading frame 103 |
79 |
0.97 |
| chr22_44422802_44422980 | 0.24 |
PARVB |
parvin, beta |
2734 |
0.33 |
| chr5_94982218_94982535 | 0.24 |
RFESD |
Rieske (Fe-S) domain containing |
82 |
0.97 |
| chr6_139350164_139350486 | 0.24 |
ABRACL |
ABRA C-terminal like |
346 |
0.91 |
| chr6_24775111_24775564 | 0.23 |
GMNN |
geminin, DNA replication inhibitor |
171 |
0.94 |
| chr15_75628080_75628612 | 0.23 |
COMMD4 |
COMM domain containing 4 |
15 |
0.96 |
| chr8_1940470_1940621 | 0.23 |
KBTBD11 |
kelch repeat and BTB (POZ) domain containing 11 |
18501 |
0.24 |
| chr15_87763305_87763456 | 0.23 |
RP11-133L19.1 |
|
231723 |
0.02 |
| chr20_61916082_61916233 | 0.23 |
ENSG00000266104 |
. |
2003 |
0.18 |
| chr20_62530272_62530423 | 0.23 |
DNAJC5 |
DnaJ (Hsp40) homolog, subfamily C, member 5 |
3812 |
0.09 |
| chr11_62538286_62538869 | 0.23 |
TAF6L |
TAF6-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor, 65kDa |
198 |
0.69 |
| chr10_112117242_112117393 | 0.23 |
SMNDC1 |
survival motor neuron domain containing 1 |
52608 |
0.13 |
| chr9_100684535_100684760 | 0.23 |
C9orf156 |
chromosome 9 open reading frame 156 |
122 |
0.95 |
| chr2_231090336_231091061 | 0.22 |
SP140 |
SP140 nuclear body protein |
219 |
0.54 |
| chr2_38871082_38871242 | 0.22 |
GALM |
galactose mutarotase (aldose 1-epimerase) |
21890 |
0.15 |
| chr4_3529804_3529955 | 0.22 |
LRPAP1 |
low density lipoprotein receptor-related protein associated protein 1 |
4407 |
0.19 |
| chr22_18252608_18252759 | 0.22 |
BID |
BH3 interacting domain death agonist |
4099 |
0.21 |
| chr17_63053075_63053367 | 0.22 |
GNA13 |
guanine nucleotide binding protein (G protein), alpha 13 |
264 |
0.91 |
| chr19_35615719_35615873 | 0.22 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
5529 |
0.1 |
| chr2_240323140_240323402 | 0.22 |
AC062017.1 |
Uncharacterized protein |
141 |
0.86 |
| chr7_100292135_100292392 | 0.22 |
GIGYF1 |
GRB10 interacting GYF protein 1 |
5192 |
0.11 |
| chr9_6703919_6704260 | 0.21 |
KDM4C |
lysine (K)-specific demethylase 4C |
16774 |
0.21 |
| chr9_135996853_135997045 | 0.21 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
386 |
0.81 |
| chr19_13215586_13215874 | 0.21 |
LYL1 |
lymphoblastic leukemia derived sequence 1 |
1755 |
0.21 |
| chr8_27950757_27951396 | 0.21 |
ELP3 |
elongator acetyltransferase complex subunit 3 |
112 |
0.97 |
| chrX_70918097_70918602 | 0.21 |
ENSG00000221684 |
. |
60956 |
0.11 |
| chr2_97439727_97440064 | 0.21 |
CNNM4 |
cyclin M4 |
13256 |
0.14 |
| chr1_153508703_153508968 | 0.21 |
S100A6 |
S100 calcium binding protein A6 |
115 |
0.91 |
| chr9_98278987_98279233 | 0.21 |
PTCH1 |
patched 1 |
137 |
0.95 |
| chr8_17193981_17194132 | 0.21 |
MTMR7 |
myotubularin related protein 7 |
24874 |
0.21 |
| chr10_69644313_69644464 | 0.21 |
SIRT1 |
sirtuin 1 |
39 |
0.98 |
| chr22_25815435_25815828 | 0.20 |
LRP5L |
low density lipoprotein receptor-related protein 5-like |
14287 |
0.17 |
| chr2_240302618_240303103 | 0.20 |
HDAC4 |
histone deacetylase 4 |
19783 |
0.17 |
| chr2_1077533_1077684 | 0.20 |
AC114808.3 |
|
13581 |
0.26 |
| chr6_150326465_150326645 | 0.20 |
RAET1K |
retinoic acid early transcript 1K pseudogene |
262 |
0.9 |
| chr11_64648039_64648569 | 0.20 |
EHD1 |
EH-domain containing 1 |
1132 |
0.31 |
| chr16_71879481_71879729 | 0.20 |
ATXN1L |
ataxin 1-like |
289 |
0.47 |
| chr12_113623337_113623491 | 0.20 |
RITA1 |
RBPJ interacting and tubulin associated 1 |
67 |
0.58 |
| chr3_172312310_172312617 | 0.20 |
AC007919.2 |
HCG1787166; PRO1163; Uncharacterized protein |
49020 |
0.13 |
| chr14_20929873_20930024 | 0.20 |
TMEM55B |
transmembrane protein 55B |
177 |
0.87 |
| chr15_44083848_44084237 | 0.20 |
SERF2 |
small EDRK-rich factor 2 |
2 |
0.94 |
| chrX_129040013_129040508 | 0.20 |
UTP14A |
UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) |
93 |
0.97 |
| chr10_81962972_81963123 | 0.20 |
ANXA11 |
annexin A11 |
1200 |
0.51 |
| chr4_39355902_39356119 | 0.20 |
RFC1 |
replication factor C (activator 1) 1, 145kDa |
11959 |
0.16 |
| chr13_33000514_33000892 | 0.20 |
N4BP2L1 |
NEDD4 binding protein 2-like 1 |
1448 |
0.44 |
| chrX_102631036_102631748 | 0.20 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
57 |
0.97 |
| chr3_49044093_49044679 | 0.20 |
WDR6 |
WD repeat domain 6 |
109 |
0.91 |
| chr4_1195166_1195336 | 0.20 |
SPON2 |
spondin 2, extracellular matrix protein |
3625 |
0.16 |
| chrX_100603814_100604224 | 0.20 |
TIMM8A |
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
165 |
0.92 |
| chr3_184078420_184078571 | 0.20 |
CLCN2 |
chloride channel, voltage-sensitive 2 |
895 |
0.31 |
| chr7_140098327_140098552 | 0.20 |
SLC37A3 |
solute carrier family 37, member 3 |
90 |
0.96 |
| chr19_47633889_47634040 | 0.20 |
SAE1 |
SUMO1 activating enzyme subunit 1 |
75 |
0.97 |
| chr16_30075509_30076103 | 0.20 |
ALDOA |
aldolase A, fructose-bisphosphate |
12 |
0.94 |
| chr20_48731883_48732067 | 0.19 |
UBE2V1 |
ubiquitin-conjugating enzyme E2 variant 1 |
516 |
0.78 |
| chr19_17236963_17237284 | 0.19 |
MYO9B |
myosin IXB |
24595 |
0.1 |
| chr19_4829242_4829393 | 0.19 |
TICAM1 |
toll-like receptor adaptor molecule 1 |
2399 |
0.18 |
| chr1_153918448_153918692 | 0.19 |
DENND4B |
DENN/MADD domain containing 4B |
602 |
0.54 |
| chr1_53168822_53169232 | 0.19 |
COA7 |
cytochrome c oxidase assembly factor 7 |
4989 |
0.16 |
| chr16_87887371_87887580 | 0.19 |
SLC7A5 |
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
9770 |
0.17 |
| chr19_10398277_10398487 | 0.19 |
ICAM4 |
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
702 |
0.35 |
| chr16_17463239_17463649 | 0.19 |
XYLT1 |
xylosyltransferase I |
101294 |
0.09 |
| chr18_77153881_77154299 | 0.19 |
NFATC1 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 |
1766 |
0.42 |
| chr9_132816560_132816877 | 0.19 |
GPR107 |
G protein-coupled receptor 107 |
733 |
0.67 |
| chr5_94890508_94890791 | 0.19 |
TTC37 |
tetratricopeptide repeat domain 37 |
1 |
0.63 |
| chr15_67813744_67814059 | 0.19 |
C15orf61 |
chromosome 15 open reading frame 61 |
132 |
0.97 |
| chr7_127033071_127033491 | 0.19 |
ZNF800 |
zinc finger protein 800 |
509 |
0.84 |
| chr10_23631569_23631720 | 0.19 |
RP11-371A19.2 |
|
1242 |
0.34 |
| chr1_153939522_153939993 | 0.19 |
SLC39A1 |
solute carrier family 39 (zinc transporter), member 1 |
340 |
0.51 |
| chr22_40307152_40307436 | 0.19 |
GRAP2 |
GRB2-related adaptor protein 2 |
10181 |
0.17 |
| chr17_32906311_32906556 | 0.19 |
C17orf102 |
chromosome 17 open reading frame 102 |
45 |
0.97 |
| chr2_190627024_190627494 | 0.19 |
OSGEPL1-AS1 |
OSGEPL1 antisense RNA 1 |
171 |
0.56 |
| chr2_202987834_202988283 | 0.19 |
AC079354.3 |
|
6277 |
0.17 |
| chr11_62538890_62539107 | 0.19 |
RP11-727F15.11 |
|
80 |
0.6 |
| chr6_35744217_35744431 | 0.18 |
CLPSL2 |
colipase-like 2 |
68 |
0.96 |
| chr6_169612787_169612938 | 0.18 |
XXyac-YX65C7_A.2 |
|
487 |
0.87 |
| chr17_59299974_59300148 | 0.18 |
RP11-136H19.1 |
|
86736 |
0.08 |
| chr20_35575234_35575857 | 0.18 |
SAMHD1 |
SAM domain and HD domain 1 |
4566 |
0.25 |
| chr7_48018318_48018852 | 0.18 |
HUS1 |
HUS1 checkpoint homolog (S. pombe) |
516 |
0.79 |
| chr16_66914092_66914355 | 0.18 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
41 |
0.95 |
| chr17_68183563_68183714 | 0.18 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
17962 |
0.2 |
| chr4_164471433_164471640 | 0.18 |
ENSG00000264535 |
. |
27092 |
0.19 |
| chr13_21635697_21635989 | 0.18 |
LATS2 |
large tumor suppressor kinase 2 |
157 |
0.95 |
| chr2_219736729_219736880 | 0.18 |
WNT10A |
wingless-type MMTV integration site family, member 10A |
8281 |
0.12 |
| chr19_1439067_1439250 | 0.18 |
RPS15 |
ribosomal protein S15 |
352 |
0.66 |
| chr2_46727092_46727710 | 0.18 |
RP11-417F21.2 |
|
531 |
0.73 |
| chr7_2393496_2393740 | 0.18 |
EIF3B |
eukaryotic translation initiation factor 3, subunit B |
103 |
0.78 |
| chr19_33166568_33166725 | 0.18 |
RGS9BP |
regulator of G protein signaling 9 binding protein |
333 |
0.61 |
| chr17_7745282_7745470 | 0.18 |
KDM6B |
lysine (K)-specific demethylase 6B |
2096 |
0.17 |
| chr8_41907411_41908748 | 0.18 |
KAT6A |
K(lysine) acetyltransferase 6A |
1426 |
0.43 |
| chr19_2616954_2617105 | 0.18 |
CTC-265F19.2 |
|
5169 |
0.16 |
| chr11_63706516_63706697 | 0.18 |
NAA40 |
N(alpha)-acetyltransferase 40, NatD catalytic subunit |
114 |
0.94 |
| chr8_103820133_103820297 | 0.18 |
AZIN1 |
antizyme inhibitor 1 |
50115 |
0.12 |
| chr5_159866335_159866555 | 0.18 |
PTTG1 |
pituitary tumor-transforming 1 |
17528 |
0.13 |
| chr20_18547706_18547912 | 0.18 |
DTD1 |
D-tyrosyl-tRNA deacylase 1 |
20728 |
0.11 |
| chr3_119811799_119812619 | 0.18 |
GSK3B |
glycogen synthase kinase 3 beta |
304 |
0.9 |
| chr13_80881448_80881599 | 0.17 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
32271 |
0.23 |
| chr10_104223217_104223368 | 0.17 |
TMEM180 |
transmembrane protein 180 |
2122 |
0.16 |
| chr6_135857324_135857475 | 0.17 |
AHI1 |
Abelson helper integration site 1 |
38485 |
0.19 |
| chr9_34646531_34647052 | 0.17 |
GALT |
galactose-1-phosphate uridylyltransferase |
131 |
0.9 |
| chr9_36572612_36572804 | 0.17 |
MELK |
maternal embryonic leucine zipper kinase |
151 |
0.98 |
| chr21_43933401_43933705 | 0.17 |
SLC37A1 |
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
471 |
0.74 |
| chr7_21938421_21938604 | 0.17 |
CDCA7L |
cell division cycle associated 7-like |
5365 |
0.3 |
| chr7_1100114_1100265 | 0.17 |
RP11-449P15.1 |
|
1292 |
0.26 |
| chr10_76073985_76074136 | 0.17 |
ADK |
adenosine kinase |
137539 |
0.04 |
| chr14_105530971_105531214 | 0.17 |
GPR132 |
G protein-coupled receptor 132 |
675 |
0.7 |
| chr21_46237823_46238060 | 0.17 |
SUMO3 |
small ubiquitin-like modifier 3 |
25 |
0.96 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.4 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.1 | 0.6 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.1 | 0.4 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.5 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.1 | 0.2 | GO:0051097 | negative regulation of helicase activity(GO:0051097) |
| 0.1 | 0.2 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.1 | 0.3 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.1 | 0.1 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.1 | 0.2 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.1 | 0.2 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.1 | 0.2 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.1 | 0.2 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0015809 | arginine transport(GO:0015809) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0043441 | acetoacetic acid biosynthetic process(GO:0043441) |
| 0.0 | 0.1 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.2 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.7 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.2 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.2 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.0 | 0.4 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.6 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.3 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.2 | GO:0000132 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.2 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.2 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.6 | GO:0007339 | binding of sperm to zona pellucida(GO:0007339) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.1 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0032516 | positive regulation of phosphoprotein phosphatase activity(GO:0032516) |
| 0.0 | 0.5 | GO:0009303 | rRNA transcription(GO:0009303) |
| 0.0 | 0.2 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.3 | GO:0043496 | regulation of protein homodimerization activity(GO:0043496) |
| 0.0 | 0.1 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.3 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.4 | GO:0048169 | regulation of long-term neuronal synaptic plasticity(GO:0048169) |
| 0.0 | 0.1 | GO:0006578 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0071545 | inositol phosphate catabolic process(GO:0071545) |
| 0.0 | 0.2 | GO:0046037 | GMP metabolic process(GO:0046037) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.3 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.3 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:0071166 | ribonucleoprotein complex localization(GO:0071166) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0043320 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0052251 | induction by organism of defense response of other organism involved in symbiotic interaction(GO:0052251) |
| 0.0 | 0.3 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.0 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.3 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.6 | GO:0000289 | nuclear-transcribed mRNA poly(A) tail shortening(GO:0000289) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.0 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.0 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.1 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.2 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.1 | GO:0071027 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.0 | GO:0046100 | hypoxanthine metabolic process(GO:0046100) hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.2 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0006084 | acetyl-CoA metabolic process(GO:0006084) |
| 0.0 | 0.1 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.0 | GO:0043173 | nucleotide salvage(GO:0043173) |
| 0.0 | 0.1 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.4 | GO:0005980 | glycogen catabolic process(GO:0005980) |
| 0.0 | 0.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0046543 | thelarche(GO:0042695) development of secondary female sexual characteristics(GO:0046543) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0006107 | oxaloacetate metabolic process(GO:0006107) |
| 0.0 | 0.1 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0032878 | regulation of establishment or maintenance of cell polarity(GO:0032878) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:0051383 | kinetochore organization(GO:0051383) |
| 0.0 | 0.6 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.0 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.3 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) negative regulation of cellular response to insulin stimulus(GO:1900077) |
| 0.0 | 0.1 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.1 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.1 | GO:0070059 | intrinsic apoptotic signaling pathway in response to endoplasmic reticulum stress(GO:0070059) |
| 0.0 | 0.1 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0034969 | histone arginine methylation(GO:0034969) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0097501 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.3 | GO:0051865 | protein autoubiquitination(GO:0051865) |
| 0.0 | 0.1 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.1 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0048525 | negative regulation of viral process(GO:0048525) negative regulation of viral life cycle(GO:1903901) |
| 0.0 | 0.1 | GO:0061462 | protein targeting to lysosome(GO:0006622) protein targeting to vacuole(GO:0006623) protein localization to lysosome(GO:0061462) protein localization to vacuole(GO:0072665) establishment of protein localization to vacuole(GO:0072666) |
| 0.0 | 0.1 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0000266 | mitochondrial fission(GO:0000266) |
| 0.0 | 0.0 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0006900 | membrane budding(GO:0006900) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.0 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.1 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.1 | GO:0006515 | misfolded or incompletely synthesized protein catabolic process(GO:0006515) |
| 0.0 | 0.0 | GO:2000189 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.0 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.0 | GO:0048668 | collateral sprouting(GO:0048668) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.0 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:0060396 | growth hormone receptor signaling pathway(GO:0060396) cellular response to growth hormone stimulus(GO:0071378) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.2 | GO:0002474 | antigen processing and presentation of peptide antigen via MHC class I(GO:0002474) |
| 0.0 | 0.1 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.0 | GO:0032239 | regulation of nucleobase-containing compound transport(GO:0032239) regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0001881 | receptor recycling(GO:0001881) |
| 0.0 | 0.1 | GO:0034138 | toll-like receptor 3 signaling pathway(GO:0034138) |
| 0.0 | 0.2 | GO:1904031 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.0 | GO:0051458 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.0 | GO:0000726 | non-recombinational repair(GO:0000726) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0043330 | response to exogenous dsRNA(GO:0043330) |
| 0.0 | 0.2 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 1.0 | GO:0018196 | peptidyl-asparagine modification(GO:0018196) protein N-linked glycosylation via asparagine(GO:0018279) |
| 0.0 | 0.9 | GO:0006754 | ATP biosynthetic process(GO:0006754) |
| 0.0 | 0.1 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0051024 | positive regulation of immunoglobulin secretion(GO:0051024) |
| 0.0 | 0.0 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.0 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0009226 | nucleotide-sugar biosynthetic process(GO:0009226) |
| 0.0 | 0.1 | GO:0045651 | positive regulation of macrophage differentiation(GO:0045651) |
| 0.0 | 0.1 | GO:0016265 | obsolete death(GO:0016265) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.1 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.0 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.0 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.4 | GO:0008156 | negative regulation of DNA replication(GO:0008156) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0055070 | copper ion homeostasis(GO:0055070) |
| 0.0 | 1.7 | GO:0006184 | obsolete GTP catabolic process(GO:0006184) |
| 0.0 | 0.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.0 | GO:0019884 | antigen processing and presentation of exogenous antigen(GO:0019884) |
| 0.0 | 0.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0042976 | activation of Janus kinase activity(GO:0042976) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.2 | GO:0032648 | regulation of interferon-beta production(GO:0032648) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.2 | GO:0018904 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.2 | GO:0045736 | negative regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045736) negative regulation of cyclin-dependent protein kinase activity(GO:1904030) |
| 0.0 | 0.0 | GO:0070584 | mitochondrion morphogenesis(GO:0070584) |
| 0.0 | 0.0 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.0 | 0.0 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.2 | GO:0032201 | telomere maintenance via semi-conservative replication(GO:0032201) |
| 0.0 | 0.0 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.0 | 0.0 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.0 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:0016572 | histone phosphorylation(GO:0016572) |
| 0.0 | 0.1 | GO:0048247 | lymphocyte chemotaxis(GO:0048247) lymphocyte migration(GO:0072676) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.1 | GO:0006388 | RNA splicing, via endonucleolytic cleavage and ligation(GO:0000394) tRNA splicing, via endonucleolytic cleavage and ligation(GO:0006388) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0097031 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.0 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.0 | GO:0016137 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0000060 | protein import into nucleus, translocation(GO:0000060) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) mature B cell differentiation(GO:0002335) |
| 0.0 | 0.0 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.7 | GO:0051028 | mRNA transport(GO:0051028) |
| 0.0 | 0.3 | GO:0006120 | mitochondrial electron transport, NADH to ubiquinone(GO:0006120) |
| 0.0 | 0.1 | GO:0031163 | iron-sulfur cluster assembly(GO:0016226) metallo-sulfur cluster assembly(GO:0031163) |
| 0.0 | 0.0 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.2 | GO:0006611 | protein export from nucleus(GO:0006611) |
| 0.0 | 0.1 | GO:0008333 | endosome to lysosome transport(GO:0008333) |
| 0.0 | 0.1 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.0 | 0.0 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.2 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.0 | GO:0048799 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.6 | GO:0034134 | toll-like receptor 1 signaling pathway(GO:0034130) toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 1.9 | GO:0000398 | RNA splicing, via transesterification reactions with bulged adenosine as nucleophile(GO:0000377) mRNA splicing, via spliceosome(GO:0000398) |
| 0.0 | 0.1 | GO:0032367 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.0 | GO:0006729 | tetrahydrobiopterin biosynthetic process(GO:0006729) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0018107 | peptidyl-threonine phosphorylation(GO:0018107) |
| 0.0 | 0.0 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.0 | GO:0006552 | leucine catabolic process(GO:0006552) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.1 | 0.5 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.1 | 0.3 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.1 | 0.2 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.1 | 0.3 | GO:0016589 | NURF complex(GO:0016589) |
| 0.1 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 0.7 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.4 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.4 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.6 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.0 | GO:0000778 | condensed nuclear chromosome kinetochore(GO:0000778) |
| 0.0 | 0.2 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0046540 | U4/U6 x U5 tri-snRNP complex(GO:0046540) spliceosomal tri-snRNP complex(GO:0097526) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.3 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.3 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.3 | GO:0030914 | STAGA complex(GO:0030914) |
| 0.0 | 0.5 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.0 | GO:0000779 | condensed chromosome, centromeric region(GO:0000779) |
| 0.0 | 0.2 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.4 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.4 | GO:0005844 | polysome(GO:0005844) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.0 | GO:0035327 | transcriptionally active chromatin(GO:0035327) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.5 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.5 | GO:0042645 | mitochondrial nucleoid(GO:0042645) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.2 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0043230 | extracellular organelle(GO:0043230) |
| 0.0 | 0.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.5 | GO:0015030 | Cajal body(GO:0015030) |
| 0.0 | 0.5 | GO:0008287 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.4 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.0 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.7 | GO:0000776 | kinetochore(GO:0000776) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.3 | GO:0031526 | brush border membrane(GO:0031526) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.6 | GO:0005643 | nuclear pore(GO:0005643) |
| 0.0 | 0.0 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.2 | GO:0000314 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.0 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.1 | 1.0 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.1 | 0.2 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.1 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.4 | GO:0070513 | death domain binding(GO:0070513) |
| 0.1 | 0.2 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.1 | 0.2 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.1 | 0.3 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.1 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.2 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.1 | 0.3 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.1 | 0.4 | GO:0005536 | glucose binding(GO:0005536) |
| 0.1 | 0.2 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.3 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.6 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.2 | GO:0016944 | obsolete RNA polymerase II transcription elongation factor activity(GO:0016944) |
| 0.0 | 0.2 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.3 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.4 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.1 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.0 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0003829 | beta-1,3-galactosyl-O-glycosyl-glycoprotein beta-1,6-N-acetylglucosaminyltransferase activity(GO:0003829) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.0 | 0.2 | GO:0000982 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.2 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.6 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.2 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.4 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0051787 | misfolded protein binding(GO:0051787) |
| 0.0 | 0.7 | GO:0004521 | endoribonuclease activity(GO:0004521) |
| 0.0 | 0.3 | GO:0004004 | ATP-dependent RNA helicase activity(GO:0004004) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 1.0 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.5 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.1 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.1 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.0 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.0 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0017089 | glycolipid transporter activity(GO:0017089) |
| 0.0 | 0.0 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.0 | GO:0016274 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.0 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.4 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.5 | GO:0043028 | cysteine-type endopeptidase regulator activity involved in apoptotic process(GO:0043028) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.3 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0009922 | fatty acid elongase activity(GO:0009922) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0017169 | CDP-alcohol phosphatidyltransferase activity(GO:0017169) |
| 0.0 | 0.1 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0016679 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.3 | GO:0019843 | rRNA binding(GO:0019843) |
| 0.0 | 0.0 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.0 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.0 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0061630 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.0 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.0 | GO:0019237 | centromeric DNA binding(GO:0019237) |
| 0.0 | 0.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0046969 | histone deacetylase activity (H3-K9 specific)(GO:0032129) NAD-dependent histone deacetylase activity (H3-K9 specific)(GO:0046969) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.3 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.1 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.0 | GO:0009008 | DNA-methyltransferase activity(GO:0009008) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.3 | GO:0016776 | phosphotransferase activity, phosphate group as acceptor(GO:0016776) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.0 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.4 | GO:0003954 | NADH dehydrogenase activity(GO:0003954) NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.0 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.1 | GO:0019206 | nucleoside kinase activity(GO:0019206) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.1 | 0.1 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.5 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.2 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.5 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.5 | PID TNF PATHWAY | TNF receptor signaling pathway |
| 0.0 | 0.5 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.0 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 1.2 | PID MYC ACTIV PATHWAY | Validated targets of C-MYC transcriptional activation |
| 0.0 | 0.0 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.0 | 0.5 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.3 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.2 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 0.3 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.2 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.1 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.2 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.6 | PID CMYB PATHWAY | C-MYB transcription factor network |
| 0.0 | 0.3 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.4 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.4 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.0 | REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | Genes involved in trans-Golgi Network Vesicle Budding |
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.4 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.8 | REACTOME THROMBOXANE SIGNALLING THROUGH TP RECEPTOR | Genes involved in Thromboxane signalling through TP receptor |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.4 | REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | Genes involved in Glycogen breakdown (glycogenolysis) |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.7 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME TRANSPORT OF RIBONUCLEOPROTEINS INTO THE HOST NUCLEUS | Genes involved in Transport of Ribonucleoproteins into the Host Nucleus |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.2 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.4 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.1 | REACTOME FORMATION OF TRANSCRIPTION COUPLED NER TC NER REPAIR COMPLEX | Genes involved in Formation of transcription-coupled NER (TC-NER) repair complex |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME REGULATORY RNA PATHWAYS | Genes involved in Regulatory RNA pathways |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME INTERACTIONS OF VPR WITH HOST CELLULAR PROTEINS | Genes involved in Interactions of Vpr with host cellular proteins |
| 0.0 | 0.2 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.9 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.8 | REACTOME FORMATION OF THE TERNARY COMPLEX AND SUBSEQUENTLY THE 43S COMPLEX | Genes involved in Formation of the ternary complex, and subsequently, the 43S complex |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.5 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.9 | REACTOME DEPOSITION OF NEW CENPA CONTAINING NUCLEOSOMES AT THE CENTROMERE | Genes involved in Deposition of New CENPA-containing Nucleosomes at the Centromere |
| 0.0 | 0.4 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.3 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.0 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.5 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.6 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 1.2 | REACTOME MRNA SPLICING | Genes involved in mRNA Splicing |
| 0.0 | 0.2 | REACTOME METABOLISM OF NON CODING RNA | Genes involved in Metabolism of non-coding RNA |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.1 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.2 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.1 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.0 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.6 | REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | Genes involved in p75 NTR receptor-mediated signalling |
| 0.0 | 0.2 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |