Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
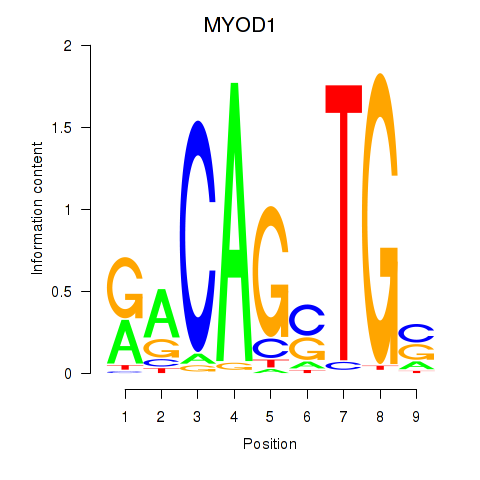
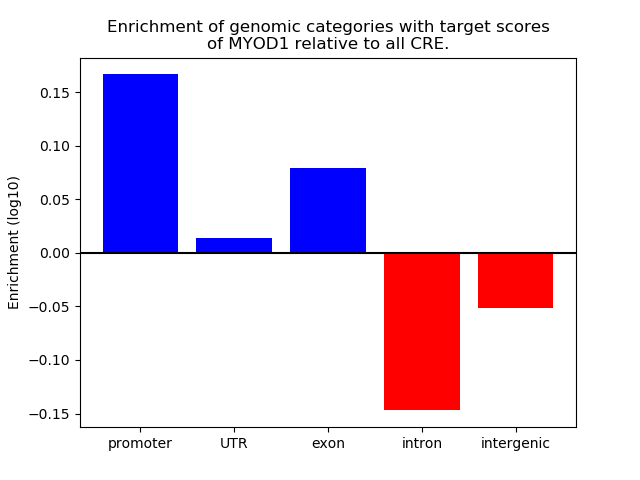
Results for MYOD1
Z-value: 0.28
Transcription factors associated with MYOD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MYOD1
|
ENSG00000129152.3 | myogenic differentiation 1 |
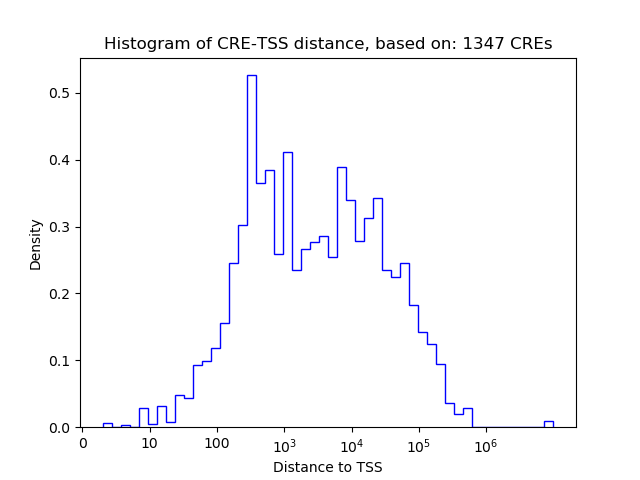
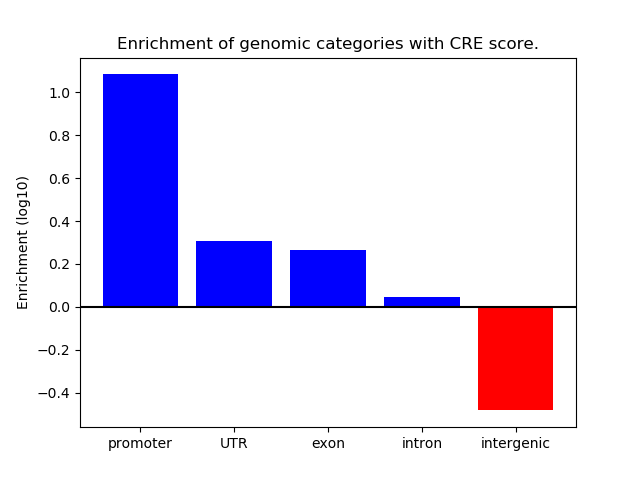
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_17741506_17741657 | MYOD1 | 466 | 0.817085 | 0.66 | 5.4e-02 | Click! |
| chr11_17742116_17742267 | MYOD1 | 1076 | 0.521110 | 0.34 | 3.7e-01 | Click! |
Activity of the MYOD1 motif across conditions
Conditions sorted by the z-value of the MYOD1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
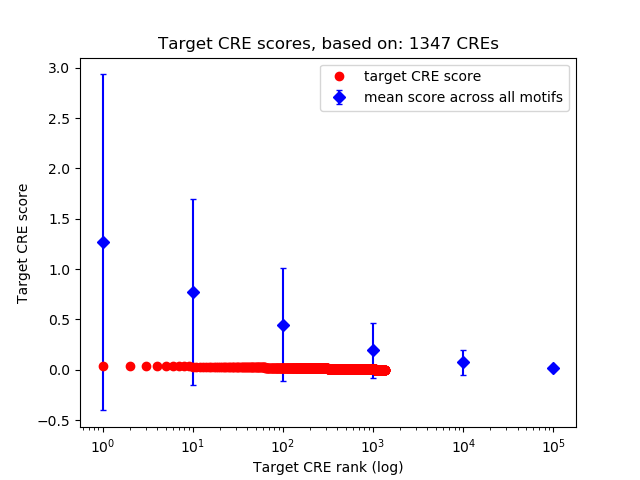
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_48630851_48631002 | 0.04 |
COL7A1 |
collagen, type VII, alpha 1 |
1667 |
0.2 |
| chr16_56945671_56945822 | 0.04 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
20214 |
0.11 |
| chr5_166722273_166722538 | 0.04 |
CTB-180C19.1 |
|
1127 |
0.47 |
| chr22_27041102_27041357 | 0.04 |
CRYBA4 |
crystallin, beta A4 |
23301 |
0.17 |
| chr11_23250751_23251044 | 0.04 |
ENSG00000264478 |
. |
242362 |
0.02 |
| chr10_104420612_104420763 | 0.03 |
TRIM8 |
tripartite motif containing 8 |
16043 |
0.16 |
| chr3_14197843_14197994 | 0.03 |
AC093495.4 |
|
11428 |
0.15 |
| chr10_3895167_3895318 | 0.03 |
KLF6 |
Kruppel-like factor 6 |
67769 |
0.12 |
| chr15_40632587_40633009 | 0.03 |
C15orf52 |
chromosome 15 open reading frame 52 |
370 |
0.7 |
| chr3_129512823_129512974 | 0.03 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
361 |
0.87 |
| chr16_88316429_88316580 | 0.03 |
ZNF469 |
zinc finger protein 469 |
177375 |
0.03 |
| chr17_39779390_39779541 | 0.03 |
KRT17 |
keratin 17 |
55 |
0.95 |
| chr17_45929276_45929427 | 0.03 |
SP6 |
Sp6 transcription factor |
792 |
0.47 |
| chr1_20707585_20707736 | 0.03 |
RP4-745E8.2 |
|
57980 |
0.11 |
| chr1_27709656_27709807 | 0.03 |
CD164L2 |
CD164 sialomucin-like 2 |
62 |
0.96 |
| chr3_66023514_66023733 | 0.03 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
360 |
0.9 |
| chr1_224697591_224697742 | 0.03 |
WDR26 |
WD repeat domain 26 |
72931 |
0.09 |
| chr9_94633472_94633623 | 0.03 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
77615 |
0.11 |
| chr12_125105824_125106359 | 0.03 |
NCOR2 |
nuclear receptor corepressor 2 |
54081 |
0.17 |
| chr5_150021079_150021262 | 0.03 |
SYNPO |
synaptopodin |
930 |
0.5 |
| chr14_105536231_105536382 | 0.03 |
GPR132 |
G protein-coupled receptor 132 |
4524 |
0.22 |
| chr17_81031335_81031486 | 0.03 |
METRNL |
meteorin, glial cell differentiation regulator-like |
6157 |
0.23 |
| chr13_22257522_22257673 | 0.03 |
FGF9 |
fibroblast growth factor 9 |
12075 |
0.25 |
| chr9_127622833_127623140 | 0.03 |
RPL35 |
ribosomal protein L35 |
1208 |
0.3 |
| chr11_84028149_84028546 | 0.03 |
DLG2 |
discs, large homolog 2 (Drosophila) |
35 |
0.99 |
| chr15_33300884_33301035 | 0.03 |
ENSG00000212415 |
. |
38031 |
0.18 |
| chr12_113797502_113797653 | 0.03 |
SLC8B1 |
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
279 |
0.85 |
| chr9_117050441_117050592 | 0.03 |
ORM1 |
orosomucoid 1 |
34820 |
0.15 |
| chr9_34651822_34652525 | 0.03 |
IL11RA |
interleukin 11 receptor, alpha |
16 |
0.94 |
| chr10_120739922_120740073 | 0.03 |
RP11-498J9.2 |
|
23399 |
0.15 |
| chr17_35278104_35278255 | 0.03 |
RP11-445F12.1 |
|
15742 |
0.13 |
| chr5_131594297_131594770 | 0.03 |
PDLIM4 |
PDZ and LIM domain 4 |
1133 |
0.45 |
| chr4_144435743_144436361 | 0.03 |
SMARCA5-AS1 |
SMARCA5 antisense RNA 1 |
264 |
0.9 |
| chr3_159572270_159572495 | 0.03 |
SCHIP1 |
schwannomin interacting protein 1 |
1654 |
0.39 |
| chr11_115630920_115631071 | 0.03 |
ENSG00000239153 |
. |
133092 |
0.06 |
| chr11_6948113_6948733 | 0.03 |
ZNF215 |
zinc finger protein 215 |
670 |
0.62 |
| chr12_122235813_122236047 | 0.03 |
RHOF |
ras homolog family member F (in filopodia) |
2653 |
0.22 |
| chr11_614388_614539 | 0.03 |
IRF7 |
interferon regulatory factor 7 |
1163 |
0.26 |
| chr2_131672660_131672811 | 0.03 |
ARHGEF4 |
Rho guanine nucleotide exchange factor (GEF) 4 |
1489 |
0.42 |
| chr16_70729706_70729857 | 0.03 |
VAC14 |
Vac14 homolog (S. cerevisiae) |
285 |
0.88 |
| chr11_20048907_20049058 | 0.03 |
NAV2 |
neuron navigator 2 |
4346 |
0.24 |
| chr16_48641622_48641773 | 0.03 |
N4BP1 |
NEDD4 binding protein 1 |
2423 |
0.29 |
| chr10_99331899_99332050 | 0.03 |
ANKRD2 |
ankyrin repeat domain 2 (stretch responsive muscle) |
224 |
0.89 |
| chrX_38729814_38730081 | 0.03 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
66811 |
0.12 |
| chr17_73717686_73717837 | 0.03 |
ITGB4 |
integrin, beta 4 |
184 |
0.9 |
| chr9_130908187_130908338 | 0.02 |
LCN2 |
lipocalin 2 |
3088 |
0.12 |
| chr6_155568937_155569088 | 0.02 |
RP11-477D19.2 |
|
8846 |
0.2 |
| chr12_6484544_6484695 | 0.02 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
24 |
0.57 |
| chr11_46696724_46696875 | 0.02 |
ATG13 |
autophagy related 13 |
11249 |
0.13 |
| chr5_171380881_171381289 | 0.02 |
FBXW11 |
F-box and WD repeat domain containing 11 |
23675 |
0.23 |
| chr9_139574265_139574564 | 0.02 |
AGPAT2 |
1-acylglycerol-3-phosphate O-acyltransferase 2 |
7434 |
0.08 |
| chr15_93344718_93345000 | 0.02 |
CTD-2313J17.1 |
|
6938 |
0.18 |
| chr14_105813436_105813587 | 0.02 |
PACS2 |
phosphofurin acidic cluster sorting protein 2 |
10749 |
0.15 |
| chr16_1153076_1153227 | 0.02 |
C1QTNF8 |
C1q and tumor necrosis factor related protein 8 |
6907 |
0.13 |
| chr12_6420537_6420688 | 0.02 |
PLEKHG6 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
513 |
0.73 |
| chr5_139721787_139722024 | 0.02 |
HBEGF |
heparin-binding EGF-like growth factor |
4283 |
0.14 |
| chr6_154781868_154782019 | 0.02 |
CNKSR3 |
CNKSR family member 3 |
17682 |
0.28 |
| chr2_127822596_127822803 | 0.02 |
BIN1 |
bridging integrator 1 |
41878 |
0.16 |
| chr5_106904194_106904345 | 0.02 |
EFNA5 |
ephrin-A5 |
102059 |
0.08 |
| chr17_55131735_55131912 | 0.02 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
30630 |
0.13 |
| chr4_90768990_90769141 | 0.02 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
9599 |
0.21 |
| chr18_20046308_20046621 | 0.02 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
48586 |
0.17 |
| chr14_70346080_70346697 | 0.02 |
SMOC1 |
SPARC related modular calcium binding 1 |
245 |
0.95 |
| chr10_75346557_75346885 | 0.02 |
USP54 |
ubiquitin specific peptidase 54 |
4375 |
0.16 |
| chr12_110723019_110723319 | 0.02 |
ATP2A2 |
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
3683 |
0.28 |
| chr16_85075183_85075477 | 0.02 |
KIAA0513 |
KIAA0513 |
13920 |
0.21 |
| chr17_46908918_46909142 | 0.02 |
CALCOCO2 |
calcium binding and coiled-coil domain 2 |
440 |
0.75 |
| chr4_95987148_95987299 | 0.02 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
14393 |
0.32 |
| chr3_112964540_112964768 | 0.02 |
BOC |
BOC cell adhesion associated, oncogene regulated |
29456 |
0.19 |
| chr1_43151193_43151344 | 0.02 |
YBX1 |
Y box binding protein 1 |
2610 |
0.24 |
| chr18_8799798_8800187 | 0.02 |
RP11-674N23.4 |
|
1662 |
0.4 |
| chr11_64012846_64013110 | 0.02 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
310 |
0.51 |
| chr12_49883832_49884081 | 0.02 |
KCNH3 |
potassium voltage-gated channel, subfamily H (eag-related), member 3 |
48984 |
0.1 |
| chr8_103770507_103770658 | 0.02 |
ENSG00000266799 |
. |
24635 |
0.2 |
| chr2_221145028_221145179 | 0.02 |
ENSG00000221199 |
. |
88504 |
0.1 |
| chr5_92585448_92586206 | 0.02 |
ENSG00000237187 |
. |
172292 |
0.04 |
| chr2_220115453_220115604 | 0.02 |
TUBA4A |
tubulin, alpha 4a |
2187 |
0.12 |
| chr19_19052300_19052451 | 0.02 |
HOMER3 |
homer homolog 3 (Drosophila) |
305 |
0.76 |
| chr20_2795159_2795349 | 0.02 |
TMEM239 |
CDNA FLJ26142 fis, clone TST04526; Transmembrane protein 239; Uncharacterized protein |
360 |
0.47 |
| chr11_1567498_1567649 | 0.02 |
DUSP8 |
dual specificity phosphatase 8 |
19593 |
0.12 |
| chr1_209393485_209393990 | 0.02 |
ENSG00000230937 |
. |
211741 |
0.02 |
| chr1_22463869_22464020 | 0.02 |
WNT4 |
wingless-type MMTV integration site family, member 4 |
5515 |
0.23 |
| chr16_3057908_3058074 | 0.02 |
LA16c-380H5.2 |
|
2889 |
0.08 |
| chr6_41673906_41674057 | 0.02 |
TFEB |
transcription factor EB |
303 |
0.85 |
| chr3_62247222_62247373 | 0.02 |
ENSG00000241472 |
. |
1118 |
0.62 |
| chr11_46708337_46708488 | 0.02 |
ARHGAP1 |
Rho GTPase activating protein 1 |
9237 |
0.13 |
| chr10_126429198_126429868 | 0.02 |
FAM53B |
family with sequence similarity 53, member B |
2006 |
0.29 |
| chr5_154209933_154210084 | 0.02 |
ENSG00000263361 |
. |
990 |
0.5 |
| chr8_55048378_55048680 | 0.02 |
MRPL15 |
mitochondrial ribosomal protein L15 |
692 |
0.71 |
| chr10_11873191_11873480 | 0.02 |
PROSER2 |
proline and serine-rich protein 2 |
347 |
0.9 |
| chr2_43242041_43242199 | 0.02 |
ENSG00000207087 |
. |
76512 |
0.11 |
| chr13_109924709_109925156 | 0.02 |
MYO16-AS1 |
MYO16 antisense RNA 1 |
71101 |
0.13 |
| chr17_38178315_38178466 | 0.02 |
MED24 |
mediator complex subunit 24 |
1023 |
0.38 |
| chr9_130180174_130180325 | 0.02 |
ZNF79 |
zinc finger protein 79 |
6412 |
0.14 |
| chr7_127804563_127804714 | 0.02 |
ENSG00000207705 |
. |
43286 |
0.14 |
| chr1_197885819_197885970 | 0.02 |
LHX9 |
LIM homeobox 9 |
595 |
0.8 |
| chr1_95005094_95005314 | 0.02 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
1989 |
0.46 |
| chr1_46654018_46654169 | 0.02 |
POMGNT1 |
protein O-linked mannose N-acetylglucosaminyltransferase 1 (beta 1,2-) |
9054 |
0.12 |
| chr3_50203978_50204129 | 0.02 |
ENSG00000207922 |
. |
6706 |
0.12 |
| chr15_56351412_56351563 | 0.02 |
ENSG00000239703 |
. |
29308 |
0.2 |
| chr2_51222998_51223149 | 0.02 |
NRXN1 |
neurexin 1 |
32338 |
0.18 |
| chr5_98397075_98397538 | 0.02 |
ENSG00000200351 |
. |
124855 |
0.06 |
| chr1_1240794_1241080 | 0.02 |
ACAP3 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 3 |
2332 |
0.11 |
| chr2_62702703_62702972 | 0.02 |
ENSG00000241625 |
. |
15476 |
0.23 |
| chr2_85956322_85956473 | 0.02 |
ENSG00000199687 |
. |
937 |
0.49 |
| chr14_75085047_75085411 | 0.02 |
LTBP2 |
latent transforming growth factor beta binding protein 2 |
5923 |
0.21 |
| chr1_3382586_3382737 | 0.02 |
ARHGEF16 |
Rho guanine nucleotide exchange factor (GEF) 16 |
92 |
0.97 |
| chr4_183769117_183769268 | 0.02 |
ENSG00000221186 |
. |
21811 |
0.22 |
| chr11_451343_451494 | 0.02 |
PTDSS2 |
phosphatidylserine synthase 2 |
1138 |
0.29 |
| chr18_30049396_30050442 | 0.02 |
GAREM |
GRB2 associated, regulator of MAPK1 |
476 |
0.87 |
| chr19_2079674_2079825 | 0.02 |
MOB3A |
MOB kinase activator 3A |
1031 |
0.38 |
| chr3_20106255_20106406 | 0.02 |
KAT2B |
K(lysine) acetyltransferase 2B |
24815 |
0.17 |
| chr2_218875419_218875830 | 0.02 |
TNS1 |
tensin 1 |
7906 |
0.19 |
| chr7_79081753_79081967 | 0.02 |
ENSG00000234456 |
. |
338 |
0.8 |
| chr6_119255358_119255671 | 0.02 |
MCM9 |
minichromosome maintenance complex component 9 |
127 |
0.82 |
| chr15_41062187_41062634 | 0.02 |
C15orf62 |
chromosome 15 open reading frame 62 |
251 |
0.86 |
| chr19_49654515_49654666 | 0.02 |
HRC |
histidine rich calcium binding protein |
2102 |
0.15 |
| chr4_125065453_125065672 | 0.02 |
ANKRD50 |
ankyrin repeat domain 50 |
568325 |
0.0 |
| chr20_62289694_62289895 | 0.02 |
RTEL1 |
regulator of telomere elongation helicase 1 |
81 |
0.94 |
| chr14_54948388_54948539 | 0.02 |
GMFB |
glia maturation factor, beta |
6913 |
0.21 |
| chr17_62837136_62837287 | 0.02 |
PLEKHM1P |
pleckstrin homology domain containing, family M (with RUN domain) member 1 pseudogene |
3968 |
0.14 |
| chr14_100069286_100069437 | 0.02 |
CCDC85C |
coiled-coil domain containing 85C |
1002 |
0.53 |
| chr4_146445784_146445935 | 0.02 |
SMAD1-AS1 |
SMAD1 antisense RNA 1 |
7589 |
0.21 |
| chr5_175587605_175587756 | 0.02 |
FAM153B |
family with sequence similarity 153, member B |
75771 |
0.08 |
| chr11_61049274_61049514 | 0.02 |
VWCE |
von Willebrand factor C and EGF domains |
8116 |
0.13 |
| chr7_47674871_47675022 | 0.02 |
C7orf65 |
chromosome 7 open reading frame 65 |
19896 |
0.23 |
| chr3_43256020_43256171 | 0.02 |
ENSG00000222331 |
. |
3732 |
0.28 |
| chr10_28537061_28537300 | 0.02 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
9514 |
0.27 |
| chr5_110846365_110846666 | 0.02 |
STARD4-AS1 |
STARD4 antisense RNA 1 |
1409 |
0.37 |
| chr1_201438452_201438736 | 0.02 |
PHLDA3 |
pleckstrin homology-like domain, family A, member 3 |
229 |
0.92 |
| chr16_2557182_2557815 | 0.02 |
ATP6V0C |
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
6373 |
0.06 |
| chr5_175980994_175981145 | 0.02 |
ENSG00000207420 |
. |
2636 |
0.17 |
| chr13_44887324_44887479 | 0.02 |
SERP2 |
stress-associated endoplasmic reticulum protein family member 2 |
60577 |
0.14 |
| chr14_23315995_23316146 | 0.02 |
ENSG00000212335 |
. |
5867 |
0.08 |
| chr22_30601478_30601684 | 0.02 |
RP3-438O4.4 |
|
1517 |
0.31 |
| chr10_1094442_1094708 | 0.02 |
IDI1 |
isopentenyl-diphosphate delta isomerase 1 |
238 |
0.82 |
| chr19_13992257_13992611 | 0.02 |
NANOS3 |
nanos homolog 3 (Drosophila) |
4371 |
0.1 |
| chr4_26789379_26789589 | 0.02 |
STIM2 |
stromal interaction molecule 2 |
69816 |
0.11 |
| chr15_78330943_78331094 | 0.02 |
ENSG00000221476 |
. |
145 |
0.94 |
| chr12_120667761_120668330 | 0.02 |
PXN |
paxillin |
3398 |
0.15 |
| chrX_9754895_9755291 | 0.02 |
SHROOM2 |
shroom family member 2 |
597 |
0.54 |
| chr17_56595689_56596135 | 0.02 |
MTMR4 |
myotubularin related protein 4 |
646 |
0.52 |
| chr20_19192275_19192426 | 0.02 |
SLC24A3 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 |
940 |
0.53 |
| chr1_230223344_230223516 | 0.02 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
20412 |
0.23 |
| chr19_3365786_3365951 | 0.02 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
679 |
0.7 |
| chr5_146933859_146934303 | 0.02 |
DPYSL3 |
dihydropyrimidinase-like 3 |
44462 |
0.19 |
| chr1_232622150_232622301 | 0.02 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
24062 |
0.27 |
| chr8_100796366_100796517 | 0.02 |
RP11-402L5.1 |
|
14974 |
0.26 |
| chr1_40423471_40423622 | 0.02 |
MFSD2A |
major facilitator superfamily domain containing 2A |
2724 |
0.22 |
| chr6_114084680_114084971 | 0.02 |
ENSG00000253091 |
. |
17842 |
0.21 |
| chr8_11204724_11204991 | 0.02 |
AF131216.5 |
|
154 |
0.93 |
| chr11_67478742_67478899 | 0.02 |
ENSG00000266451 |
. |
23259 |
0.12 |
| chr7_76957466_76957741 | 0.02 |
GSAP |
gamma-secretase activating protein |
2040 |
0.4 |
| chr12_46384039_46384292 | 0.02 |
SCAF11 |
SR-related CTD-associated factor 11 |
183 |
0.97 |
| chr15_39871853_39872515 | 0.02 |
THBS1 |
thrombospondin 1 |
1096 |
0.52 |
| chr2_19547772_19548046 | 0.02 |
ENSG00000266738 |
. |
281 |
0.94 |
| chr18_77616537_77616791 | 0.02 |
KCNG2 |
potassium voltage-gated channel, subfamily G, member 2 |
7004 |
0.27 |
| chr1_54778827_54778978 | 0.02 |
RP5-997D24.3 |
|
27824 |
0.16 |
| chr11_63971893_63972044 | 0.02 |
FERMT3 |
fermitin family member 3 |
2182 |
0.12 |
| chr11_124615580_124615731 | 0.02 |
RP11-677M14.2 |
|
578 |
0.58 |
| chr14_26016303_26016454 | 0.02 |
ENSG00000212270 |
. |
259540 |
0.02 |
| chr3_187926447_187926702 | 0.02 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
588 |
0.8 |
| chr19_13263975_13264220 | 0.02 |
CTC-250I14.6 |
|
429 |
0.65 |
| chr11_44614749_44614953 | 0.02 |
CD82 |
CD82 molecule |
3555 |
0.24 |
| chr1_150849308_150849506 | 0.02 |
ARNT |
aryl hydrocarbon receptor nuclear translocator |
163 |
0.93 |
| chr21_33945347_33945498 | 0.02 |
ENSG00000252950 |
. |
10037 |
0.14 |
| chr15_69919016_69919167 | 0.02 |
ENSG00000238870 |
. |
104070 |
0.07 |
| chr7_5488255_5488406 | 0.02 |
TNRC18 |
trinucleotide repeat containing 18 |
23285 |
0.11 |
| chr6_139870784_139870935 | 0.02 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
175102 |
0.03 |
| chr9_38047346_38047861 | 0.02 |
SHB |
Src homology 2 domain containing adaptor protein B |
21605 |
0.23 |
| chr17_48712666_48713008 | 0.02 |
ABCC3 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 3 |
619 |
0.6 |
| chr17_48600636_48600787 | 0.02 |
MYCBPAP |
MYCBP associated protein |
370 |
0.75 |
| chr7_98853393_98853544 | 0.02 |
KPNA7 |
karyopherin alpha 7 (importin alpha 8) |
48339 |
0.12 |
| chr5_176798986_176799137 | 0.02 |
RGS14 |
regulator of G-protein signaling 14 |
5117 |
0.1 |
| chr1_179095553_179095704 | 0.02 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
16551 |
0.19 |
| chr2_96812420_96812577 | 0.02 |
DUSP2 |
dual specificity phosphatase 2 |
1319 |
0.38 |
| chr17_76123581_76124282 | 0.02 |
TMC6 |
transmembrane channel-like 6 |
780 |
0.49 |
| chr1_16480648_16480799 | 0.02 |
RP11-276H7.2 |
|
983 |
0.38 |
| chr8_107668314_107668465 | 0.02 |
RP11-649G15.2 |
|
1387 |
0.39 |
| chr15_25102737_25102888 | 0.02 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
1114 |
0.44 |
| chr5_17898081_17898232 | 0.02 |
ENSG00000212278 |
. |
338076 |
0.01 |
| chrX_106958420_106958715 | 0.02 |
TSC22D3 |
TSC22 domain family, member 3 |
1064 |
0.59 |
| chr22_32929726_32929936 | 0.02 |
LL22NC03-104C7.1 |
|
49455 |
0.12 |
| chr13_109625213_109625364 | 0.02 |
MYO16 |
myosin XVI |
86771 |
0.1 |
| chr16_84210540_84210779 | 0.02 |
DNAAF1 |
dynein, axonemal, assembly factor 1 |
912 |
0.43 |
| chr12_6492920_6493170 | 0.02 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
154 |
0.92 |
| chr13_39260620_39261051 | 0.02 |
FREM2 |
FRAS1 related extracellular matrix protein 2 |
431 |
0.9 |
| chr6_82604485_82604797 | 0.02 |
ENSG00000206886 |
. |
130900 |
0.05 |
| chrX_78621891_78622920 | 0.02 |
ITM2A |
integral membrane protein 2A |
451 |
0.91 |
| chr17_66179526_66179677 | 0.02 |
LRRC37A16P |
leucine rich repeat containing 37, member A16, pseudogene |
30992 |
0.15 |
| chr11_130284349_130284500 | 0.02 |
ADAMTS8 |
ADAM metallopeptidase with thrombospondin type 1 motif, 8 |
14464 |
0.23 |
| chr20_34652778_34652969 | 0.02 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
26553 |
0.16 |
| chr1_212838734_212839243 | 0.02 |
ENSG00000207491 |
. |
26618 |
0.12 |
| chr2_232537261_232537412 | 0.02 |
ENSG00000239202 |
. |
26352 |
0.14 |
| chr1_15500593_15500744 | 0.02 |
C1orf195 |
chromosome 1 open reading frame 195 |
2855 |
0.29 |
| chr5_2659090_2659241 | 0.02 |
IRX2 |
iroquois homeobox 2 |
92604 |
0.09 |
| chr8_23145924_23146455 | 0.02 |
R3HCC1 |
R3H domain and coiled-coil containing 1 |
254 |
0.9 |
| chr18_12946769_12946940 | 0.02 |
SEH1L |
SEH1-like (S. cerevisiae) |
278 |
0.91 |
| chr7_149468529_149468760 | 0.02 |
ZNF467 |
zinc finger protein 467 |
1664 |
0.4 |
| chr2_241377476_241377740 | 0.02 |
GPC1 |
glypican 1 |
2520 |
0.24 |
Network of associatons between targets according to the STRING database.