Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
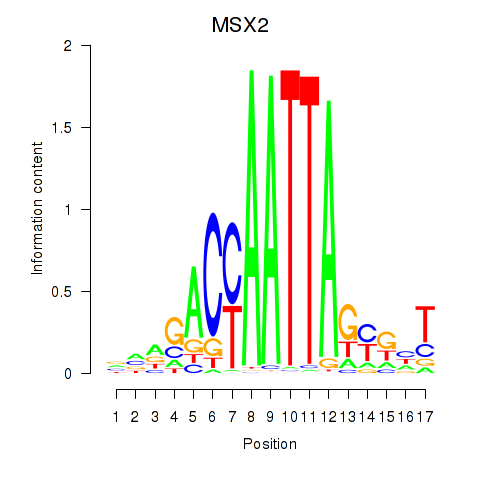
Results for MSX2
Z-value: 0.61
Transcription factors associated with MSX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MSX2
|
ENSG00000120149.7 | msh homeobox 2 |
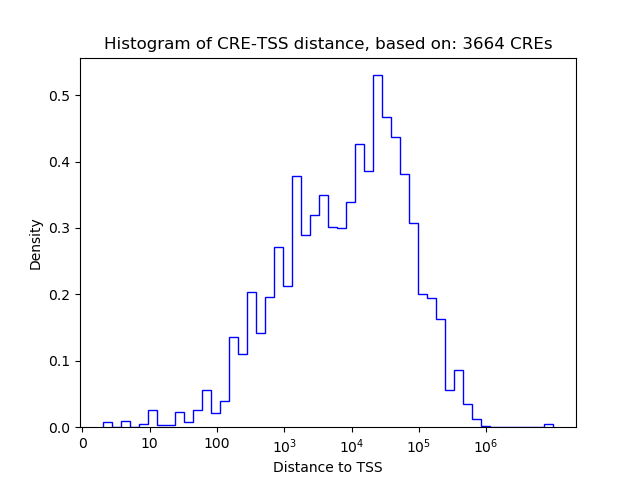
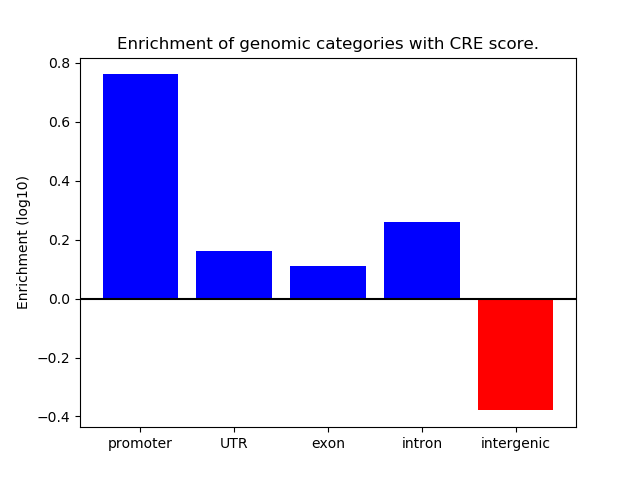
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_174150731_174150989 | MSX2 | 676 | 0.785257 | 0.67 | 4.7e-02 | Click! |
| chr5_174148598_174148749 | MSX2 | 2863 | 0.355218 | 0.67 | 4.7e-02 | Click! |
| chr5_174152116_174152407 | MSX2 | 656 | 0.792778 | 0.65 | 6.0e-02 | Click! |
| chr5_174151487_174151917 | MSX2 | 97 | 0.980311 | 0.64 | 6.2e-02 | Click! |
| chr5_174147841_174148039 | MSX2 | 3596 | 0.324666 | 0.62 | 7.2e-02 | Click! |
Activity of the MSX2 motif across conditions
Conditions sorted by the z-value of the MSX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
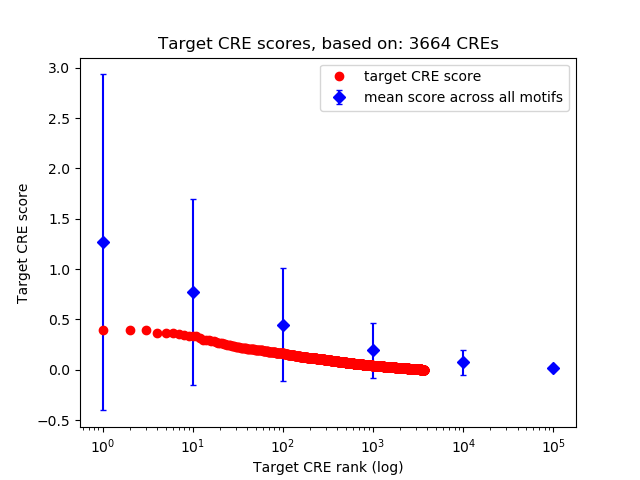
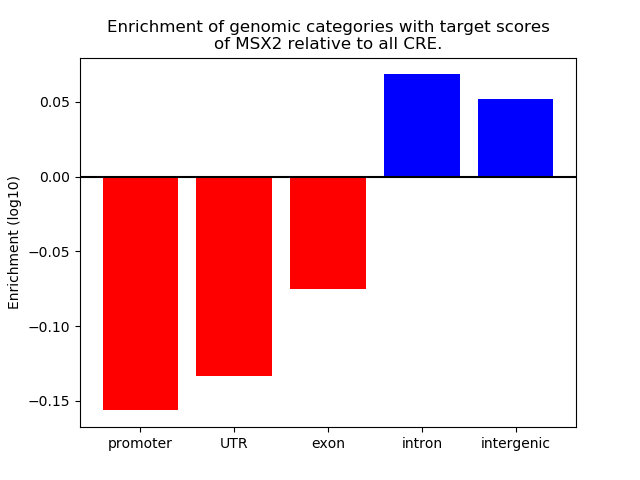
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_69454777_69455129 | 0.39 |
CCND1 |
cyclin D1 |
902 |
0.63 |
| chr10_96161850_96162001 | 0.39 |
TBC1D12 |
TBC1 domain family, member 12 |
336 |
0.91 |
| chr8_37350646_37351319 | 0.39 |
RP11-150O12.6 |
|
23557 |
0.24 |
| chr10_52750817_52751095 | 0.36 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
11 |
0.99 |
| chr10_112290027_112290231 | 0.36 |
DUSP5 |
dual specificity phosphatase 5 |
32533 |
0.13 |
| chr13_77466816_77466967 | 0.36 |
KCTD12 |
potassium channel tetramerization domain containing 12 |
6351 |
0.21 |
| chr5_96000350_96000501 | 0.35 |
CAST |
calpastatin |
1668 |
0.37 |
| chr2_23603304_23603524 | 0.34 |
KLHL29 |
kelch-like family member 29 |
4674 |
0.35 |
| chr6_41472777_41473161 | 0.34 |
RP11-328M4.2 |
|
40843 |
0.13 |
| chr13_76336103_76336254 | 0.34 |
LMO7 |
LIM domain 7 |
1381 |
0.54 |
| chr5_95294796_95295032 | 0.33 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
2638 |
0.26 |
| chr1_79043181_79043547 | 0.32 |
IFI44L |
interferon-induced protein 44-like |
42243 |
0.17 |
| chr2_101728731_101728882 | 0.30 |
ENSG00000265860 |
. |
18775 |
0.19 |
| chr11_56181990_56182141 | 0.30 |
OR5R1 |
olfactory receptor, family 5, subfamily R, member 1 |
3643 |
0.18 |
| chr10_6763782_6764322 | 0.30 |
PRKCQ |
protein kinase C, theta |
141789 |
0.05 |
| chr1_30417491_30417642 | 0.29 |
ENSG00000222787 |
. |
59817 |
0.17 |
| chr18_55888880_55889307 | 0.28 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
290 |
0.93 |
| chr12_56917781_56918152 | 0.28 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
2183 |
0.23 |
| chr9_72745702_72745853 | 0.27 |
MAMDC2-AS1 |
MAMDC2 antisense RNA 1 |
17001 |
0.22 |
| chr14_43058348_43058499 | 0.27 |
CTD-2307P3.1 |
|
226237 |
0.02 |
| chr2_163171316_163171467 | 0.26 |
IFIH1 |
interferon induced with helicase C domain 1 |
3803 |
0.25 |
| chr17_41668751_41668949 | 0.26 |
ETV4 |
ets variant 4 |
11862 |
0.16 |
| chr7_82070146_82070432 | 0.26 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
2742 |
0.43 |
| chr3_113462542_113462739 | 0.25 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
2001 |
0.3 |
| chr11_101917896_101918104 | 0.24 |
C11orf70 |
chromosome 11 open reading frame 70 |
174 |
0.94 |
| chr2_188418561_188419245 | 0.24 |
AC007319.1 |
|
27 |
0.69 |
| chr9_125836272_125836843 | 0.24 |
MIR600HG |
MIR600 host gene (non-protein coding) |
36887 |
0.13 |
| chr6_28415243_28415394 | 0.23 |
ZSCAN23 |
zinc finger and SCAN domain containing 23 |
4074 |
0.2 |
| chr11_122049732_122050124 | 0.23 |
ENSG00000207994 |
. |
26912 |
0.16 |
| chr18_6884956_6885107 | 0.23 |
RP11-146G7.2 |
|
10008 |
0.19 |
| chr1_160176529_160176680 | 0.23 |
PEA15 |
phosphoprotein enriched in astrocytes 15 |
1393 |
0.2 |
| chr4_81228803_81229090 | 0.23 |
C4orf22 |
chromosome 4 open reading frame 22 |
27928 |
0.2 |
| chr16_84652917_84653068 | 0.22 |
COTL1 |
coactosin-like 1 (Dictyostelium) |
1309 |
0.42 |
| chr7_70201396_70202343 | 0.22 |
AUTS2 |
autism susceptibility candidate 2 |
7744 |
0.34 |
| chr20_36040358_36040543 | 0.22 |
SRC |
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
27897 |
0.21 |
| chr1_54964656_54964938 | 0.21 |
ENSG00000265404 |
. |
5544 |
0.2 |
| chr1_210074416_210074567 | 0.21 |
SYT14 |
synaptotagmin XIV |
37047 |
0.16 |
| chr10_119303944_119304573 | 0.21 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
321 |
0.88 |
| chr9_91349159_91349310 | 0.21 |
ENSG00000265873 |
. |
11586 |
0.3 |
| chr10_119302142_119302346 | 0.21 |
EMX2 |
empty spiracles homeobox 2 |
265 |
0.64 |
| chr9_124799161_124799312 | 0.21 |
TTLL11 |
tubulin tyrosine ligase-like family, member 11 |
56649 |
0.12 |
| chr20_61884855_61885070 | 0.21 |
NKAIN4 |
Na+/K+ transporting ATPase interacting 4 |
173 |
0.91 |
| chr1_8084803_8084954 | 0.21 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
1465 |
0.44 |
| chr10_126198812_126198963 | 0.21 |
LHPP |
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
48475 |
0.13 |
| chr11_27491638_27491912 | 0.21 |
RP11-159H22.2 |
|
1501 |
0.32 |
| chr11_120271835_120271998 | 0.20 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
15913 |
0.2 |
| chr17_48588630_48588781 | 0.20 |
MYCBPAP |
MYCBP associated protein |
2722 |
0.15 |
| chr5_168323546_168323697 | 0.20 |
CTB-174D11.2 |
|
96384 |
0.07 |
| chr1_198413244_198413395 | 0.20 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
96485 |
0.09 |
| chr7_157476784_157476935 | 0.20 |
AC006003.3 |
|
56289 |
0.13 |
| chr2_174267051_174267205 | 0.20 |
CDCA7 |
cell division cycle associated 7 |
47528 |
0.19 |
| chr1_201464859_201465010 | 0.20 |
CSRP1 |
cysteine and glycine-rich protein 1 |
767 |
0.61 |
| chr7_45961953_45962825 | 0.20 |
IGFBP3 |
insulin-like growth factor binding protein 3 |
916 |
0.6 |
| chr4_39639074_39639261 | 0.20 |
SMIM14 |
small integral membrane protein 14 |
1346 |
0.42 |
| chr1_56106402_56106768 | 0.20 |
ENSG00000272051 |
. |
155940 |
0.04 |
| chr2_8135764_8135915 | 0.20 |
ENSG00000221255 |
. |
418867 |
0.01 |
| chr2_216299709_216300678 | 0.19 |
FN1 |
fibronectin 1 |
597 |
0.55 |
| chr4_26344236_26344574 | 0.19 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
357 |
0.93 |
| chr10_30237773_30238416 | 0.19 |
KIAA1462 |
KIAA1462 |
110359 |
0.07 |
| chr8_102513775_102514058 | 0.19 |
GRHL2 |
grainyhead-like 2 (Drosophila) |
8930 |
0.22 |
| chr9_89952337_89952906 | 0.19 |
ENSG00000212421 |
. |
77256 |
0.11 |
| chr5_154465051_154465202 | 0.18 |
ENSG00000221131 |
. |
60766 |
0.13 |
| chr9_133722351_133722531 | 0.18 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
11988 |
0.21 |
| chr3_114598031_114598292 | 0.18 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
120043 |
0.06 |
| chr4_57984994_57985389 | 0.18 |
IGFBP7 |
insulin-like growth factor binding protein 7 |
8640 |
0.18 |
| chr20_52352609_52352999 | 0.18 |
ENSG00000238468 |
. |
67507 |
0.12 |
| chr3_110132535_110132686 | 0.18 |
ENSG00000221206 |
. |
138202 |
0.05 |
| chr10_114552420_114552740 | 0.18 |
RP11-57H14.4 |
|
30669 |
0.21 |
| chr3_116583114_116583333 | 0.18 |
ENSG00000265433 |
. |
14009 |
0.25 |
| chr13_27190840_27190991 | 0.18 |
WASF3-AS1 |
WASF3 antisense RNA 1 |
24586 |
0.23 |
| chr5_3597777_3597928 | 0.18 |
IRX1 |
iroquois homeobox 1 |
1684 |
0.41 |
| chr10_15762100_15762293 | 0.18 |
ITGA8 |
integrin, alpha 8 |
72 |
0.99 |
| chr2_119188658_119188833 | 0.18 |
INSIG2 |
insulin induced gene 2 |
342695 |
0.01 |
| chr3_196080166_196080317 | 0.18 |
ENSG00000212146 |
. |
11655 |
0.1 |
| chr16_2315946_2316097 | 0.18 |
RNPS1 |
RNA binding protein S1, serine-rich domain |
503 |
0.49 |
| chr6_73262911_73263125 | 0.18 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
68502 |
0.12 |
| chr16_54968352_54968503 | 0.18 |
IRX5 |
iroquois homeobox 5 |
2443 |
0.45 |
| chr6_15505200_15505483 | 0.17 |
DTNBP1 |
dystrobrevin binding protein 1 |
43252 |
0.19 |
| chr1_182811531_182811809 | 0.17 |
DHX9 |
DEAH (Asp-Glu-Ala-His) box helicase 9 |
3166 |
0.28 |
| chr5_80444015_80444393 | 0.17 |
CTD-2193P3.2 |
|
33533 |
0.2 |
| chr1_210424215_210424840 | 0.17 |
SERTAD4-AS1 |
SERTAD4 antisense RNA 1 |
17135 |
0.21 |
| chr17_12569246_12569498 | 0.17 |
MYOCD |
myocardin |
66 |
0.98 |
| chr9_107688440_107688591 | 0.17 |
RP11-217B7.2 |
|
1319 |
0.43 |
| chr3_185446261_185446412 | 0.17 |
C3orf65 |
chromosome 3 open reading frame 65 |
15256 |
0.21 |
| chr2_511397_511548 | 0.17 |
TMEM18 |
transmembrane protein 18 |
164303 |
0.03 |
| chr1_161932087_161932899 | 0.17 |
OLFML2B |
olfactomedin-like 2B |
22529 |
0.22 |
| chr12_67669324_67669475 | 0.17 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
5718 |
0.35 |
| chr16_73092840_73093092 | 0.17 |
ZFHX3 |
zinc finger homeobox 3 |
631 |
0.78 |
| chr10_85954003_85954712 | 0.17 |
CDHR1 |
cadherin-related family member 1 |
53 |
0.97 |
| chr8_23202144_23202555 | 0.17 |
ENSG00000253837 |
. |
8628 |
0.17 |
| chr14_36409550_36409712 | 0.17 |
RP11-116N8.1 |
|
42443 |
0.16 |
| chr12_66220556_66220804 | 0.17 |
HMGA2 |
high mobility group AT-hook 2 |
1777 |
0.39 |
| chr2_58272799_58272950 | 0.17 |
VRK2 |
vaccinia related kinase 2 |
855 |
0.7 |
| chr14_81789696_81789847 | 0.17 |
STON2 |
stonin 2 |
46494 |
0.18 |
| chr1_154840943_154841324 | 0.17 |
KCNN3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
338 |
0.86 |
| chr15_33154166_33154394 | 0.17 |
FMN1 |
formin 1 |
26175 |
0.19 |
| chr3_169189329_169189480 | 0.17 |
RP11-641D5.2 |
|
23749 |
0.25 |
| chr22_43400050_43400213 | 0.16 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
1685 |
0.39 |
| chr7_14598843_14598994 | 0.16 |
ENSG00000221428 |
. |
216113 |
0.02 |
| chr3_55062884_55063062 | 0.16 |
LRTM1 |
leucine-rich repeats and transmembrane domains 1 |
61858 |
0.15 |
| chr1_198393639_198393891 | 0.16 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
116039 |
0.07 |
| chr9_3794141_3794308 | 0.16 |
RP11-252M18.3 |
|
81360 |
0.1 |
| chr15_33135515_33135666 | 0.16 |
FMN1 |
formin 1 |
44865 |
0.14 |
| chr10_33248509_33248959 | 0.16 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
1574 |
0.52 |
| chr2_200446222_200446758 | 0.16 |
SATB2 |
SATB homeobox 2 |
110501 |
0.07 |
| chr1_172327450_172327703 | 0.16 |
ENSG00000206684 |
. |
8104 |
0.18 |
| chr4_83810146_83810528 | 0.16 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
1834 |
0.28 |
| chr1_16007655_16007806 | 0.16 |
PLEKHM2 |
pleckstrin homology domain containing, family M (with RUN domain) member 2 |
3097 |
0.14 |
| chr22_46476561_46476775 | 0.15 |
FLJ27365 |
hsa-mir-4763 |
476 |
0.66 |
| chr18_20612011_20612377 | 0.15 |
ENSG00000223023 |
. |
7635 |
0.17 |
| chr6_33589286_33590289 | 0.15 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
626 |
0.65 |
| chr11_79793585_79793736 | 0.15 |
ENSG00000221551 |
. |
390237 |
0.01 |
| chr5_38807697_38807893 | 0.15 |
RP11-122C5.3 |
|
24113 |
0.22 |
| chr10_63596511_63596662 | 0.15 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
64473 |
0.13 |
| chrX_78002931_78003376 | 0.15 |
LPAR4 |
lysophosphatidic acid receptor 4 |
72 |
0.99 |
| chr5_170763491_170764141 | 0.15 |
ENSG00000265740 |
. |
22305 |
0.14 |
| chr2_132120817_132121347 | 0.15 |
FAR2P4 |
fatty acyl CoA reductase 2 pseudogene 4 |
66666 |
0.1 |
| chr12_54418548_54418910 | 0.15 |
HOXC6 |
homeobox C6 |
3413 |
0.09 |
| chr11_12849556_12849989 | 0.15 |
RP11-47J17.3 |
|
4558 |
0.23 |
| chr4_176622049_176622200 | 0.15 |
GPM6A |
glycoprotein M6A |
86414 |
0.1 |
| chr7_121944638_121944844 | 0.15 |
FEZF1 |
FEZ family zinc finger 1 |
182 |
0.61 |
| chr10_119132010_119132432 | 0.15 |
PDZD8 |
PDZ domain containing 8 |
2757 |
0.3 |
| chr15_47476270_47477144 | 0.15 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
409 |
0.87 |
| chr21_17909068_17909746 | 0.15 |
ENSG00000207638 |
. |
2002 |
0.33 |
| chr3_188841418_188841569 | 0.15 |
TPRG1 |
tumor protein p63 regulated 1 |
23583 |
0.27 |
| chr6_106246069_106246220 | 0.15 |
ENSG00000200198 |
. |
106104 |
0.08 |
| chr12_115135696_115136278 | 0.15 |
TBX3 |
T-box 3 |
14018 |
0.21 |
| chr16_11655655_11655806 | 0.14 |
LITAF |
lipopolysaccharide-induced TNF factor |
24499 |
0.15 |
| chr6_28442401_28443147 | 0.14 |
ZSCAN23 |
zinc finger and SCAN domain containing 23 |
31530 |
0.13 |
| chr18_53253101_53253291 | 0.14 |
TCF4 |
transcription factor 4 |
1 |
0.99 |
| chr14_74724486_74725063 | 0.14 |
VSX2 |
visual system homeobox 2 |
18599 |
0.14 |
| chr7_146980619_146980770 | 0.14 |
ENSG00000221442 |
. |
94519 |
0.09 |
| chr3_192288894_192289136 | 0.14 |
ENSG00000238902 |
. |
37114 |
0.17 |
| chr2_71772491_71772642 | 0.14 |
DYSF |
dysferlin |
78734 |
0.11 |
| chr17_39874099_39874250 | 0.14 |
ENSG00000201920 |
. |
232 |
0.86 |
| chr10_74436092_74436294 | 0.14 |
MCU |
mitochondrial calcium uniporter |
15696 |
0.15 |
| chr4_66301222_66301373 | 0.14 |
EPHA5 |
EPH receptor A5 |
234356 |
0.02 |
| chr3_168825494_168825645 | 0.14 |
MECOM |
MDS1 and EVI1 complex locus |
20253 |
0.29 |
| chr15_101899572_101899723 | 0.14 |
PCSK6 |
proprotein convertase subtilisin/kexin type 6 |
6820 |
0.2 |
| chr1_120255525_120255935 | 0.14 |
PHGDH |
phosphoglycerate dehydrogenase |
1220 |
0.51 |
| chr5_82771308_82771459 | 0.14 |
VCAN |
versican |
3639 |
0.35 |
| chr13_60348574_60349034 | 0.14 |
DIAPH3-AS1 |
DIAPH3 antisense RNA 1 |
238081 |
0.02 |
| chr3_114790766_114791082 | 0.14 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
702 |
0.82 |
| chr7_19141522_19141673 | 0.14 |
AC003986.6 |
|
10500 |
0.16 |
| chr1_112604147_112604298 | 0.14 |
KCND3 |
potassium voltage-gated channel, Shal-related subfamily, member 3 |
72445 |
0.11 |
| chr6_18028807_18028958 | 0.13 |
KIF13A |
kinesin family member 13A |
41028 |
0.18 |
| chr22_36232810_36232961 | 0.13 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
3380 |
0.35 |
| chr3_27367755_27367906 | 0.13 |
NEK10 |
NIMA-related kinase 10 |
34782 |
0.16 |
| chr10_6836277_6836922 | 0.13 |
PRKCQ |
protein kinase C, theta |
214336 |
0.02 |
| chr3_25472946_25473178 | 0.13 |
RARB |
retinoic acid receptor, beta |
3260 |
0.35 |
| chr3_100923421_100923572 | 0.13 |
ENSG00000238312 |
. |
2820 |
0.37 |
| chr14_21564786_21565064 | 0.13 |
ZNF219 |
zinc finger protein 219 |
1874 |
0.17 |
| chr12_120246427_120246712 | 0.13 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
5382 |
0.27 |
| chr1_231997217_231997368 | 0.13 |
DISC1-IT1 |
DISC1 intronic transcript 1 (non-protein coding) |
64288 |
0.14 |
| chr21_30502420_30502571 | 0.13 |
MAP3K7CL |
MAP3K7 C-terminal like |
311 |
0.86 |
| chr11_128811477_128811628 | 0.13 |
TP53AIP1 |
tumor protein p53 regulated apoptosis inducing protein 1 |
1270 |
0.42 |
| chr11_111667514_111667665 | 0.13 |
PPP2R1B |
protein phosphatase 2, regulatory subunit A, beta |
30438 |
0.12 |
| chr1_149378983_149379172 | 0.13 |
FCGR1C |
Fc fragment of IgG, high affinity Ic, receptor (CD64), pseudogene |
9720 |
0.13 |
| chr4_14169430_14169581 | 0.13 |
ENSG00000252092 |
. |
509254 |
0.0 |
| chr5_147836900_147837051 | 0.13 |
CTD-2283N19.1 |
|
26605 |
0.18 |
| chr5_164797749_164797900 | 0.13 |
ENSG00000252794 |
. |
238622 |
0.02 |
| chr1_70509689_70509840 | 0.13 |
RP11-181B18.1 |
|
12859 |
0.27 |
| chr11_34648519_34648670 | 0.13 |
EHF |
ets homologous factor |
2771 |
0.38 |
| chr18_31157392_31158104 | 0.13 |
ASXL3 |
additional sex combs like 3 (Drosophila) |
848 |
0.76 |
| chr8_99961176_99961327 | 0.13 |
OSR2 |
odd-skipped related transciption factor 2 |
631 |
0.7 |
| chr14_89507425_89507846 | 0.13 |
FOXN3 |
forkhead box N3 |
139453 |
0.05 |
| chr13_33858125_33858279 | 0.13 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
1690 |
0.4 |
| chr12_80838762_80838913 | 0.13 |
PTPRQ |
protein tyrosine phosphatase, receptor type, Q |
711 |
0.78 |
| chr7_102532238_102532586 | 0.13 |
LRRC17 |
leucine rich repeat containing 17 |
21026 |
0.16 |
| chr19_34113333_34113659 | 0.13 |
CHST8 |
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 8 |
619 |
0.82 |
| chr22_46471692_46472277 | 0.13 |
FLJ27365 |
hsa-mir-4763 |
4208 |
0.11 |
| chr10_126846587_126846738 | 0.13 |
CTBP2 |
C-terminal binding protein 2 |
623 |
0.83 |
| chr10_104306894_104307045 | 0.13 |
ENSG00000242311 |
. |
21247 |
0.12 |
| chr7_5620896_5621047 | 0.13 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
11468 |
0.14 |
| chr9_78870959_78871162 | 0.13 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
67505 |
0.13 |
| chr17_75567549_75568025 | 0.13 |
SEPT9 |
septin 9 |
89792 |
0.07 |
| chr14_99720527_99720678 | 0.13 |
AL109767.1 |
|
8683 |
0.22 |
| chr1_151960835_151961083 | 0.13 |
S100A10 |
S100 calcium binding protein A10 |
4095 |
0.17 |
| chr8_95447406_95448205 | 0.13 |
FSBP |
fibrinogen silencer binding protein |
1365 |
0.47 |
| chr17_39065845_39065996 | 0.12 |
AC004231.2 |
|
11762 |
0.09 |
| chr18_59619337_59619488 | 0.12 |
RNF152 |
ring finger protein 152 |
57948 |
0.16 |
| chr10_95224937_95225088 | 0.12 |
MYOF |
myoferlin |
16939 |
0.19 |
| chr13_95952806_95953583 | 0.12 |
ABCC4 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 4 |
395 |
0.86 |
| chr5_58055562_58056142 | 0.12 |
RP11-479O16.1 |
|
24754 |
0.24 |
| chr15_50648671_50648965 | 0.12 |
ENSG00000244879 |
. |
150 |
0.91 |
| chr22_46483352_46483503 | 0.12 |
FLJ27365 |
hsa-mir-4763 |
1544 |
0.22 |
| chr14_54423071_54423266 | 0.12 |
BMP4 |
bone morphogenetic protein 4 |
361 |
0.91 |
| chr20_14317138_14317687 | 0.12 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
842 |
0.69 |
| chr7_5637609_5637772 | 0.12 |
FSCN1 |
fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) |
3373 |
0.21 |
| chr5_158036732_158037177 | 0.12 |
CTD-2363C16.1 |
|
373060 |
0.01 |
| chr7_107640774_107640963 | 0.12 |
ENSG00000238297 |
. |
441 |
0.76 |
| chr14_22330961_22331112 | 0.12 |
ENSG00000222776 |
. |
82251 |
0.1 |
| chr14_103541034_103541185 | 0.12 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
17310 |
0.15 |
| chr19_1745649_1745800 | 0.12 |
ONECUT3 |
one cut homeobox 3 |
6648 |
0.14 |
| chr2_48675517_48675668 | 0.12 |
PPP1R21 |
protein phosphatase 1, regulatory subunit 21 |
7324 |
0.23 |
| chr15_96878465_96878729 | 0.12 |
ENSG00000222651 |
. |
2107 |
0.24 |
| chr17_78194664_78195096 | 0.12 |
SGSH |
N-sulfoglucosamine sulfohydrolase |
158 |
0.63 |
| chr1_200865204_200865355 | 0.12 |
C1orf106 |
chromosome 1 open reading frame 106 |
1330 |
0.42 |
| chr1_46714502_46714653 | 0.12 |
RAD54L |
RAD54-like (S. cerevisiae) |
1136 |
0.42 |
| chr17_11640186_11640337 | 0.12 |
ENSG00000263623 |
. |
4520 |
0.33 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.3 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0048875 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.0 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.0 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.0 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.2 | GO:1903846 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0045836 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.1 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.0 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.0 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.0 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) regulation of NFAT protein import into nucleus(GO:0051532) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.0 | GO:1900078 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.1 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.0 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.4 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.2 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.3 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.4 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.2 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.3 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME P130CAS LINKAGE TO MAPK SIGNALING FOR INTEGRINS | Genes involved in p130Cas linkage to MAPK signaling for integrins |
| 0.0 | 0.4 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |