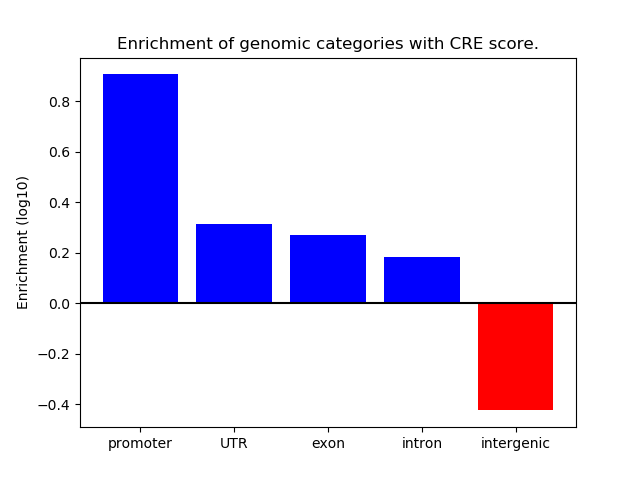
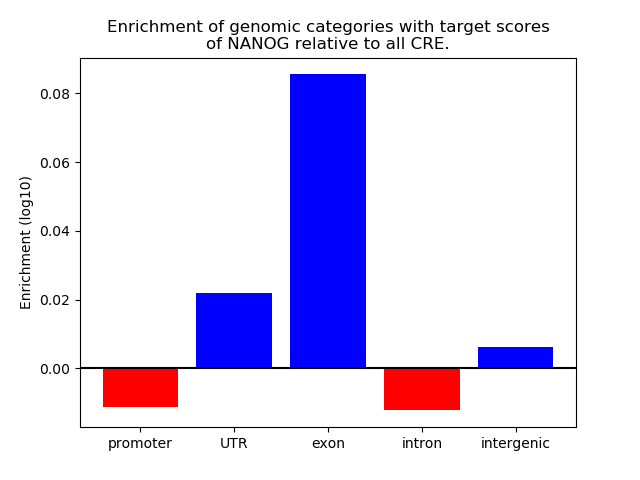
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
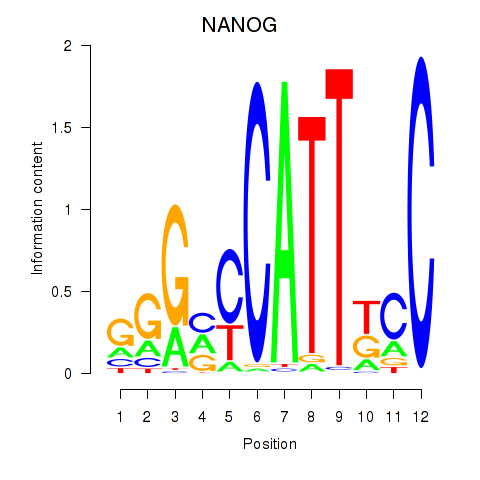
Results for NANOG
Z-value: 1.23
Transcription factors associated with NANOG
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NANOG
|
ENSG00000111704.6 | Nanog homeobox |
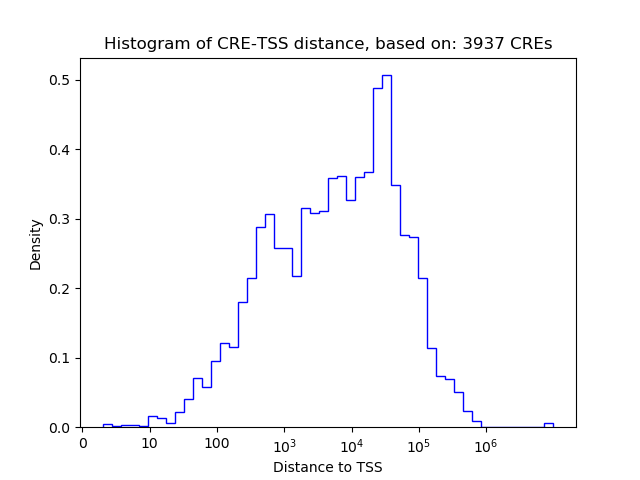
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_7945798_7945949 | NANOG | 3662 | 0.136175 | 0.80 | 1.0e-02 | Click! |
Activity of the NANOG motif across conditions
Conditions sorted by the z-value of the NANOG motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
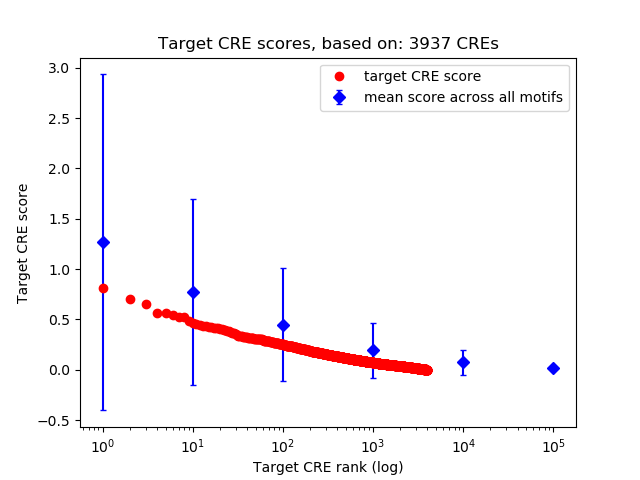
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_114180362_114180910 | 0.82 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
2095 |
0.32 |
| chr16_2174847_2175247 | 0.70 |
RP11-304L19.2 |
|
5839 |
0.06 |
| chr7_27212262_27212557 | 0.66 |
HOXA10 |
homeobox A10 |
1516 |
0.14 |
| chr19_6076963_6077580 | 0.57 |
CTC-232P5.3 |
|
9307 |
0.15 |
| chr8_104153660_104153959 | 0.57 |
C8orf56 |
chromosome 8 open reading frame 56 |
106 |
0.89 |
| chr10_49812568_49813319 | 0.55 |
ARHGAP22 |
Rho GTPase activating protein 22 |
54 |
0.98 |
| chr17_55980324_55980725 | 0.53 |
CUEDC1 |
CUE domain containing 1 |
226 |
0.93 |
| chr2_60779607_60780575 | 0.52 |
BCL11A |
B-cell CLL/lymphoma 11A (zinc finger protein) |
449 |
0.87 |
| chr19_33784339_33784950 | 0.49 |
CTD-2540B15.11 |
|
6196 |
0.14 |
| chr4_16900577_16901013 | 0.46 |
LDB2 |
LIM domain binding 2 |
363 |
0.93 |
| chr11_121800761_121801203 | 0.45 |
ENSG00000252556 |
. |
74081 |
0.11 |
| chr19_46916092_46916684 | 0.44 |
CCDC8 |
coiled-coil domain containing 8 |
453 |
0.75 |
| chr8_18871547_18872047 | 0.43 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
601 |
0.78 |
| chr8_142141679_142142152 | 0.43 |
RP11-809O17.1 |
|
1855 |
0.33 |
| chr15_40626168_40626616 | 0.43 |
ENSG00000252714 |
. |
2543 |
0.12 |
| chr7_128337244_128337509 | 0.43 |
ENSG00000201041 |
. |
117 |
0.79 |
| chr7_30199428_30199624 | 0.42 |
AC007036.5 |
|
2379 |
0.26 |
| chr2_142888867_142889088 | 0.41 |
LRP1B |
low density lipoprotein receptor-related protein 1B |
293 |
0.93 |
| chr7_100776904_100777303 | 0.41 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
6724 |
0.11 |
| chr1_209381259_209381734 | 0.41 |
ENSG00000230937 |
. |
223982 |
0.02 |
| chr2_225775342_225775493 | 0.40 |
DOCK10 |
dedicator of cytokinesis 10 |
36365 |
0.23 |
| chr8_62624685_62625179 | 0.40 |
ASPH |
aspartate beta-hydroxylase |
2152 |
0.32 |
| chr11_120083079_120083522 | 0.39 |
OAF |
OAF homolog (Drosophila) |
1291 |
0.42 |
| chr5_77943226_77943942 | 0.38 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
1064 |
0.69 |
| chr4_16899723_16900066 | 0.38 |
LDB2 |
LIM domain binding 2 |
291 |
0.95 |
| chr11_92499156_92499668 | 0.38 |
RP11-203F8.1 |
|
621 |
0.79 |
| chr4_159092030_159092251 | 0.36 |
RP11-597D13.9 |
|
236 |
0.78 |
| chr4_30718989_30719310 | 0.36 |
PCDH7 |
protocadherin 7 |
2888 |
0.4 |
| chr3_185463378_185463630 | 0.36 |
ENSG00000265470 |
. |
22188 |
0.19 |
| chr5_108992370_108992546 | 0.36 |
ENSG00000266090 |
. |
28823 |
0.18 |
| chr20_47897294_47897664 | 0.35 |
ENSG00000212304 |
. |
259 |
0.71 |
| chr10_49893410_49893567 | 0.34 |
WDFY4 |
WDFY family member 4 |
567 |
0.8 |
| chr3_13040834_13041042 | 0.34 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
12402 |
0.27 |
| chr14_23504597_23504748 | 0.33 |
PSMB5 |
proteasome (prosome, macropain) subunit, beta type, 5 |
233 |
0.81 |
| chr9_128170108_128170336 | 0.33 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
76558 |
0.09 |
| chr21_43882491_43882642 | 0.33 |
SLC37A1 |
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
33562 |
0.1 |
| chr5_158444877_158445028 | 0.33 |
CTD-2363C16.2 |
|
32138 |
0.18 |
| chr11_118079199_118079350 | 0.32 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
4326 |
0.17 |
| chr11_69258827_69259069 | 0.32 |
CCND1 |
cyclin D1 |
196907 |
0.02 |
| chr3_29554516_29555025 | 0.32 |
RBMS3-AS2 |
RBMS3 antisense RNA 2 |
103776 |
0.08 |
| chr10_60273912_60274168 | 0.32 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
1140 |
0.65 |
| chr5_149534173_149534381 | 0.32 |
PDGFRB |
platelet-derived growth factor receptor, beta polypeptide |
913 |
0.52 |
| chr7_67384045_67384316 | 0.32 |
ENSG00000265600 |
. |
562513 |
0.0 |
| chr9_111578370_111578673 | 0.32 |
ACTL7B |
actin-like 7B |
40718 |
0.16 |
| chr20_47340203_47340354 | 0.31 |
ENSG00000251876 |
. |
15707 |
0.28 |
| chr4_144280055_144280346 | 0.31 |
ENSG00000265623 |
. |
15587 |
0.2 |
| chr12_123215443_123215626 | 0.31 |
HCAR1 |
hydroxycarboxylic acid receptor 1 |
144 |
0.94 |
| chr1_121271248_121271399 | 0.31 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
335386 |
0.01 |
| chr7_65746937_65747265 | 0.31 |
TPST1 |
tyrosylprotein sulfotransferase 1 |
60510 |
0.11 |
| chr16_4993031_4993210 | 0.31 |
PPL |
periplakin |
5984 |
0.15 |
| chr22_24862876_24863027 | 0.31 |
UPB1 |
ureidopropionase, beta |
255 |
0.91 |
| chr15_68564352_68564564 | 0.31 |
FEM1B |
fem-1 homolog b (C. elegans) |
5683 |
0.19 |
| chr13_103478539_103479046 | 0.31 |
ENSG00000222301 |
. |
6449 |
0.13 |
| chr1_86036403_86036586 | 0.31 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
7439 |
0.2 |
| chr4_81187181_81187534 | 0.31 |
FGF5 |
fibroblast growth factor 5 |
396 |
0.89 |
| chr12_121136486_121136637 | 0.30 |
RP11-173P15.3 |
|
743 |
0.49 |
| chr7_23507689_23508280 | 0.30 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
2102 |
0.28 |
| chr4_173708323_173708474 | 0.30 |
ENSG00000241652 |
. |
354396 |
0.01 |
| chr12_32740907_32741058 | 0.30 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
10478 |
0.26 |
| chr9_124725889_124726074 | 0.30 |
TTLL11 |
tubulin tyrosine ligase-like family, member 11 |
129904 |
0.04 |
| chr7_100464814_100466140 | 0.29 |
TRIP6 |
thyroid hormone receptor interactor 6 |
717 |
0.47 |
| chr14_24783731_24783882 | 0.29 |
LTB4R |
leukotriene B4 receptor |
100 |
0.9 |
| chr19_7159886_7160037 | 0.29 |
ENSG00000266483 |
. |
545 |
0.75 |
| chr3_134369662_134369917 | 0.29 |
KY |
kyphoscoliosis peptidase |
71 |
0.98 |
| chr5_71768722_71768873 | 0.29 |
RP11-389C8.2 |
|
30583 |
0.19 |
| chr18_59001001_59001554 | 0.29 |
CDH20 |
cadherin 20, type 2 |
229 |
0.96 |
| chr11_61594474_61594781 | 0.29 |
FADS2 |
fatty acid desaturase 2 |
237 |
0.86 |
| chr8_116463616_116464117 | 0.29 |
TRPS1 |
trichorhinophalangeal syndrome I |
40582 |
0.2 |
| chr14_64664030_64664199 | 0.28 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
4797 |
0.28 |
| chr14_91797432_91797583 | 0.28 |
ENSG00000265856 |
. |
2550 |
0.31 |
| chr3_138581962_138582285 | 0.28 |
PIK3CB |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit beta |
28343 |
0.2 |
| chr6_7202628_7202802 | 0.28 |
ENSG00000201483 |
. |
14796 |
0.19 |
| chr10_13746137_13746452 | 0.28 |
ENSG00000222235 |
. |
684 |
0.58 |
| chr22_45630973_45631282 | 0.28 |
KIAA0930 |
KIAA0930 |
1031 |
0.52 |
| chr8_132828485_132828822 | 0.28 |
EFR3A |
EFR3 homolog A (S. cerevisiae) |
87682 |
0.1 |
| chr14_101292536_101293492 | 0.28 |
AL117190.2 |
|
2523 |
0.08 |
| chr12_10902635_10902853 | 0.27 |
YBX3 |
Y box binding protein 3 |
26833 |
0.13 |
| chr9_13278250_13278433 | 0.27 |
RP11-272P10.2 |
|
845 |
0.51 |
| chr9_94065254_94065575 | 0.27 |
AUH |
AU RNA binding protein/enoyl-CoA hydratase |
58763 |
0.16 |
| chr7_70201396_70202343 | 0.27 |
AUTS2 |
autism susceptibility candidate 2 |
7744 |
0.34 |
| chr15_89686892_89687043 | 0.27 |
ENSG00000239151 |
. |
22957 |
0.16 |
| chr17_21279729_21280949 | 0.27 |
KCNJ12 |
potassium inwardly-rectifying channel, subfamily J, member 12 |
830 |
0.69 |
| chr12_132313238_132313523 | 0.27 |
MMP17 |
matrix metallopeptidase 17 (membrane-inserted) |
295 |
0.89 |
| chr5_83678519_83678875 | 0.26 |
EDIL3 |
EGF-like repeats and discoidin I-like domains 3 |
1507 |
0.37 |
| chr11_1777334_1777604 | 0.26 |
CTSD |
cathepsin D |
1293 |
0.24 |
| chr1_21649315_21649466 | 0.26 |
ECE1 |
endothelin converting enzyme 1 |
22607 |
0.18 |
| chr3_114405799_114405950 | 0.26 |
ENSG00000264623 |
. |
56498 |
0.15 |
| chr15_94830020_94830171 | 0.26 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
11335 |
0.32 |
| chr19_55684291_55685412 | 0.26 |
SYT5 |
synaptotagmin V |
1083 |
0.28 |
| chr18_77551387_77551694 | 0.26 |
KCNG2 |
potassium voltage-gated channel, subfamily G, member 2 |
72128 |
0.1 |
| chr16_54969072_54969449 | 0.26 |
IRX5 |
iroquois homeobox 5 |
3276 |
0.4 |
| chr9_95297440_95297591 | 0.26 |
ECM2 |
extracellular matrix protein 2, female organ and adipocyte specific |
739 |
0.65 |
| chr2_180726238_180726902 | 0.26 |
ZNF385B |
zinc finger protein 385B |
338 |
0.8 |
| chr1_44513542_44514157 | 0.26 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
16715 |
0.14 |
| chr8_29685440_29685777 | 0.26 |
ENSG00000221003 |
. |
100513 |
0.07 |
| chr1_8918227_8918467 | 0.26 |
ENO1-IT1 |
ENO1 intronic transcript 1 (non-protein coding) |
19719 |
0.12 |
| chr2_36585229_36585663 | 0.25 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
1832 |
0.5 |
| chr5_95562456_95562808 | 0.25 |
ENSG00000206997 |
. |
16701 |
0.27 |
| chr14_52707563_52708006 | 0.25 |
PTGDR |
prostaglandin D2 receptor (DP) |
26647 |
0.22 |
| chr21_37305025_37305176 | 0.25 |
FKSG68 |
|
34373 |
0.16 |
| chr15_90580732_90580981 | 0.25 |
ZNF710 |
zinc finger protein 710 |
30390 |
0.11 |
| chr5_33892545_33892868 | 0.25 |
ADAMTS12 |
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
409 |
0.85 |
| chr6_152085810_152086045 | 0.25 |
ESR1 |
estrogen receptor 1 |
39639 |
0.18 |
| chr7_81405901_81406052 | 0.25 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
6222 |
0.33 |
| chr1_172352648_172352805 | 0.25 |
DNM3 |
dynamin 3 |
4568 |
0.19 |
| chr18_46460407_46461027 | 0.25 |
SMAD7 |
SMAD family member 7 |
14158 |
0.24 |
| chr10_123922760_123923208 | 0.25 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
43 |
0.99 |
| chr11_111412763_111412914 | 0.24 |
LAYN |
layilin |
561 |
0.64 |
| chr17_60781005_60781559 | 0.24 |
RP11-156L14.1 |
|
2285 |
0.25 |
| chr1_31971829_31971980 | 0.24 |
ENSG00000206981 |
. |
1379 |
0.42 |
| chr7_27182401_27182640 | 0.24 |
HOXA5 |
homeobox A5 |
767 |
0.32 |
| chr11_16023597_16024065 | 0.24 |
CTD-3096P4.1 |
|
20905 |
0.28 |
| chr17_2658644_2658831 | 0.24 |
ENSG00000265566 |
. |
1611 |
0.28 |
| chr6_113198454_113198633 | 0.24 |
ENSG00000201386 |
. |
94152 |
0.09 |
| chr3_188841606_188841757 | 0.24 |
TPRG1 |
tumor protein p63 regulated 1 |
23771 |
0.27 |
| chr20_48600008_48600312 | 0.24 |
SNAI1 |
snail family zinc finger 1 |
624 |
0.68 |
| chr18_42169569_42169984 | 0.24 |
SETBP1 |
SET binding protein 1 |
90362 |
0.1 |
| chr16_85783565_85784591 | 0.23 |
C16orf74 |
chromosome 16 open reading frame 74 |
479 |
0.7 |
| chr10_16992831_16993197 | 0.23 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
33180 |
0.22 |
| chr12_117036919_117037510 | 0.23 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
23558 |
0.24 |
| chr16_3095827_3096553 | 0.23 |
MMP25 |
matrix metallopeptidase 25 |
492 |
0.51 |
| chrX_117973061_117973360 | 0.23 |
ZCCHC12 |
zinc finger, CCHC domain containing 12 |
15457 |
0.24 |
| chr19_10736478_10736985 | 0.23 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
530 |
0.65 |
| chr11_65790225_65790471 | 0.23 |
CATSPER1 |
cation channel, sperm associated 1 |
3640 |
0.11 |
| chr3_21792365_21792565 | 0.23 |
ZNF385D |
zinc finger protein 385D |
462 |
0.89 |
| chr19_51964655_51964830 | 0.23 |
SIGLEC8 |
sialic acid binding Ig-like lectin 8 |
3032 |
0.13 |
| chr11_46004459_46004616 | 0.23 |
PHF21A |
PHD finger protein 21A |
37117 |
0.13 |
| chr2_45228184_45228926 | 0.23 |
SIX2 |
SIX homeobox 2 |
8014 |
0.26 |
| chr22_43327364_43327528 | 0.23 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
14433 |
0.22 |
| chr10_30092784_30093253 | 0.23 |
SVIL |
supervillin |
68285 |
0.12 |
| chr1_56942848_56943117 | 0.23 |
ENSG00000223307 |
. |
99892 |
0.08 |
| chr6_49293859_49294324 | 0.23 |
ENSG00000252457 |
. |
18430 |
0.26 |
| chr9_118917055_118917469 | 0.23 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
1179 |
0.59 |
| chr2_86643247_86643503 | 0.23 |
KDM3A |
lysine (K)-specific demethylase 3A |
24395 |
0.16 |
| chr1_72749537_72750410 | 0.23 |
NEGR1 |
neuronal growth regulator 1 |
1556 |
0.58 |
| chr3_188139297_188139660 | 0.22 |
LPP-AS1 |
LPP antisense RNA 1 |
146976 |
0.04 |
| chr6_11044848_11045055 | 0.22 |
ELOVL2-AS1 |
ELOVL2 antisense RNA 1 |
25 |
0.87 |
| chr15_81377697_81377848 | 0.22 |
C15orf26 |
chromosome 15 open reading frame 26 |
13977 |
0.22 |
| chr2_220394806_220394967 | 0.22 |
CHPF |
chondroitin polymerizing factor |
12933 |
0.07 |
| chr4_73185764_73186042 | 0.22 |
RP11-373J21.1 |
|
2842 |
0.42 |
| chr15_71184312_71185443 | 0.22 |
LRRC49 |
leucine rich repeat containing 49 |
88 |
0.55 |
| chr11_73024866_73025155 | 0.22 |
ARHGEF17 |
Rho guanine nucleotide exchange factor (GEF) 17 |
2400 |
0.21 |
| chr5_178980827_178981020 | 0.22 |
RUFY1 |
RUN and FYVE domain containing 1 |
3084 |
0.24 |
| chr8_128771730_128771923 | 0.22 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
23349 |
0.22 |
| chr1_114311936_114312108 | 0.22 |
PHTF1 |
putative homeodomain transcription factor 1 |
9924 |
0.19 |
| chr5_42719011_42719269 | 0.22 |
CCDC152 |
coiled-coil domain containing 152 |
37763 |
0.19 |
| chr19_11433197_11433348 | 0.22 |
CTC-510F12.6 |
|
440 |
0.67 |
| chr9_124048192_124048723 | 0.22 |
GSN |
gelsolin |
413 |
0.59 |
| chr10_134349773_134350081 | 0.21 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
1397 |
0.46 |
| chr6_14671214_14671445 | 0.21 |
ENSG00000206960 |
. |
24563 |
0.28 |
| chr5_83017716_83018280 | 0.21 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
566 |
0.78 |
| chr10_102278895_102279852 | 0.21 |
SEC31B |
SEC31 homolog B (S. cerevisiae) |
218 |
0.92 |
| chr10_17074110_17074261 | 0.21 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
47991 |
0.17 |
| chr11_109966416_109966567 | 0.21 |
ZC3H12C |
zinc finger CCCH-type containing 12C |
2404 |
0.45 |
| chr8_145925356_145925968 | 0.21 |
AF186192.5 |
|
5257 |
0.15 |
| chr12_79916269_79916420 | 0.21 |
ENSG00000243714 |
. |
2606 |
0.3 |
| chr21_39862457_39862608 | 0.21 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
7813 |
0.32 |
| chr10_65460212_65460378 | 0.21 |
REEP3 |
receptor accessory protein 3 |
179172 |
0.03 |
| chr9_134614958_134615256 | 0.21 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
224 |
0.94 |
| chr5_14154391_14154790 | 0.21 |
TRIO |
trio Rho guanine nucleotide exchange factor |
10761 |
0.32 |
| chr22_33197948_33198143 | 0.21 |
TIMP3 |
TIMP metallopeptidase inhibitor 3 |
358 |
0.9 |
| chr11_86319379_86319598 | 0.21 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
48626 |
0.18 |
| chr19_16178450_16178920 | 0.21 |
TPM4 |
tropomyosin 4 |
175 |
0.95 |
| chr17_74543145_74543365 | 0.21 |
RP11-666A8.7 |
|
2778 |
0.11 |
| chr6_157020523_157020674 | 0.20 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
78465 |
0.1 |
| chr12_122884907_122885085 | 0.20 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
435 |
0.86 |
| chr19_16936008_16936179 | 0.20 |
SIN3B |
SIN3 transcription regulator family member B |
4118 |
0.16 |
| chr12_123713391_123713550 | 0.20 |
MPHOSPH9 |
M-phase phosphoprotein 9 |
1523 |
0.29 |
| chr4_90674612_90674925 | 0.20 |
RP11-115D19.1 |
|
72105 |
0.11 |
| chr12_6657091_6657293 | 0.20 |
IFFO1 |
intermediate filament family orphan 1 |
3874 |
0.09 |
| chr7_143078673_143079796 | 0.20 |
ZYX |
zyxin |
234 |
0.85 |
| chr22_27695536_27695687 | 0.20 |
ENSG00000200443 |
. |
263761 |
0.02 |
| chr8_56832215_56832446 | 0.20 |
ENSG00000216204 |
. |
10810 |
0.14 |
| chr10_17277189_17277453 | 0.20 |
RP11-124N14.3 |
|
489 |
0.76 |
| chr2_64837127_64837278 | 0.20 |
ENSG00000252414 |
. |
31176 |
0.17 |
| chr9_96231273_96231424 | 0.20 |
FAM120AOS |
family with sequence similarity 120A opposite strand |
15474 |
0.14 |
| chr8_28912831_28912982 | 0.20 |
CTD-2647L4.4 |
|
546 |
0.71 |
| chr15_74212019_74212306 | 0.20 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
49 |
0.97 |
| chr3_139258322_139258643 | 0.20 |
RBP1 |
retinol binding protein 1, cellular |
79 |
0.75 |
| chr9_33081206_33081450 | 0.20 |
SMU1 |
smu-1 suppressor of mec-8 and unc-52 homolog (C. elegans) |
4663 |
0.19 |
| chr1_182931948_182932121 | 0.20 |
ENSG00000264768 |
. |
3324 |
0.2 |
| chr22_24541736_24541961 | 0.20 |
CABIN1 |
calcineurin binding protein 1 |
9958 |
0.17 |
| chr20_57458028_57458245 | 0.20 |
ENSG00000225806 |
. |
5711 |
0.14 |
| chr7_23514162_23514615 | 0.20 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
4302 |
0.2 |
| chr7_37723352_37723576 | 0.20 |
GPR141 |
G protein-coupled receptor 141 |
11 |
0.5 |
| chr6_169471024_169471293 | 0.19 |
XXyac-YX65C7_A.2 |
|
142191 |
0.05 |
| chr4_14113589_14113913 | 0.19 |
ENSG00000252092 |
. |
453500 |
0.01 |
| chr5_111090218_111090459 | 0.19 |
NREP |
neuronal regeneration related protein |
1610 |
0.42 |
| chr7_134512305_134512501 | 0.19 |
CALD1 |
caldesmon 1 |
16232 |
0.27 |
| chr22_38173266_38173417 | 0.19 |
ENSG00000238569 |
. |
8192 |
0.11 |
| chr12_109246535_109246783 | 0.19 |
SSH1 |
slingshot protein phosphatase 1 |
4700 |
0.17 |
| chr1_209958694_209958845 | 0.19 |
C1orf74 |
chromosome 1 open reading frame 74 |
865 |
0.47 |
| chr10_126106510_126107495 | 0.19 |
OAT |
ornithine aminotransferase |
503 |
0.8 |
| chr16_54689333_54689511 | 0.19 |
ENSG00000264079 |
. |
106114 |
0.08 |
| chr9_38026581_38026893 | 0.19 |
SHB |
Src homology 2 domain containing adaptor protein B |
42471 |
0.17 |
| chr2_106014491_106015411 | 0.19 |
FHL2 |
four and a half LIM domains 2 |
556 |
0.78 |
| chr4_91049201_91049889 | 0.19 |
CCSER1 |
coiled-coil serine-rich protein 1 |
809 |
0.78 |
| chr4_170190845_170191352 | 0.19 |
SH3RF1 |
SH3 domain containing ring finger 1 |
10 |
0.99 |
| chr2_113997514_113997878 | 0.19 |
RP11-65I12.1 |
|
611 |
0.6 |
| chr16_476332_477479 | 0.19 |
RAB11FIP3 |
RAB11 family interacting protein 3 (class II) |
526 |
0.62 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.2 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0071218 | cellular response to misfolded protein(GO:0071218) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.0 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.1 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.3 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.2 | GO:0033604 | negative regulation of catecholamine secretion(GO:0033604) |
| 0.0 | 0.1 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0072191 | ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0032344 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.0 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.4 | GO:0016925 | protein sumoylation(GO:0016925) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.2 | GO:0036296 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.1 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.2 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.0 | GO:1902339 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0097531 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0034442 | plasma lipoprotein particle oxidation(GO:0034441) regulation of lipoprotein oxidation(GO:0034442) negative regulation of lipoprotein oxidation(GO:0034443) regulation of plasma lipoprotein particle oxidation(GO:0034444) negative regulation of plasma lipoprotein particle oxidation(GO:0034445) lipoprotein modification(GO:0042160) lipoprotein oxidation(GO:0042161) |
| 0.0 | 0.2 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.1 | GO:0006268 | DNA unwinding involved in DNA replication(GO:0006268) |
| 0.0 | 0.1 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.2 | GO:0032784 | regulation of DNA-templated transcription, elongation(GO:0032784) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0072077 | mesenchymal to epithelial transition involved in metanephros morphogenesis(GO:0003337) renal vesicle morphogenesis(GO:0072077) metanephric renal vesicle morphogenesis(GO:0072283) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.0 | GO:1901534 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.2 | GO:0032770 | positive regulation of monooxygenase activity(GO:0032770) |
| 0.0 | 0.0 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.3 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0060014 | granulosa cell differentiation(GO:0060014) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.0 | GO:0035930 | corticosteroid hormone secretion(GO:0035930) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) |
| 0.0 | 0.0 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.0 | GO:0036465 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.0 | GO:0002686 | negative regulation of leukocyte migration(GO:0002686) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0099518 | vesicle transport along microtubule(GO:0047496) vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.1 | GO:0030949 | positive regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030949) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.7 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.3 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.2 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.4 | GO:0030018 | Z disc(GO:0030018) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.1 | 0.3 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.3 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.1 | 0.3 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.2 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.5 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.2 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.3 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.2 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.2 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.3 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0008200 | ion channel inhibitor activity(GO:0008200) |
| 0.0 | 0.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0002060 | purine nucleobase binding(GO:0002060) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 1.1 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.6 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
| 0.0 | 0.2 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.4 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.0 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.1 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |