Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
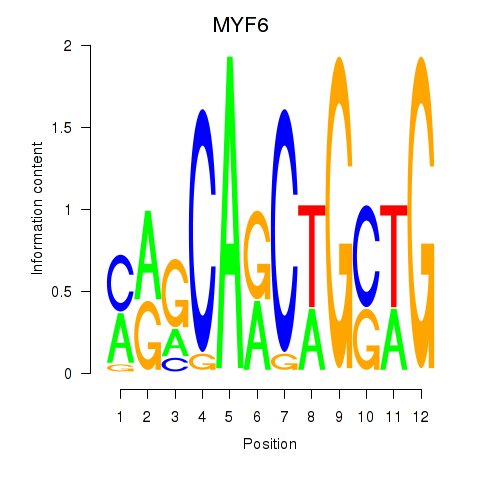
Results for MYF6
Z-value: 0.87
Transcription factors associated with MYF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYF6
|
ENSG00000111046.3 | myogenic factor 6 |
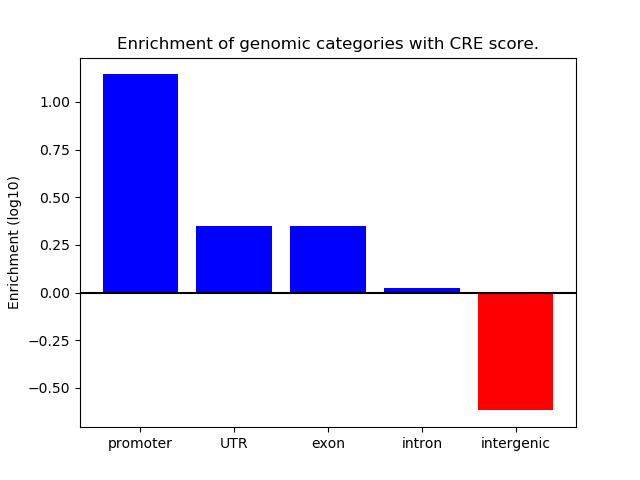
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_81092154_81092305 | MYF6 | 9048 | 0.211378 | -0.71 | 3.0e-02 | Click! |
| chr12_81102183_81102935 | MYF6 | 1282 | 0.485596 | -0.51 | 1.6e-01 | Click! |
| chr12_81064372_81064523 | MYF6 | 36830 | 0.146841 | -0.44 | 2.4e-01 | Click! |
| chr12_81096783_81097042 | MYF6 | 4365 | 0.241896 | -0.34 | 3.7e-01 | Click! |
| chr12_81097059_81097356 | MYF6 | 4070 | 0.247165 | -0.33 | 3.8e-01 | Click! |
Activity of the MYF6 motif across conditions
Conditions sorted by the z-value of the MYF6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
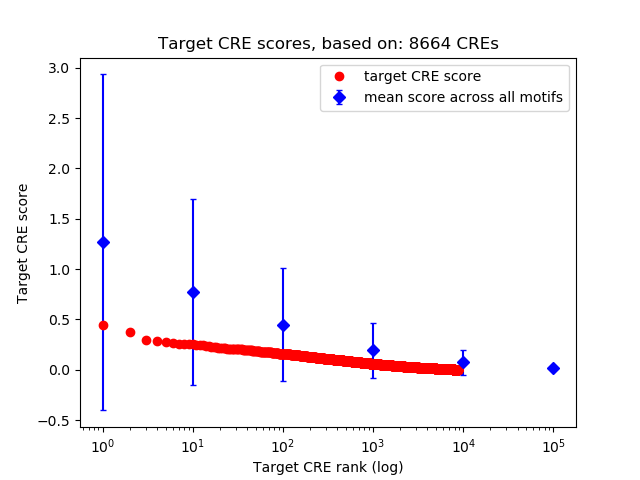
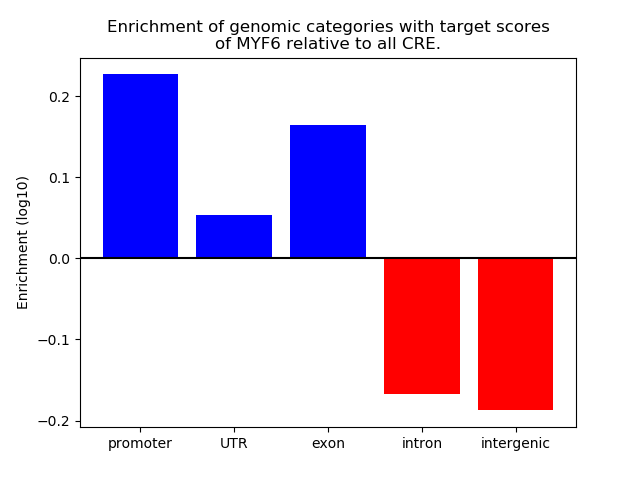
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_441089_441413 | 0.45 |
ENSG00000243562 |
. |
558 |
0.44 |
| chr1_2347262_2347748 | 0.37 |
PEX10 |
peroxisomal biogenesis factor 10 |
2269 |
0.17 |
| chr1_174768895_174769046 | 0.30 |
RABGAP1L |
RAB GTPase activating protein 1-like |
100 |
0.98 |
| chr17_80260072_80260536 | 0.29 |
CD7 |
CD7 molecule |
15124 |
0.1 |
| chr2_191745621_191745919 | 0.28 |
GLS |
glutaminase |
210 |
0.9 |
| chr11_1482189_1482340 | 0.26 |
BRSK2 |
BR serine/threonine kinase 2 |
10349 |
0.17 |
| chrX_39186546_39186697 | 0.26 |
ENSG00000207122 |
. |
246083 |
0.02 |
| chr12_76424698_76425269 | 0.26 |
PHLDA1 |
pleckstrin homology-like domain, family A, member 1 |
401 |
0.66 |
| chr14_61788916_61789580 | 0.26 |
PRKCH |
protein kinase C, eta |
134 |
0.7 |
| chr11_58873962_58874353 | 0.25 |
FAM111B |
family with sequence similarity 111, member B |
501 |
0.79 |
| chr16_68798920_68799109 | 0.25 |
ENSG00000200558 |
. |
22631 |
0.14 |
| chr11_46367528_46367907 | 0.25 |
DGKZ |
diacylglycerol kinase, zeta |
730 |
0.6 |
| chr18_2967390_2967541 | 0.24 |
RP11-737O24.1 |
|
449 |
0.78 |
| chr6_209971_210123 | 0.24 |
DUSP22 |
dual specificity phosphatase 22 |
81583 |
0.11 |
| chr22_21738155_21738638 | 0.23 |
RIMBP3B |
RIMS binding protein 3B |
733 |
0.51 |
| chr3_29495707_29495858 | 0.23 |
ENSG00000216169 |
. |
84870 |
0.1 |
| chr5_156535881_156536093 | 0.23 |
HAVCR2 |
hepatitis A virus cellular receptor 2 |
146 |
0.94 |
| chr22_20460794_20461266 | 0.23 |
RIMBP3 |
RIMS binding protein 3 |
756 |
0.52 |
| chr3_12689624_12689775 | 0.22 |
ENSG00000239140 |
. |
15466 |
0.17 |
| chr5_180017657_180017884 | 0.22 |
SCGB3A1 |
secretoglobin, family 3A, member 1 |
770 |
0.62 |
| chr19_6060885_6061036 | 0.22 |
RFX2 |
regulatory factor X, 2 (influences HLA class II expression) |
3666 |
0.18 |
| chr22_39963541_39963692 | 0.22 |
CACNA1I |
calcium channel, voltage-dependent, T type, alpha 1I subunit |
3142 |
0.23 |
| chr22_21904755_21905186 | 0.21 |
RIMBP3C |
RIMS binding protein 3C |
150 |
0.88 |
| chr16_14380187_14380356 | 0.21 |
ENSG00000201075 |
. |
10909 |
0.18 |
| chr17_62009073_62009653 | 0.21 |
CD79B |
CD79b molecule, immunoglobulin-associated beta |
258 |
0.86 |
| chr18_13376586_13376737 | 0.21 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
5892 |
0.17 |
| chr11_441985_442183 | 0.21 |
ANO9 |
anoctamin 9 |
73 |
0.92 |
| chr2_68966105_68966288 | 0.21 |
ARHGAP25 |
Rho GTPase activating protein 25 |
4182 |
0.3 |
| chr19_5132131_5132282 | 0.21 |
CTC-482H14.5 |
|
45924 |
0.14 |
| chr12_112036778_112037095 | 0.21 |
ATXN2 |
ataxin 2 |
218 |
0.66 |
| chr14_22554936_22555087 | 0.21 |
ENSG00000238634 |
. |
55876 |
0.15 |
| chr21_36217976_36218127 | 0.20 |
RUNX1 |
runt-related transcription factor 1 |
41429 |
0.2 |
| chr1_116096403_116096554 | 0.20 |
ENSG00000239984 |
. |
52614 |
0.13 |
| chr13_26624895_26625081 | 0.20 |
SHISA2 |
shisa family member 2 |
181 |
0.97 |
| chr7_130007608_130007759 | 0.20 |
CPA5 |
carboxypeptidase A5 |
4401 |
0.15 |
| chr16_68771339_68771490 | 0.20 |
CDH1 |
cadherin 1, type 1, E-cadherin (epithelial) |
159 |
0.94 |
| chr8_128988445_128988635 | 0.20 |
ENSG00000221771 |
. |
15661 |
0.16 |
| chr12_19592806_19593080 | 0.20 |
AEBP2 |
AE binding protein 2 |
310 |
0.92 |
| chr11_2320607_2320858 | 0.20 |
C11orf21 |
chromosome 11 open reading frame 21 |
2411 |
0.18 |
| chr1_100819629_100819780 | 0.20 |
CDC14A |
cell division cycle 14A |
1199 |
0.48 |
| chr8_62626758_62627054 | 0.19 |
ASPH |
aspartate beta-hydroxylase |
178 |
0.78 |
| chr5_35857770_35858187 | 0.19 |
IL7R |
interleukin 7 receptor |
984 |
0.58 |
| chr22_24824729_24824978 | 0.19 |
ADORA2A |
adenosine A2a receptor |
1323 |
0.44 |
| chr12_7068113_7068279 | 0.19 |
ENSG00000207713 |
. |
4666 |
0.06 |
| chr8_21770420_21771232 | 0.19 |
DOK2 |
docking protein 2, 56kDa |
348 |
0.88 |
| chr20_62369505_62369786 | 0.19 |
RP4-583P15.14 |
|
22 |
0.92 |
| chr17_38250724_38250875 | 0.19 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
6179 |
0.12 |
| chr18_43266884_43267229 | 0.19 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
20949 |
0.17 |
| chr19_19737943_19738234 | 0.19 |
LPAR2 |
lysophosphatidic acid receptor 2 |
521 |
0.66 |
| chr1_201438452_201438736 | 0.18 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
229 |
0.92 |
| chr1_2455307_2455458 | 0.18 |
PANK4 |
pantothenate kinase 4 |
2653 |
0.14 |
| chrX_78403286_78403508 | 0.18 |
GPR174 |
G protein-coupled receptor 174 |
23072 |
0.28 |
| chr1_185528978_185529129 | 0.18 |
ENSG00000207108 |
. |
70607 |
0.11 |
| chr1_26644827_26645344 | 0.18 |
UBXN11 |
UBX domain protein 11 |
231 |
0.76 |
| chr14_93043697_93043848 | 0.18 |
RIN3 |
Ras and Rab interactor 3 |
63624 |
0.13 |
| chr7_150148052_150148382 | 0.18 |
GIMAP8 |
GTPase, IMAP family member 8 |
499 |
0.78 |
| chr11_122568313_122568557 | 0.18 |
ENSG00000239079 |
. |
28584 |
0.19 |
| chr16_1031099_1031287 | 0.18 |
AC009041.2 |
|
59 |
0.53 |
| chr7_45076135_45076286 | 0.18 |
CCM2 |
cerebral cavernous malformation 2 |
8939 |
0.15 |
| chr8_65607910_65608061 | 0.18 |
RP11-1D12.1 |
|
39031 |
0.2 |
| chr16_3628507_3629057 | 0.18 |
NLRC3 |
NLR family, CARD domain containing 3 |
1381 |
0.35 |
| chr2_148779045_148779339 | 0.18 |
ORC4 |
origin recognition complex, subunit 4 |
45 |
0.92 |
| chr22_28194756_28194907 | 0.18 |
MN1 |
meningioma (disrupted in balanced translocation) 1 |
2655 |
0.36 |
| chr1_36023709_36024187 | 0.18 |
NCDN |
neurochondrin |
159 |
0.86 |
| chr13_78271067_78271525 | 0.18 |
SLAIN1 |
SLAIN motif family, member 1 |
727 |
0.51 |
| chr17_14204490_14204770 | 0.18 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
230 |
0.95 |
| chr12_258585_258736 | 0.18 |
RP11-598F7.4 |
|
325 |
0.85 |
| chr1_90468959_90469110 | 0.17 |
ZNF326 |
zinc finger protein 326 |
8307 |
0.24 |
| chr1_27847952_27848103 | 0.17 |
RP1-159A19.4 |
|
4289 |
0.2 |
| chr8_145003826_145003977 | 0.17 |
PLEC |
plectin |
2722 |
0.16 |
| chr17_1478951_1479102 | 0.17 |
PITPNA |
phosphatidylinositol transfer protein, alpha |
12916 |
0.11 |
| chr1_43415165_43415361 | 0.17 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
9237 |
0.19 |
| chr1_160547005_160547156 | 0.17 |
CD84 |
CD84 molecule |
2183 |
0.25 |
| chr2_5768317_5769063 | 0.17 |
AC108025.2 |
|
62488 |
0.12 |
| chr13_50645613_50645841 | 0.17 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
10580 |
0.16 |
| chr4_48655659_48656132 | 0.17 |
FRYL |
FRY-like |
19468 |
0.24 |
| chr16_1392661_1392945 | 0.17 |
BAIAP3 |
BAI1-associated protein 3 |
1866 |
0.16 |
| chr1_110333243_110333394 | 0.17 |
EPS8L3 |
EPS8-like 3 |
26669 |
0.13 |
| chr11_108408827_108408978 | 0.17 |
EXPH5 |
exophilin 5 |
32 |
0.98 |
| chr4_16219327_16219478 | 0.17 |
TAPT1 |
transmembrane anterior posterior transformation 1 |
8692 |
0.2 |
| chr2_112940458_112940634 | 0.17 |
FBLN7 |
fibulin 7 |
1086 |
0.59 |
| chr12_47610316_47610761 | 0.17 |
PCED1B |
PC-esterase domain containing 1B |
157 |
0.96 |
| chr4_140910160_140910334 | 0.17 |
MAML3 |
mastermind-like 3 (Drosophila) |
98126 |
0.08 |
| chrX_10514569_10514720 | 0.16 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
30351 |
0.21 |
| chr15_38854702_38854853 | 0.16 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
2059 |
0.32 |
| chr2_242843781_242843932 | 0.16 |
AC131097.4 |
Protein LOC285095 |
846 |
0.53 |
| chr19_6590606_6591184 | 0.16 |
CD70 |
CD70 molecule |
255 |
0.89 |
| chr14_22947996_22948147 | 0.16 |
ENSG00000251002 |
. |
2207 |
0.16 |
| chr14_106855204_106855368 | 0.16 |
IGHV3-37 |
immunoglobulin heavy variable 3-37 (pseudogene) |
2260 |
0.07 |
| chr8_24855476_24855627 | 0.16 |
CTD-2168K21.2 |
|
41416 |
0.17 |
| chr1_200882421_200882682 | 0.16 |
C1orf106 |
chromosome 1 open reading frame 106 |
18602 |
0.16 |
| chr7_150180608_150181504 | 0.16 |
GIMAP7 |
GTPase, IMAP family member 7 |
30862 |
0.13 |
| chr7_152209916_152210186 | 0.16 |
ENSG00000221454 |
. |
73745 |
0.08 |
| chr15_44958533_44958684 | 0.16 |
PATL2 |
protein associated with topoisomerase II homolog 2 (yeast) |
2075 |
0.24 |
| chr12_120033969_120034120 | 0.16 |
TMEM233 |
transmembrane protein 233 |
2780 |
0.25 |
| chr16_28996496_28996848 | 0.16 |
LAT |
linker for activation of T cells |
11 |
0.94 |
| chr2_10472002_10472153 | 0.16 |
HPCAL1 |
hippocalcin-like 1 |
28251 |
0.15 |
| chr5_176784926_176785308 | 0.16 |
RGS14 |
regulator of G-protein signaling 14 |
279 |
0.82 |
| chr20_31071096_31071265 | 0.16 |
C20orf112 |
chromosome 20 open reading frame 112 |
94 |
0.97 |
| chr8_117168069_117168220 | 0.16 |
ENSG00000221793 |
. |
19146 |
0.28 |
| chr21_44834894_44835045 | 0.16 |
SIK1 |
salt-inducible kinase 1 |
12039 |
0.28 |
| chr16_28634523_28635087 | 0.16 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
69 |
0.96 |
| chr18_10131788_10132277 | 0.16 |
ENSG00000263630 |
. |
126838 |
0.05 |
| chr11_5735144_5735295 | 0.16 |
TRIM22 |
tripartite motif containing 22 |
5781 |
0.13 |
| chr8_121744015_121744417 | 0.16 |
RP11-713M15.1 |
|
29277 |
0.22 |
| chr5_1315874_1316496 | 0.16 |
ENSG00000263670 |
. |
6693 |
0.18 |
| chr2_16083664_16083952 | 0.16 |
MYCNOS |
MYCN opposite strand |
1437 |
0.36 |
| chr13_28696488_28696678 | 0.16 |
PAN3-AS1 |
PAN3 antisense RNA 1 |
15747 |
0.17 |
| chr2_131673224_131673375 | 0.16 |
ARHGEF4 |
Rho guanine nucleotide exchange factor (GEF) 4 |
925 |
0.59 |
| chr20_34078435_34078749 | 0.16 |
RP3-477O4.14 |
|
151 |
0.92 |
| chr9_92094215_92094379 | 0.15 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
508 |
0.83 |
| chr2_71687821_71687972 | 0.15 |
DYSF |
dysferlin |
5936 |
0.27 |
| chr1_207244089_207244240 | 0.15 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
5815 |
0.14 |
| chr8_128492794_128493088 | 0.15 |
CASC8 |
cancer susceptibility candidate 8 (non-protein coding) |
1443 |
0.55 |
| chr6_144655565_144656031 | 0.15 |
RP1-91J24.3 |
|
1361 |
0.51 |
| chrX_68725182_68725333 | 0.15 |
FAM155B |
family with sequence similarity 155, member B |
173 |
0.88 |
| chr4_144435743_144436361 | 0.15 |
SMARCA5-AS1 |
SMARCA5 antisense RNA 1 |
264 |
0.9 |
| chr5_147184374_147184525 | 0.15 |
JAKMIP2 |
janus kinase and microtubule interacting protein 2 |
22111 |
0.17 |
| chr1_173376102_173376280 | 0.15 |
RP11-296O14.3 |
|
35045 |
0.18 |
| chr14_61811311_61811517 | 0.15 |
PRKCH |
protein kinase C, eta |
372 |
0.88 |
| chr11_19302572_19302855 | 0.15 |
E2F8 |
E2F transcription factor 8 |
39546 |
0.14 |
| chr13_110780795_110780946 | 0.15 |
ENSG00000265885 |
. |
30372 |
0.24 |
| chr6_143228096_143228637 | 0.15 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
37972 |
0.2 |
| chr11_134201877_134202251 | 0.15 |
GLB1L2 |
galactosidase, beta 1-like 2 |
108 |
0.97 |
| chr10_105211794_105212415 | 0.15 |
CALHM2 |
calcium homeostasis modulator 2 |
29 |
0.88 |
| chr6_31540520_31540671 | 0.15 |
LTA |
lymphotoxin alpha |
535 |
0.44 |
| chr19_872645_872938 | 0.15 |
CFD |
complement factor D (adipsin) |
13101 |
0.07 |
| chr17_55557828_55557979 | 0.15 |
RP11-118E18.2 |
|
43055 |
0.16 |
| chr17_27375692_27375843 | 0.15 |
PIPOX |
pipecolic acid oxidase |
4498 |
0.14 |
| chr21_44466329_44466480 | 0.15 |
CBS |
cystathionine-beta-synthase |
12647 |
0.18 |
| chr9_130738368_130738519 | 0.15 |
FAM102A |
family with sequence similarity 102, member A |
4349 |
0.13 |
| chr1_233430570_233431212 | 0.15 |
PCNXL2 |
pecanex-like 2 (Drosophila) |
568 |
0.83 |
| chr11_1865337_1865488 | 0.15 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
3988 |
0.11 |
| chr7_153749645_153749898 | 0.15 |
DPP6 |
dipeptidyl-peptidase 6 |
6 |
0.98 |
| chr5_1793533_1793754 | 0.15 |
MRPL36 |
mitochondrial ribosomal protein L36 |
6221 |
0.19 |
| chr9_136004184_136004335 | 0.15 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
529 |
0.71 |
| chr12_56732840_56732991 | 0.15 |
IL23A |
interleukin 23, alpha subunit p19 |
252 |
0.8 |
| chr12_113906014_113906165 | 0.15 |
RP11-82C23.2 |
|
3719 |
0.19 |
| chrX_7061516_7061667 | 0.15 |
ENSG00000264268 |
. |
4310 |
0.26 |
| chr10_3665379_3665530 | 0.14 |
RP11-184A2.3 |
|
127805 |
0.06 |
| chr10_30993356_30993652 | 0.14 |
SVILP1 |
supervillin pseudogene 1 |
8654 |
0.27 |
| chr17_47572677_47573321 | 0.14 |
NGFR |
nerve growth factor receptor |
344 |
0.86 |
| chr17_47819247_47819793 | 0.14 |
FAM117A |
family with sequence similarity 117, member A |
17631 |
0.14 |
| chr1_245851125_245851276 | 0.14 |
RP11-522M21.3 |
|
11420 |
0.28 |
| chr15_69652737_69652888 | 0.14 |
ENSG00000252972 |
. |
7112 |
0.13 |
| chr2_85106634_85106918 | 0.14 |
TRABD2A |
TraB domain containing 2A |
1430 |
0.42 |
| chr19_47609844_47609995 | 0.14 |
SAE1 |
SUMO1 activating enzyme subunit 1 |
6612 |
0.16 |
| chr1_233749998_233750164 | 0.14 |
KCNK1 |
potassium channel, subfamily K, member 1 |
331 |
0.91 |
| chr17_4802078_4802229 | 0.14 |
C17orf107 |
chromosome 17 open reading frame 107 |
560 |
0.5 |
| chr1_9688741_9688898 | 0.14 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
22971 |
0.15 |
| chr5_150535420_150535844 | 0.14 |
ANXA6 |
annexin A6 |
1676 |
0.39 |
| chr1_7740584_7741157 | 0.14 |
CAMTA1 |
calmodulin binding transcription activator 1 |
55598 |
0.13 |
| chr2_97466776_97466927 | 0.14 |
ENSG00000264157 |
. |
2836 |
0.19 |
| chr15_101143012_101143328 | 0.14 |
ASB7 |
ankyrin repeat and SOCS box containing 7 |
356 |
0.69 |
| chr1_227950100_227950251 | 0.14 |
SNAP47 |
synaptosomal-associated protein, 47kDa |
14411 |
0.16 |
| chr14_91710752_91710947 | 0.14 |
GPR68 |
G protein-coupled receptor 68 |
3 |
0.97 |
| chr5_137802209_137802360 | 0.14 |
EGR1 |
early growth response 1 |
1105 |
0.47 |
| chr9_75142469_75142962 | 0.14 |
ENSG00000238402 |
. |
563 |
0.81 |
| chr3_124553965_124554410 | 0.14 |
ITGB5 |
integrin, beta 5 |
6211 |
0.23 |
| chrX_75648113_75648326 | 0.14 |
MAGEE1 |
melanoma antigen family E, 1 |
173 |
0.98 |
| chr19_10397706_10398023 | 0.14 |
ICAM4 |
intercellular adhesion molecule 4 (Landsteiner-Wiener blood group) |
184 |
0.83 |
| chr19_3338800_3339014 | 0.14 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
20654 |
0.16 |
| chr10_104941185_104941336 | 0.14 |
NT5C2 |
5'-nucleotidase, cytosolic II |
6521 |
0.23 |
| chr17_73716953_73717205 | 0.14 |
ITGB4 |
integrin, beta 4 |
329 |
0.8 |
| chr6_31540117_31540270 | 0.14 |
LTA |
lymphotoxin alpha |
133 |
0.85 |
| chr19_50063668_50063821 | 0.14 |
NOSIP |
nitric oxide synthase interacting protein |
143 |
0.88 |
| chr2_120934672_120935030 | 0.14 |
ENSG00000202046 |
. |
14970 |
0.19 |
| chr9_102058986_102059155 | 0.14 |
ENSG00000222337 |
. |
12766 |
0.24 |
| chr4_26270650_26270801 | 0.14 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
3504 |
0.38 |
| chr7_55636821_55636978 | 0.14 |
VOPP1 |
vesicular, overexpressed in cancer, prosurvival protein 1 |
2662 |
0.29 |
| chr20_57739224_57739506 | 0.14 |
ZNF831 |
zinc finger protein 831 |
26710 |
0.21 |
| chr6_32605191_32605375 | 0.14 |
HLA-DQA1 |
major histocompatibility complex, class II, DQ alpha 1 |
86 |
0.96 |
| chr22_20194565_20194781 | 0.14 |
XXbac-B444P24.8 |
|
2232 |
0.21 |
| chr9_127633043_127633194 | 0.14 |
ARPC5L |
actin related protein 2/3 complex, subunit 5-like |
1634 |
0.27 |
| chr1_228676613_228676764 | 0.14 |
RNF187 |
ring finger protein 187 |
1926 |
0.16 |
| chr5_139494643_139494794 | 0.14 |
PURA |
purine-rich element binding protein A |
1010 |
0.46 |
| chr16_11145985_11146136 | 0.13 |
RP11-66H6.3 |
|
18899 |
0.19 |
| chr4_165304775_165304980 | 0.13 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
264 |
0.96 |
| chr10_24912096_24912247 | 0.13 |
ARHGAP21 |
Rho GTPase activating protein 21 |
408 |
0.88 |
| chr11_67234655_67234806 | 0.13 |
TMEM134 |
transmembrane protein 134 |
1974 |
0.13 |
| chr2_105478176_105478327 | 0.13 |
AC018730.4 |
|
5703 |
0.14 |
| chr6_47002361_47002718 | 0.13 |
GPR110 |
G protein-coupled receptor 110 |
7491 |
0.28 |
| chr5_146257138_146257401 | 0.13 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
936 |
0.67 |
| chr9_108319921_108320392 | 0.13 |
RP11-235C23.5 |
|
154 |
0.64 |
| chr16_74640764_74640944 | 0.13 |
GLG1 |
golgi glycoprotein 1 |
138 |
0.97 |
| chr17_53341326_53341539 | 0.13 |
HLF |
hepatic leukemia factor |
941 |
0.65 |
| chr17_3598807_3599359 | 0.13 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
244 |
0.86 |
| chr4_90228457_90229530 | 0.13 |
GPRIN3 |
GPRIN family member 3 |
168 |
0.97 |
| chr2_136577729_136578391 | 0.13 |
AC011893.3 |
|
299 |
0.89 |
| chr9_95826666_95826817 | 0.13 |
SUSD3 |
sushi domain containing 3 |
4641 |
0.2 |
| chr5_137801416_137801666 | 0.13 |
EGR1 |
early growth response 1 |
362 |
0.85 |
| chr10_96996474_96996625 | 0.13 |
RP11-310E22.4 |
|
5648 |
0.25 |
| chr7_110724205_110724356 | 0.13 |
LRRN3 |
leucine rich repeat neuronal 3 |
6782 |
0.27 |
| chr7_26192093_26192429 | 0.13 |
NFE2L3 |
nuclear factor, erythroid 2-like 3 |
401 |
0.85 |
| chr17_28705982_28706205 | 0.13 |
CPD |
carboxypeptidase D |
170 |
0.94 |
| chr2_10522774_10523051 | 0.13 |
HPCAL1 |
hippocalcin-like 1 |
37235 |
0.15 |
| chr2_166650914_166651206 | 0.13 |
GALNT3 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
100 |
0.98 |
| chr22_42177525_42177912 | 0.13 |
MEI1 |
meiosis inhibitor 1 |
21 |
0.96 |
| chr1_22383056_22383282 | 0.13 |
CDC42-IT1 |
CDC42 intronic transcript 1 (non-protein coding) |
2521 |
0.18 |
| chr2_25473653_25473920 | 0.13 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
1394 |
0.47 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0072110 | glomerular mesangial cell proliferation(GO:0072110) regulation of glomerular mesangial cell proliferation(GO:0072124) |
| 0.1 | 0.2 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.1 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.1 | GO:1901722 | regulation of cell proliferation involved in kidney development(GO:1901722) |
| 0.1 | 0.3 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.2 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0010592 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.2 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.2 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.1 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.0 | GO:0090192 | regulation of glomerulus development(GO:0090192) |
| 0.0 | 0.2 | GO:0051001 | negative regulation of nitric-oxide synthase activity(GO:0051001) |
| 0.0 | 0.1 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0003170 | heart valve development(GO:0003170) heart valve morphogenesis(GO:0003179) |
| 0.0 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 0.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.1 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0060219 | camera-type eye photoreceptor cell differentiation(GO:0060219) |
| 0.0 | 0.1 | GO:2000188 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.0 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.0 | GO:0090281 | negative regulation of calcium ion import(GO:0090281) |
| 0.0 | 0.2 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0033145 | positive regulation of intracellular steroid hormone receptor signaling pathway(GO:0033145) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0051988 | regulation of attachment of spindle microtubules to kinetochore(GO:0051988) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0002327 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0016093 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.1 | GO:0060897 | neural plate anterior/posterior regionalization(GO:0021999) neural plate regionalization(GO:0060897) |
| 0.0 | 0.2 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0032462 | regulation of protein homooligomerization(GO:0032462) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.0 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.1 | GO:0034629 | cellular protein complex localization(GO:0034629) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0002902 | regulation of B cell apoptotic process(GO:0002902) |
| 0.0 | 0.0 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0008298 | intracellular mRNA localization(GO:0008298) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:1903301 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) regulation of hexokinase activity(GO:1903299) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.1 | GO:0006553 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.0 | GO:0040009 | regulation of growth rate(GO:0040009) |
| 0.0 | 0.2 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.0 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.0 | GO:0006565 | L-serine catabolic process(GO:0006565) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.2 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.4 | GO:0006270 | DNA replication initiation(GO:0006270) |
| 0.0 | 0.1 | GO:0018126 | protein hydroxylation(GO:0018126) |
| 0.0 | 0.1 | GO:0045061 | thymic T cell selection(GO:0045061) |
| 0.0 | 0.0 | GO:0051608 | histamine transport(GO:0051608) |
| 0.0 | 0.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0032366 | intracellular sterol transport(GO:0032366) intracellular cholesterol transport(GO:0032367) |
| 0.0 | 0.0 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) auditory receptor cell fate determination(GO:0042668) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.0 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.0 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.1 | GO:0000731 | DNA synthesis involved in DNA repair(GO:0000731) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0008291 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.0 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.1 | GO:0045879 | negative regulation of smoothened signaling pathway(GO:0045879) |
| 0.0 | 0.0 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0035246 | peptidyl-arginine N-methylation(GO:0035246) |
| 0.0 | 0.0 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.1 | GO:0033235 | positive regulation of protein sumoylation(GO:0033235) |
| 0.0 | 0.0 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0016558 | protein import into peroxisome matrix(GO:0016558) |
| 0.0 | 0.1 | GO:0007020 | microtubule nucleation(GO:0007020) |
| 0.0 | 0.0 | GO:0021702 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) cerebellar Purkinje cell layer formation(GO:0021694) cerebellar Purkinje cell differentiation(GO:0021702) |
| 0.0 | 0.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0000722 | telomere maintenance via recombination(GO:0000722) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.0 | GO:0061316 | negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cardioblast cell fate specification(GO:0009997) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) canonical Wnt signaling pathway involved in heart development(GO:0061316) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.0 | 0.1 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.1 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.0 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.1 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.0 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.2 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.3 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.2 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.2 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.2 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.0 | 0.1 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0097506 | uracil DNA N-glycosylase activity(GO:0004844) deaminated base DNA N-glycosylase activity(GO:0097506) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.1 | GO:0051393 | alpha-actinin binding(GO:0051393) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.0 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.7 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.3 | GO:0004129 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.0 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.0 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.0 | GO:0004748 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.9 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.0 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.3 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH2 | Genes involved in Signaling by NOTCH2 |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.1 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.0 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |