Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
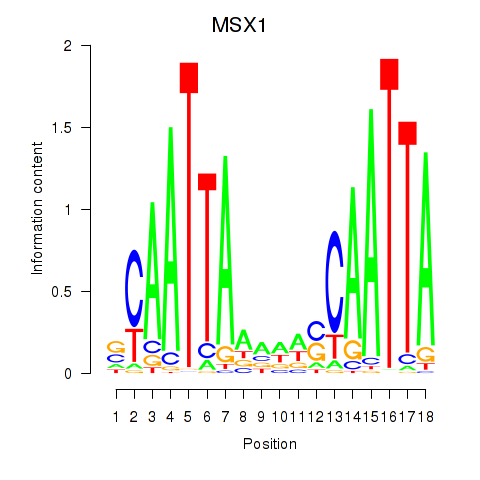
Results for MSX1
Z-value: 0.80
Transcription factors associated with MSX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MSX1
|
ENSG00000163132.6 | msh homeobox 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_4873247_4873398 | MSX1 | 11929 | 0.255507 | -0.61 | 8.0e-02 | Click! |
| chr4_4765683_4765834 | MSX1 | 95635 | 0.079346 | -0.53 | 1.4e-01 | Click! |
| chr4_4873038_4873189 | MSX1 | 11720 | 0.256232 | -0.51 | 1.6e-01 | Click! |
| chr4_4854523_4854767 | MSX1 | 6748 | 0.287695 | 0.50 | 1.7e-01 | Click! |
| chr4_4861125_4861397 | MSX1 | 132 | 0.976461 | 0.47 | 2.0e-01 | Click! |
Activity of the MSX1 motif across conditions
Conditions sorted by the z-value of the MSX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
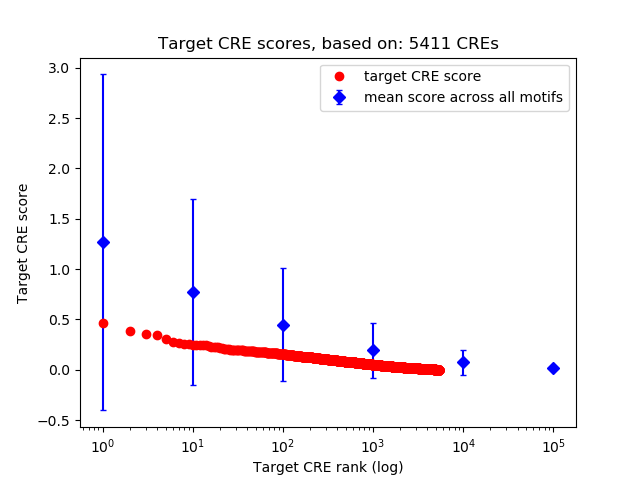
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_38172411_38172686 | 0.47 |
POSTN |
periostin, osteoblast specific factor |
315 |
0.95 |
| chr3_129468296_129468447 | 0.38 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
44888 |
0.13 |
| chr6_1613704_1614304 | 0.35 |
FOXC1 |
forkhead box C1 |
3323 |
0.37 |
| chr6_1612835_1613496 | 0.34 |
FOXC1 |
forkhead box C1 |
2484 |
0.42 |
| chr1_234675056_234675207 | 0.31 |
ENSG00000212144 |
. |
53890 |
0.11 |
| chr3_189932576_189932727 | 0.28 |
ENSG00000212489 |
. |
49339 |
0.13 |
| chr3_29327656_29328192 | 0.27 |
RBMS3-AS3 |
RBMS3 antisense RNA 3 |
4293 |
0.26 |
| chr9_4865980_4866131 | 0.26 |
AL158147.2 |
HCG2011465; Uncharacterized protein |
6795 |
0.16 |
| chr7_158609578_158609780 | 0.25 |
ESYT2 |
extended synaptotagmin-like protein 2 |
12484 |
0.24 |
| chr1_172313169_172313650 | 0.25 |
ENSG00000252354 |
. |
3874 |
0.23 |
| chr8_10268256_10268407 | 0.25 |
RP11-981G7.1 |
|
22851 |
0.2 |
| chr7_116347101_116347278 | 0.25 |
MET |
met proto-oncogene |
8050 |
0.26 |
| chr15_93429972_93430811 | 0.25 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
3865 |
0.22 |
| chr11_14375529_14375805 | 0.24 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
379 |
0.91 |
| chr18_19866097_19866248 | 0.24 |
ENSG00000238907 |
. |
25633 |
0.19 |
| chr5_106791734_106791885 | 0.23 |
EFNA5 |
ephrin-A5 |
214519 |
0.02 |
| chr21_36256046_36256553 | 0.23 |
RUNX1 |
runt-related transcription factor 1 |
3181 |
0.38 |
| chr8_83181283_83181434 | 0.23 |
SNX16 |
sorting nexin 16 |
426257 |
0.01 |
| chr3_30289901_30290052 | 0.23 |
ENSG00000199927 |
. |
56051 |
0.15 |
| chr9_10292066_10292217 | 0.22 |
PTPRD |
protein tyrosine phosphatase, receptor type, D |
258351 |
0.02 |
| chr2_225672771_225672922 | 0.22 |
DOCK10 |
dedicator of cytokinesis 10 |
8523 |
0.32 |
| chr2_21445047_21445198 | 0.21 |
TDRD15 |
tudor domain containing 15 |
98333 |
0.09 |
| chr9_114506322_114506473 | 0.21 |
C9orf84 |
chromosome 9 open reading frame 84 |
15416 |
0.21 |
| chr4_61665381_61665532 | 0.21 |
ENSG00000265829 |
. |
122881 |
0.06 |
| chr6_155544928_155545289 | 0.20 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
7008 |
0.22 |
| chr21_29172171_29172322 | 0.20 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
833414 |
0.0 |
| chr16_1069426_1069639 | 0.20 |
SOX8 |
SRY (sex determining region Y)-box 8 |
37724 |
0.09 |
| chr10_8635845_8635996 | 0.20 |
ENSG00000212505 |
. |
62874 |
0.16 |
| chr9_81871537_81871846 | 0.20 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
314997 |
0.01 |
| chr5_94951954_94952105 | 0.20 |
GPR150 |
G protein-coupled receptor 150 |
3753 |
0.21 |
| chr5_26332801_26332952 | 0.20 |
ENSG00000222560 |
. |
319878 |
0.01 |
| chr11_95906977_95907128 | 0.20 |
ENSG00000266192 |
. |
167550 |
0.03 |
| chr8_32407136_32407287 | 0.20 |
NRG1 |
neuregulin 1 |
966 |
0.71 |
| chrY_17603350_17603501 | 0.20 |
ENSG00000252664 |
. |
571221 |
0.0 |
| chr22_46479348_46479691 | 0.19 |
FLJ27365 |
hsa-mir-4763 |
2364 |
0.16 |
| chr10_80549668_80549819 | 0.19 |
ENSG00000223243 |
. |
39384 |
0.22 |
| chr3_134091064_134091340 | 0.19 |
AMOTL2 |
angiomotin like 2 |
448 |
0.85 |
| chr3_49447935_49448923 | 0.19 |
RHOA |
ras homolog family member A |
932 |
0.32 |
| chr15_93999319_93999470 | 0.19 |
ENSG00000212063 |
. |
169842 |
0.04 |
| chr4_43019583_43019734 | 0.19 |
GRXCR1 |
glutaredoxin, cysteine rich 1 |
124374 |
0.06 |
| chr4_47491811_47491962 | 0.19 |
ATP10D |
ATPase, class V, type 10D |
4570 |
0.24 |
| chr1_221330964_221331115 | 0.19 |
HLX |
H2.0-like homeobox |
276455 |
0.01 |
| chr4_14728626_14728777 | 0.18 |
ENSG00000202449 |
. |
36367 |
0.24 |
| chr12_54089934_54090280 | 0.18 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
18306 |
0.14 |
| chr21_25900554_25900705 | 0.18 |
ENSG00000232512 |
. |
572816 |
0.0 |
| chr7_113054663_113054814 | 0.18 |
TSRM |
Uncharacterized protein; Zinc finger domain-related protein TSRM |
36389 |
0.24 |
| chr1_113447611_113447762 | 0.18 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
30924 |
0.18 |
| chr17_2879197_2879355 | 0.18 |
CTD-3060P21.1 |
|
10087 |
0.23 |
| chr9_74377079_74377230 | 0.18 |
TMEM2 |
transmembrane protein 2 |
6148 |
0.31 |
| chr11_39124455_39124606 | 0.18 |
ENSG00000201591 |
. |
158127 |
0.04 |
| chr1_244504765_244505074 | 0.18 |
C1orf100 |
chromosome 1 open reading frame 100 |
11018 |
0.25 |
| chr4_184558081_184558479 | 0.18 |
ENSG00000252157 |
. |
16549 |
0.14 |
| chr6_69548767_69548918 | 0.18 |
BAI3 |
brain-specific angiogenesis inhibitor 3 |
203583 |
0.03 |
| chr10_20208323_20208474 | 0.18 |
PLXDC2 |
plexin domain containing 2 |
103230 |
0.07 |
| chr16_82664556_82664707 | 0.18 |
CDH13 |
cadherin 13 |
3933 |
0.35 |
| chr7_55638778_55639418 | 0.18 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
463 |
0.8 |
| chr10_63843987_63844138 | 0.18 |
ENSG00000221094 |
. |
31912 |
0.19 |
| chr4_146467351_146467609 | 0.18 |
SMAD1-AS1 |
SMAD1 antisense RNA 1 |
29210 |
0.17 |
| chr2_189198683_189198972 | 0.18 |
ENSG00000207951 |
. |
36608 |
0.17 |
| chr15_67374785_67374936 | 0.17 |
SMAD3 |
SMAD family member 3 |
16060 |
0.25 |
| chr2_134432207_134432889 | 0.17 |
ENSG00000200708 |
. |
78886 |
0.11 |
| chr2_113545549_113545700 | 0.17 |
IL1A |
interleukin 1, alpha |
3457 |
0.21 |
| chr20_43975692_43975843 | 0.17 |
SDC4 |
syndecan 4 |
1297 |
0.32 |
| chr15_64813512_64813804 | 0.17 |
ZNF609 |
zinc finger protein 609 |
22167 |
0.15 |
| chr6_1798343_1798634 | 0.17 |
FOXC1 |
forkhead box C1 |
187807 |
0.03 |
| chr1_189407668_189407819 | 0.17 |
ENSG00000252553 |
. |
227643 |
0.02 |
| chr2_214003351_214003502 | 0.17 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
9927 |
0.3 |
| chr17_1478282_1478467 | 0.17 |
PITPNA |
phosphatidylinositol transfer protein, alpha |
12264 |
0.11 |
| chr12_128113659_128113866 | 0.17 |
ENSG00000199584 |
. |
9770 |
0.33 |
| chr2_113958350_113958501 | 0.17 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
4568 |
0.16 |
| chr3_58359199_58359350 | 0.17 |
PXK |
PX domain containing serine/threonine kinase |
22127 |
0.17 |
| chr4_169553357_169553551 | 0.17 |
PALLD |
palladin, cytoskeletal associated protein |
686 |
0.73 |
| chr7_37349873_37350077 | 0.17 |
ELMO1 |
engulfment and cell motility 1 |
32392 |
0.18 |
| chr1_120191467_120191618 | 0.17 |
ZNF697 |
zinc finger protein 697 |
1146 |
0.52 |
| chr4_85553592_85553743 | 0.17 |
CDS1 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
49535 |
0.15 |
| chr2_118855601_118855752 | 0.17 |
INSIG2 |
insulin induced gene 2 |
9626 |
0.24 |
| chrX_2507647_2507798 | 0.17 |
CD99P1 |
CD99 molecule pseudogene 1 |
19734 |
0.22 |
| chrY_2457647_2457798 | 0.17 |
ENSG00000251841 |
. |
195068 |
0.03 |
| chr21_41095530_41095681 | 0.17 |
IGSF5 |
immunoglobulin superfamily, member 5 |
21729 |
0.21 |
| chr11_12132484_12132785 | 0.17 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
496 |
0.87 |
| chr1_44792092_44792243 | 0.17 |
ERI3 |
ERI1 exoribonuclease family member 3 |
3452 |
0.22 |
| chr19_19514440_19514591 | 0.17 |
CTB-184G21.3 |
|
1103 |
0.38 |
| chr11_102183446_102183978 | 0.17 |
ENSG00000212466 |
. |
741 |
0.64 |
| chr21_36771263_36771477 | 0.16 |
ENSG00000211590 |
. |
321643 |
0.01 |
| chr2_175708769_175708920 | 0.16 |
CHN1 |
chimerin 1 |
2289 |
0.38 |
| chr5_95564073_95564224 | 0.16 |
ENSG00000206997 |
. |
18217 |
0.26 |
| chr14_51242372_51242693 | 0.16 |
NIN |
ninein (GSK3B interacting protein) |
9019 |
0.17 |
| chr3_145437289_145437440 | 0.16 |
RP11-274H2.5 |
|
342102 |
0.01 |
| chr3_43735748_43735899 | 0.16 |
ANO10 |
anoctamin 10 |
2737 |
0.3 |
| chr8_102898890_102899041 | 0.16 |
ENSG00000238372 |
. |
39377 |
0.2 |
| chr17_5268697_5268848 | 0.16 |
RP11-420A6.2 |
|
6149 |
0.15 |
| chr4_8702215_8702366 | 0.16 |
CPZ |
carboxypeptidase Z |
98593 |
0.07 |
| chr9_4137636_4137913 | 0.16 |
GLIS3 |
GLIS family zinc finger 3 |
7419 |
0.28 |
| chr21_18884161_18884312 | 0.16 |
CXADR |
coxsackie virus and adenovirus receptor |
464 |
0.77 |
| chr15_71391266_71391417 | 0.16 |
THSD4 |
thrombospondin, type I, domain containing 4 |
2050 |
0.37 |
| chr2_52029885_52030036 | 0.16 |
ENSG00000222692 |
. |
367604 |
0.01 |
| chr6_39772789_39772940 | 0.16 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
12070 |
0.26 |
| chr5_19885020_19885171 | 0.16 |
CDH18 |
cadherin 18, type 2 |
1286 |
0.64 |
| chr1_230223080_230223231 | 0.16 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
20137 |
0.23 |
| chr3_15482302_15482608 | 0.16 |
EAF1-AS1 |
EAF1 antisense RNA 1 |
354 |
0.48 |
| chr20_52214556_52214707 | 0.16 |
ZNF217 |
zinc finger protein 217 |
4253 |
0.23 |
| chr9_78870959_78871162 | 0.16 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
67505 |
0.13 |
| chr5_32991481_32991632 | 0.15 |
AC026703.1 |
|
202611 |
0.03 |
| chr13_19683905_19684056 | 0.15 |
ENSG00000252287 |
. |
21353 |
0.16 |
| chr5_95457320_95457471 | 0.15 |
ENSG00000207578 |
. |
42553 |
0.17 |
| chr1_181455866_181456017 | 0.15 |
CACNA1E |
calcium channel, voltage-dependent, R type, alpha 1E subunit |
3060 |
0.4 |
| chr4_38754112_38754445 | 0.15 |
ENSG00000222230 |
. |
6235 |
0.19 |
| chr20_49110116_49110267 | 0.15 |
PTPN1 |
protein tyrosine phosphatase, non-receptor type 1 |
16700 |
0.18 |
| chr7_101457714_101458422 | 0.15 |
CUX1 |
cut-like homeobox 1 |
891 |
0.64 |
| chr15_73926288_73926474 | 0.15 |
NPTN |
neuroplastin |
94 |
0.98 |
| chr5_142217391_142217542 | 0.15 |
ARHGAP26-AS1 |
ARHGAP26 antisense RNA 1 |
31009 |
0.21 |
| chr1_116222239_116222390 | 0.15 |
VANGL1 |
VANGL planar cell polarity protein 1 |
28291 |
0.19 |
| chr18_24083489_24083640 | 0.15 |
KCTD1 |
potassium channel tetramerization domain containing 1 |
43399 |
0.17 |
| chr7_116416437_116416588 | 0.15 |
MET |
met proto-oncogene |
717 |
0.73 |
| chr7_27190645_27191214 | 0.15 |
HOXA-AS3 |
HOXA cluster antisense RNA 3 |
1258 |
0.18 |
| chr8_27983413_27983564 | 0.15 |
ELP3 |
elongator acetyltransferase complex subunit 3 |
32524 |
0.17 |
| chr14_69015576_69015827 | 0.15 |
CTD-2325P2.4 |
|
79461 |
0.1 |
| chr5_142775880_142776681 | 0.15 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
4137 |
0.35 |
| chr22_22204157_22204308 | 0.15 |
MAPK1 |
mitogen-activated protein kinase 1 |
17498 |
0.14 |
| chr21_15922641_15922820 | 0.15 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
4066 |
0.28 |
| chr4_160248318_160248469 | 0.15 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
4385 |
0.29 |
| chr10_97154342_97154493 | 0.15 |
SORBS1 |
sorbin and SH3 domain containing 1 |
21035 |
0.25 |
| chr12_62115825_62115976 | 0.15 |
FAM19A2 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A2 |
105114 |
0.08 |
| chr5_94384750_94385033 | 0.15 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
31700 |
0.22 |
| chr18_61447047_61447198 | 0.14 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
1911 |
0.39 |
| chr2_206543328_206543479 | 0.14 |
NRP2 |
neuropilin 2 |
3821 |
0.34 |
| chr1_172290167_172290671 | 0.14 |
ENSG00000252354 |
. |
26864 |
0.17 |
| chr4_35782309_35782460 | 0.14 |
ENSG00000206665 |
. |
284864 |
0.01 |
| chr1_189628853_189629004 | 0.14 |
ENSG00000252553 |
. |
6458 |
0.35 |
| chr3_187467694_187467973 | 0.14 |
BCL6 |
B-cell CLL/lymphoma 6 |
4318 |
0.26 |
| chr12_69731656_69731807 | 0.14 |
LYZ |
lysozyme |
10390 |
0.17 |
| chr16_87522864_87523015 | 0.14 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
2712 |
0.29 |
| chr11_119089001_119089152 | 0.14 |
CBL |
Cbl proto-oncogene, E3 ubiquitin protein ligase |
12324 |
0.1 |
| chr5_44620644_44620795 | 0.14 |
ENSG00000263556 |
. |
95573 |
0.09 |
| chr4_157873991_157874142 | 0.14 |
PDGFC |
platelet derived growth factor C |
17989 |
0.21 |
| chr2_161331149_161331300 | 0.14 |
RBMS1 |
RNA binding motif, single stranded interacting protein 1 |
18805 |
0.24 |
| chr4_157860820_157861106 | 0.14 |
PDGFC |
platelet derived growth factor C |
31092 |
0.19 |
| chr16_46655371_46655551 | 0.14 |
SHCBP1 |
SHC SH2-domain binding protein 1 |
77 |
0.97 |
| chr10_134513988_134514364 | 0.14 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
65152 |
0.12 |
| chr12_59434723_59435248 | 0.14 |
RP11-150C16.1 |
|
120565 |
0.06 |
| chr10_69865396_69865547 | 0.14 |
MYPN |
myopalladin |
441 |
0.84 |
| chr1_28575788_28575939 | 0.14 |
RP5-1092A3.4 |
|
7899 |
0.11 |
| chr11_104838069_104839428 | 0.14 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
1345 |
0.45 |
| chr9_111207460_111207611 | 0.14 |
ENSG00000222512 |
. |
86326 |
0.11 |
| chr1_98816414_98816565 | 0.14 |
ENSG00000221777 |
. |
22452 |
0.29 |
| chr8_93887341_93887492 | 0.14 |
ENSG00000248858 |
. |
2449 |
0.28 |
| chr18_61535025_61535176 | 0.14 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
3826 |
0.24 |
| chr17_32078540_32078804 | 0.14 |
ENSG00000221699 |
. |
122805 |
0.05 |
| chr2_26100038_26101197 | 0.14 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
718 |
0.76 |
| chr2_207987855_207988006 | 0.14 |
KLF7-IT1 |
KLF7 intronic transcript 1 (non-protein coding) |
1162 |
0.49 |
| chr15_64391220_64391371 | 0.14 |
SNX1 |
sorting nexin 1 |
3070 |
0.19 |
| chr5_80210177_80210328 | 0.14 |
CTC-459I6.1 |
|
38547 |
0.18 |
| chr9_102849500_102849651 | 0.13 |
ERP44 |
endoplasmic reticulum protein 44 |
11747 |
0.18 |
| chr12_85768780_85768931 | 0.13 |
ALX1 |
ALX homeobox 1 |
94970 |
0.09 |
| chr6_136256925_136257180 | 0.13 |
ENSG00000238921 |
. |
21159 |
0.21 |
| chr13_71667523_71667674 | 0.13 |
ENSG00000202433 |
. |
634002 |
0.0 |
| chr12_22435490_22435710 | 0.13 |
ST8SIA1 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 |
33620 |
0.22 |
| chr5_129840711_129840862 | 0.13 |
ENSG00000252514 |
. |
118413 |
0.07 |
| chr4_116464375_116464526 | 0.13 |
NDST4 |
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 |
429418 |
0.01 |
| chr9_99391498_99391649 | 0.13 |
CDC14B |
cell division cycle 14B |
9461 |
0.24 |
| chr11_37129867_37130018 | 0.13 |
RAG2 |
recombination activating gene 2 |
510156 |
0.0 |
| chr5_67586145_67586431 | 0.13 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
177 |
0.97 |
| chr1_197730742_197731002 | 0.13 |
RP11-448G4.4 |
|
4423 |
0.25 |
| chr7_82070146_82070432 | 0.13 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
2742 |
0.43 |
| chr20_24270294_24270445 | 0.13 |
ENSG00000200231 |
. |
52478 |
0.18 |
| chr19_11466484_11467017 | 0.13 |
DKFZP761J1410 |
Lipid phosphate phosphatase-related protein type 2 |
576 |
0.52 |
| chr4_181237914_181238065 | 0.13 |
NA |
NA |
> 106 |
NA |
| chr21_18785623_18785774 | 0.13 |
ENSG00000252462 |
. |
18167 |
0.18 |
| chr1_220527416_220527567 | 0.13 |
AC096644.1 |
Uncharacterized protein |
80532 |
0.09 |
| chr20_50722770_50722921 | 0.13 |
ZFP64 |
ZFP64 zinc finger protein |
662 |
0.82 |
| chr14_93720610_93720761 | 0.13 |
UBR7 |
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
35658 |
0.11 |
| chr22_24917713_24917864 | 0.13 |
AP000355.2 |
|
5312 |
0.15 |
| chr6_108886554_108887901 | 0.13 |
FOXO3 |
forkhead box O3 |
5158 |
0.32 |
| chr12_5427929_5428080 | 0.13 |
NTF3 |
neurotrophin 3 |
113275 |
0.07 |
| chr15_31734664_31735087 | 0.13 |
KLF13 |
Kruppel-like factor 13 |
76518 |
0.12 |
| chr9_21548772_21548928 | 0.13 |
MIR31HG |
MIR31 host gene (non-protein coding) |
10818 |
0.17 |
| chr10_91811914_91812065 | 0.13 |
ENSG00000222451 |
. |
111728 |
0.07 |
| chr4_177931083_177931234 | 0.13 |
ENSG00000222859 |
. |
171438 |
0.03 |
| chr3_151033417_151033568 | 0.13 |
GPR87 |
G protein-coupled receptor 87 |
1248 |
0.43 |
| chr3_42191892_42192043 | 0.13 |
TRAK1 |
trafficking protein, kinesin binding 1 |
1240 |
0.57 |
| chr18_59666248_59666399 | 0.13 |
RNF152 |
ring finger protein 152 |
104859 |
0.07 |
| chr18_3351537_3351688 | 0.13 |
TGIF1 |
TGFB-induced factor homeobox 1 |
59994 |
0.1 |
| chr10_105609571_105609851 | 0.13 |
SH3PXD2A |
SH3 and PX domains 2A |
5453 |
0.22 |
| chr6_98415844_98415995 | 0.13 |
ENSG00000238367 |
. |
56488 |
0.18 |
| chr20_49272065_49272493 | 0.13 |
RP4-530I15.6 |
|
2540 |
0.24 |
| chr6_72970806_72970957 | 0.13 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
8387 |
0.27 |
| chr9_21677527_21678485 | 0.13 |
ENSG00000244230 |
. |
21307 |
0.21 |
| chr5_153827010_153827219 | 0.13 |
SAP30L |
SAP30-like |
1009 |
0.44 |
| chr4_187686004_187686155 | 0.13 |
FAT1 |
FAT atypical cadherin 1 |
38203 |
0.2 |
| chr9_286081_286232 | 0.13 |
DOCK8 |
dedicator of cytokinesis 8 |
13086 |
0.19 |
| chr2_206678700_206678851 | 0.13 |
AC007362.3 |
|
50045 |
0.15 |
| chr3_45986008_45986159 | 0.13 |
CXCR6 |
chemokine (C-X-C motif) receptor 6 |
428 |
0.8 |
| chr10_27771901_27772052 | 0.13 |
RAB18 |
RAB18, member RAS oncogene family |
21221 |
0.22 |
| chr10_111794329_111794480 | 0.13 |
ADD3 |
adducin 3 (gamma) |
26682 |
0.18 |
| chr7_74077316_74077467 | 0.12 |
GTF2I |
general transcription factor IIi |
5034 |
0.25 |
| chr17_41281323_41281529 | 0.12 |
NBR2 |
neighbor of BRCA1 gene 2 (non-protein coding) |
1799 |
0.26 |
| chr9_26262415_26262566 | 0.12 |
ENSG00000266429 |
. |
351633 |
0.01 |
| chr2_195590152_195590303 | 0.12 |
ENSG00000252517 |
. |
61329 |
0.16 |
| chr4_48701192_48701343 | 0.12 |
FRYL |
FRY-like |
18079 |
0.24 |
| chr4_180046351_180046502 | 0.12 |
ENSG00000212191 |
. |
439316 |
0.01 |
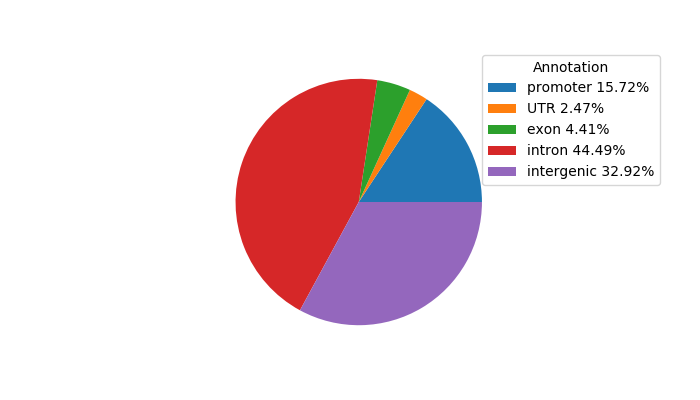
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.2 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) |
| 0.0 | 0.1 | GO:0060044 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.1 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.0 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.0 | GO:0010566 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.0 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.0 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.0 | GO:0010613 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 | 0.0 | GO:0051834 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.0 | GO:0060430 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.0 | 0.0 | GO:0030540 | female genitalia development(GO:0030540) |
| 0.0 | 0.1 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.0 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0051280 | negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |