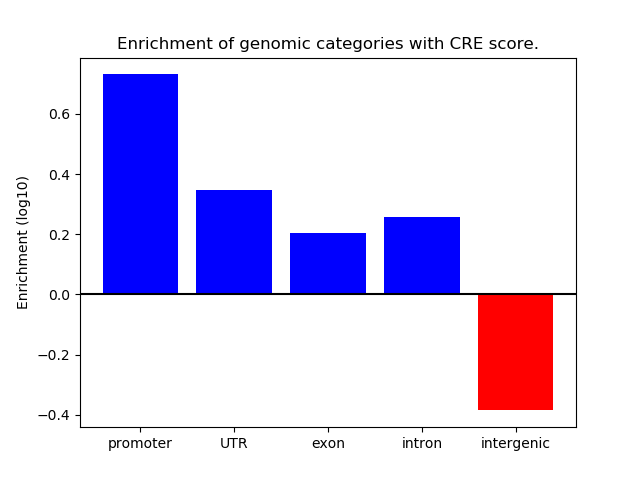
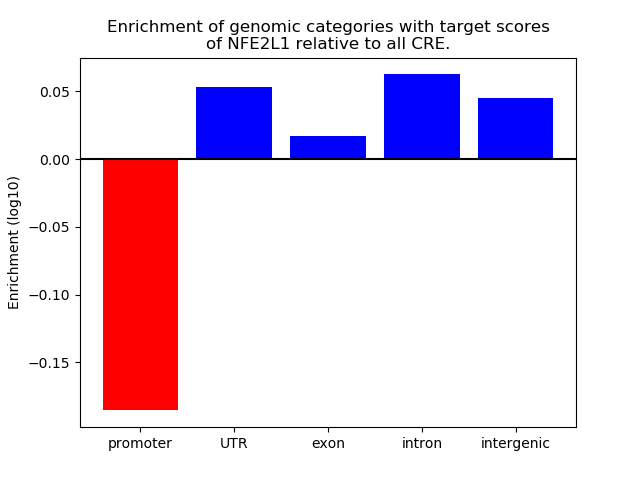
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
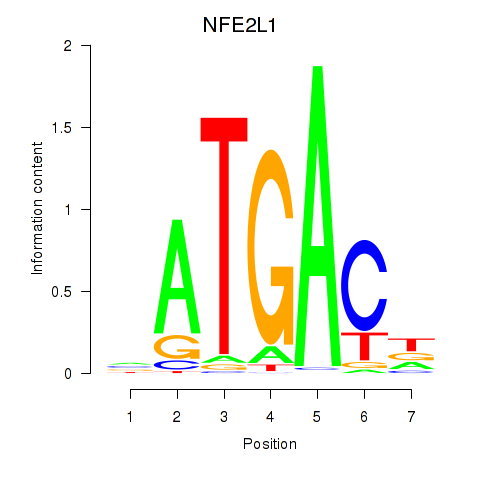
Results for NFE2L1
Z-value: 1.76
Transcription factors associated with NFE2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFE2L1
|
ENSG00000082641.11 | nuclear factor, erythroid 2 like 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_46125776_46126001 | NFE2L1 | 156 | 0.761365 | -0.96 | 3.4e-05 | Click! |
| chr17_46128688_46128930 | NFE2L1 | 2315 | 0.142764 | -0.77 | 1.6e-02 | Click! |
| chr17_46126176_46126433 | NFE2L1 | 158 | 0.853478 | -0.66 | 5.5e-02 | Click! |
| chr17_46131994_46132145 | NFE2L1 | 3 | 0.949078 | -0.64 | 6.5e-02 | Click! |
| chr17_46132310_46132461 | NFE2L1 | 103 | 0.927003 | -0.62 | 7.7e-02 | Click! |
Activity of the NFE2L1 motif across conditions
Conditions sorted by the z-value of the NFE2L1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
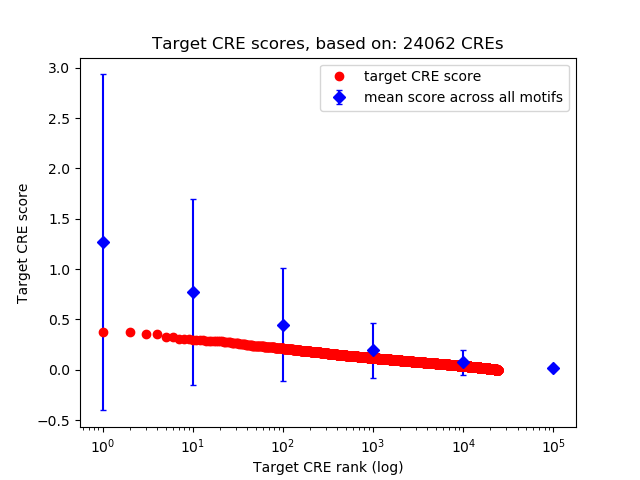
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_70847938_70848765 | 0.38 |
SRGN |
serglycin |
477 |
0.82 |
| chr1_81137024_81137175 | 0.37 |
ENSG00000266033 |
. |
341926 |
0.01 |
| chr8_5192496_5192647 | 0.35 |
ENSG00000242079 |
. |
192342 |
0.03 |
| chr9_91793130_91793737 | 0.35 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
249 |
0.95 |
| chr6_46702576_46703631 | 0.33 |
PLA2G7 |
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
24 |
0.97 |
| chr2_41837870_41838465 | 0.33 |
ENSG00000221372 |
. |
14641 |
0.29 |
| chr15_58433367_58433518 | 0.31 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
2418 |
0.3 |
| chr16_50401597_50402736 | 0.31 |
RP11-21B23.1 |
|
325 |
0.64 |
| chr17_80817564_80818630 | 0.30 |
TBCD |
tubulin folding cofactor D |
173 |
0.94 |
| chr22_34240463_34240614 | 0.30 |
LARGE |
like-glycosyltransferase |
17074 |
0.25 |
| chr1_65430726_65432146 | 0.30 |
JAK1 |
Janus kinase 1 |
751 |
0.73 |
| chr6_111408575_111409679 | 0.30 |
SLC16A10 |
solute carrier family 16 (aromatic amino acid transporter), member 10 |
346 |
0.87 |
| chr8_116470254_116470468 | 0.30 |
TRPS1 |
trichorhinophalangeal syndrome I |
34087 |
0.22 |
| chr3_147087748_147087899 | 0.29 |
RP11-649A16.1 |
|
620 |
0.73 |
| chr10_135148565_135149526 | 0.29 |
CALY |
calcyon neuron-specific vesicular protein |
1331 |
0.22 |
| chr6_138070872_138071097 | 0.29 |
ENSG00000216097 |
. |
32899 |
0.17 |
| chr18_52357258_52357685 | 0.29 |
RAB27B |
RAB27B, member RAS oncogene family |
27620 |
0.24 |
| chr2_108443697_108444171 | 0.29 |
RGPD4 |
RANBP2-like and GRIP domain containing 4 |
541 |
0.87 |
| chr14_30692792_30692943 | 0.28 |
PRKD1 |
protein kinase D1 |
31763 |
0.25 |
| chr18_22869556_22869739 | 0.28 |
CTD-2006O16.2 |
|
12443 |
0.28 |
| chr9_82226362_82226513 | 0.28 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
37833 |
0.24 |
| chr3_177320979_177321130 | 0.28 |
ENSG00000200288 |
. |
21143 |
0.27 |
| chr3_38216440_38216633 | 0.28 |
OXSR1 |
oxidative stress responsive 1 |
9540 |
0.16 |
| chr6_19465542_19465832 | 0.28 |
ENSG00000201523 |
. |
27173 |
0.27 |
| chr1_178095910_178096061 | 0.27 |
RASAL2 |
RAS protein activator like 2 |
32709 |
0.24 |
| chr15_65578383_65579365 | 0.27 |
PARP16 |
poly (ADP-ribose) polymerase family, member 16 |
307 |
0.76 |
| chr3_192549660_192549811 | 0.27 |
FGF12 |
fibroblast growth factor 12 |
64182 |
0.13 |
| chr22_46988396_46988787 | 0.27 |
GRAMD4 |
GRAM domain containing 4 |
15565 |
0.21 |
| chr1_65521422_65521573 | 0.27 |
ENSG00000265996 |
. |
2028 |
0.29 |
| chr4_72095290_72095441 | 0.27 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
42321 |
0.21 |
| chr6_125698814_125699135 | 0.26 |
RP11-735G4.1 |
|
3504 |
0.35 |
| chr1_16490997_16491148 | 0.26 |
RP11-276H7.3 |
|
5311 |
0.12 |
| chr5_72746479_72746692 | 0.26 |
FOXD1 |
forkhead box D1 |
2233 |
0.32 |
| chr4_141852958_141853109 | 0.26 |
RNF150 |
ring finger protein 150 |
65157 |
0.13 |
| chr7_105917980_105918131 | 0.25 |
NAMPT |
nicotinamide phosphoribosyltransferase |
7312 |
0.23 |
| chr6_113529861_113530012 | 0.25 |
ENSG00000201386 |
. |
237241 |
0.02 |
| chr5_148139240_148139391 | 0.25 |
ADRB2 |
adrenoceptor beta 2, surface |
66841 |
0.11 |
| chr2_191914732_191914993 | 0.25 |
ENSG00000231858 |
. |
28610 |
0.15 |
| chr2_238596342_238596493 | 0.25 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
4371 |
0.26 |
| chr4_129732983_129734245 | 0.25 |
JADE1 |
jade family PHD finger 1 |
615 |
0.83 |
| chr1_79471738_79472138 | 0.25 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
465 |
0.9 |
| chr16_74440490_74441404 | 0.25 |
CLEC18B |
C-type lectin domain family 18, member B |
14343 |
0.14 |
| chr2_201995461_201995643 | 0.24 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
963 |
0.45 |
| chr2_10587541_10588176 | 0.24 |
ODC1 |
ornithine decarboxylase 1 |
48 |
0.95 |
| chr6_35655029_35656564 | 0.24 |
FKBP5 |
FK506 binding protein 5 |
896 |
0.49 |
| chr1_32732739_32732890 | 0.24 |
LCK |
lymphocyte-specific protein tyrosine kinase |
6900 |
0.09 |
| chr5_66502810_66503002 | 0.24 |
CD180 |
CD180 molecule |
10279 |
0.29 |
| chr6_122253515_122253742 | 0.24 |
ENSG00000199932 |
. |
279166 |
0.01 |
| chr19_30205840_30206749 | 0.24 |
C19orf12 |
chromosome 19 open reading frame 12 |
133 |
0.97 |
| chr16_70221940_70222878 | 0.24 |
CLEC18C |
C-type lectin domain family 18, member C |
14349 |
0.1 |
| chr9_112955354_112955564 | 0.24 |
C9orf152 |
chromosome 9 open reading frame 152 |
15010 |
0.23 |
| chr6_27521237_27521556 | 0.24 |
ENSG00000206671 |
. |
42795 |
0.14 |
| chr14_101034010_101035263 | 0.24 |
BEGAIN |
brain-enriched guanylate kinase-associated |
192 |
0.93 |
| chr13_24146342_24146493 | 0.23 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
1614 |
0.53 |
| chr21_34602307_34603720 | 0.23 |
IFNAR2 |
interferon (alpha, beta and omega) receptor 2 |
299 |
0.87 |
| chr1_14924992_14926197 | 0.23 |
KAZN |
kazrin, periplakin interacting protein |
381 |
0.93 |
| chr13_50654211_50655703 | 0.23 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
1350 |
0.41 |
| chr5_119432272_119432494 | 0.23 |
ENSG00000251975 |
. |
240965 |
0.02 |
| chr2_47425999_47426150 | 0.23 |
CALM2 |
calmodulin 2 (phosphorylase kinase, delta) |
22334 |
0.19 |
| chr17_73774322_73775453 | 0.23 |
H3F3B |
H3 histone, family 3B (H3.3B) |
480 |
0.64 |
| chr2_133998912_133999202 | 0.23 |
AC010890.1 |
|
23483 |
0.24 |
| chr20_20840805_20840956 | 0.23 |
ENSG00000264361 |
. |
121506 |
0.06 |
| chr16_57279074_57280095 | 0.23 |
ARL2BP |
ADP-ribosylation factor-like 2 binding protein |
296 |
0.82 |
| chr8_17017795_17017946 | 0.23 |
ZDHHC2 |
zinc finger, DHHC-type containing 2 |
2438 |
0.36 |
| chr17_47329713_47329864 | 0.23 |
PHOSPHO1 |
phosphatase, orphan 1 |
21660 |
0.13 |
| chr1_169454193_169456202 | 0.23 |
SLC19A2 |
solute carrier family 19 (thiamine transporter), member 2 |
28 |
0.98 |
| chr2_38299955_38300348 | 0.23 |
CYP1B1-AS1 |
CYP1B1 antisense RNA 1 |
2640 |
0.23 |
| chr20_49138849_49139000 | 0.23 |
PTPN1 |
protein tyrosine phosphatase, non-receptor type 1 |
12004 |
0.16 |
| chr8_15646506_15646657 | 0.23 |
TUSC3 |
tumor suppressor candidate 3 |
165992 |
0.04 |
| chr7_157188129_157188280 | 0.23 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
55689 |
0.14 |
| chr17_55502106_55502257 | 0.23 |
ENSG00000263902 |
. |
12619 |
0.26 |
| chr20_3073179_3074190 | 0.23 |
ENSG00000263905 |
. |
1177 |
0.34 |
| chr12_27933175_27934389 | 0.23 |
KLHL42 |
kelch-like family member 42 |
18 |
0.73 |
| chr5_55981802_55981953 | 0.22 |
AC022431.2 |
Homo sapiens uncharacterized LOC101928448 (LOC101928448), mRNA. |
79818 |
0.09 |
| chr19_42306064_42306215 | 0.22 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
5054 |
0.13 |
| chr17_15280700_15281096 | 0.22 |
ENSG00000239888 |
. |
30803 |
0.14 |
| chr15_26041719_26041870 | 0.22 |
ENSG00000199214 |
. |
21336 |
0.18 |
| chr1_239995605_239995756 | 0.22 |
CHRM3-AS1 |
CHRM3 antisense RNA 1 |
67492 |
0.12 |
| chr16_10679196_10679376 | 0.22 |
EMP2 |
epithelial membrane protein 2 |
4731 |
0.2 |
| chr1_215183894_215184045 | 0.22 |
KCNK2 |
potassium channel, subfamily K, member 2 |
4771 |
0.37 |
| chr9_110638127_110638278 | 0.22 |
ENSG00000222459 |
. |
43057 |
0.16 |
| chr6_35465033_35465928 | 0.22 |
TEAD3 |
TEA domain family member 3 |
627 |
0.7 |
| chr8_32506055_32506353 | 0.22 |
NRG1 |
neuregulin 1 |
847 |
0.76 |
| chr13_67804057_67805835 | 0.22 |
PCDH9 |
protocadherin 9 |
478 |
0.9 |
| chr13_51483605_51485004 | 0.22 |
RNASEH2B |
ribonuclease H2, subunit B |
412 |
0.48 |
| chr5_77805493_77806187 | 0.22 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
39134 |
0.21 |
| chr4_47838472_47839750 | 0.22 |
CORIN |
corin, serine peptidase |
855 |
0.65 |
| chr12_57527559_57527864 | 0.22 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
1789 |
0.2 |
| chr5_73871785_73872054 | 0.22 |
HEXB |
hexosaminidase B (beta polypeptide) |
63929 |
0.12 |
| chr15_23931247_23932124 | 0.22 |
NDN |
necdin, melanoma antigen (MAGE) family member |
765 |
0.7 |
| chr15_99366225_99366435 | 0.21 |
ENSG00000264480 |
. |
38675 |
0.17 |
| chr19_19483852_19484190 | 0.21 |
GATAD2A |
GATA zinc finger domain containing 2A |
12614 |
0.13 |
| chr5_39423074_39423254 | 0.21 |
DAB2 |
Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) |
1806 |
0.48 |
| chr6_43139652_43140723 | 0.21 |
SRF |
serum response factor (c-fos serum response element-binding transcription factor) |
59 |
0.96 |
| chr3_148366543_148366694 | 0.21 |
AGTR1 |
angiotensin II receptor, type 1 |
48953 |
0.18 |
| chr3_175150286_175150526 | 0.21 |
ENSG00000199792 |
. |
35269 |
0.2 |
| chr13_24146543_24146694 | 0.21 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
1815 |
0.49 |
| chr2_99346083_99347252 | 0.21 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
44 |
0.98 |
| chr4_1930205_1930356 | 0.21 |
WHSC1 |
Wolf-Hirschhorn syndrome candidate 1 |
11108 |
0.16 |
| chr8_97536368_97536646 | 0.21 |
SDC2 |
syndecan 2 |
30271 |
0.22 |
| chr6_45473095_45473246 | 0.21 |
RUNX2 |
runt-related transcription factor 2 |
82948 |
0.09 |
| chr7_75986838_75988224 | 0.21 |
YWHAG |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma |
817 |
0.59 |
| chr9_130159461_130160475 | 0.21 |
SLC2A8 |
solute carrier family 2 (facilitated glucose transporter), member 8 |
305 |
0.87 |
| chr1_100504063_100504968 | 0.21 |
HIAT1 |
hippocampus abundant transcript 1 |
862 |
0.51 |
| chr16_67197260_67198336 | 0.21 |
HSF4 |
heat shock transcription factor 4 |
510 |
0.52 |
| chr13_81010799_81010950 | 0.21 |
ENSG00000202398 |
. |
67205 |
0.14 |
| chrX_46838416_46838567 | 0.21 |
JADE3 |
jade family PHD finger 3 |
66260 |
0.09 |
| chr3_43448035_43448186 | 0.21 |
SNRK-AS1 |
SNRK antisense RNA 1 |
54656 |
0.11 |
| chr17_59235574_59235725 | 0.21 |
RP11-136H19.1 |
|
22324 |
0.2 |
| chr20_11151199_11151404 | 0.21 |
C20orf187 |
chromosome 20 open reading frame 187 |
142490 |
0.05 |
| chr2_177018526_177018677 | 0.21 |
HOXD4 |
homeobox D4 |
2651 |
0.11 |
| chr11_28695349_28695500 | 0.21 |
ENSG00000222385 |
. |
412358 |
0.01 |
| chr8_67624599_67625774 | 0.21 |
SGK3 |
serum/glucocorticoid regulated kinase family, member 3 |
533 |
0.76 |
| chr8_131960988_131961496 | 0.21 |
RP11-737F9.1 |
|
13587 |
0.24 |
| chr17_53349481_53349632 | 0.21 |
RP11-515O17.2 |
|
1458 |
0.47 |
| chr12_56075239_56075869 | 0.20 |
METTL7B |
methyltransferase like 7B |
224 |
0.86 |
| chr3_36985452_36986747 | 0.20 |
TRANK1 |
tetratricopeptide repeat and ankyrin repeat containing 1 |
449 |
0.83 |
| chr2_238358341_238358492 | 0.20 |
AC112721.1 |
Uncharacterized protein |
24497 |
0.16 |
| chrX_3629951_3631442 | 0.20 |
PRKX |
protein kinase, X-linked |
953 |
0.63 |
| chr22_36608967_36609118 | 0.20 |
APOL4 |
apolipoprotein L, 4 |
8156 |
0.18 |
| chr1_66713244_66713395 | 0.20 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
10011 |
0.3 |
| chrX_118372691_118372845 | 0.20 |
PGRMC1 |
progesterone receptor membrane component 1 |
2480 |
0.36 |
| chr6_7672125_7672276 | 0.20 |
BMP6 |
bone morphogenetic protein 6 |
54830 |
0.14 |
| chr17_57920542_57920694 | 0.20 |
ENSG00000199004 |
. |
1991 |
0.25 |
| chr1_113045279_113045430 | 0.20 |
WNT2B |
wingless-type MMTV integration site family, member 2B |
6046 |
0.2 |
| chr4_82585220_82585371 | 0.20 |
RASGEF1B |
RasGEF domain family, member 1B |
192226 |
0.03 |
| chr3_87011295_87011446 | 0.20 |
VGLL3 |
vestigial like 3 (Drosophila) |
28482 |
0.26 |
| chr8_27200949_27201100 | 0.20 |
PTK2B |
protein tyrosine kinase 2 beta |
16684 |
0.2 |
| chr13_42143466_42143617 | 0.20 |
ENSG00000264190 |
. |
1010 |
0.64 |
| chr7_24988449_24988600 | 0.20 |
OSBPL3 |
oxysterol binding protein-like 3 |
30812 |
0.21 |
| chr10_129152577_129152728 | 0.20 |
FAM196A |
family with sequence similarity 196, member A |
158230 |
0.04 |
| chr12_32261770_32262117 | 0.20 |
RP11-843B15.2 |
|
1481 |
0.38 |
| chr4_79442125_79442276 | 0.20 |
ANXA3 |
annexin A3 |
30473 |
0.21 |
| chr17_32582682_32583141 | 0.20 |
AC005549.3 |
Uncharacterized protein |
605 |
0.41 |
| chr8_23396205_23396870 | 0.20 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
9964 |
0.16 |
| chr4_24472696_24473642 | 0.20 |
ENSG00000265001 |
. |
41714 |
0.15 |
| chr14_24786486_24786637 | 0.20 |
LTB4R |
leukotriene B4 receptor |
2655 |
0.1 |
| chr22_38928075_38928226 | 0.20 |
DDX17 |
DEAD (Asp-Glu-Ala-Asp) box helicase 17 |
25805 |
0.12 |
| chr8_131020883_131021034 | 0.20 |
ENSG00000264653 |
. |
259 |
0.91 |
| chr8_142138821_142140144 | 0.20 |
RP11-809O17.1 |
|
578 |
0.46 |
| chr1_217076565_217076716 | 0.20 |
ESRRG |
estrogen-related receptor gamma |
36372 |
0.24 |
| chr12_114863083_114863234 | 0.20 |
TBX5-AS1 |
TBX5 antisense RNA 1 |
14064 |
0.21 |
| chr12_20622316_20622467 | 0.20 |
RP11-284H19.1 |
|
99195 |
0.08 |
| chr10_88159807_88161210 | 0.20 |
GRID1 |
glutamate receptor, ionotropic, delta 1 |
34273 |
0.17 |
| chr3_194029323_194029474 | 0.20 |
CPN2 |
carboxypeptidase N, polypeptide 2 |
42649 |
0.14 |
| chr7_18429816_18429967 | 0.19 |
AC010082.2 |
|
40470 |
0.19 |
| chr2_216288358_216288696 | 0.19 |
FN1 |
fibronectin 1 |
12263 |
0.21 |
| chr8_16995862_16996013 | 0.19 |
ZDHHC2 |
zinc finger, DHHC-type containing 2 |
17601 |
0.23 |
| chr18_47017567_47018883 | 0.19 |
RPL17 |
ribosomal protein L17 |
15 |
0.51 |
| chr2_71856946_71857097 | 0.19 |
DYSF |
dysferlin |
163189 |
0.04 |
| chrX_44774522_44774719 | 0.19 |
ENSG00000252113 |
. |
29869 |
0.19 |
| chr10_7301046_7301197 | 0.19 |
SFMBT2 |
Scm-like with four mbt domains 2 |
149586 |
0.04 |
| chr3_81772804_81773074 | 0.19 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
19841 |
0.3 |
| chr20_30457693_30458808 | 0.19 |
DUSP15 |
dual specificity phosphatase 15 |
125 |
0.67 |
| chr21_38831523_38831674 | 0.19 |
DYRK1A |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
29489 |
0.19 |
| chr2_201978151_201978302 | 0.19 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
2601 |
0.2 |
| chr3_120626761_120627590 | 0.19 |
STXBP5L |
syntaxin binding protein 5-like |
14 |
0.99 |
| chr12_105114147_105114900 | 0.19 |
ENSG00000264295 |
. |
129112 |
0.05 |
| chr12_51636019_51636170 | 0.19 |
DAZAP2 |
DAZ associated protein 2 |
3015 |
0.18 |
| chr4_184313441_184313592 | 0.19 |
ENSG00000263395 |
. |
7810 |
0.18 |
| chr6_2111373_2111600 | 0.19 |
GMDS |
GDP-mannose 4,6-dehydratase |
64739 |
0.15 |
| chr9_96819103_96819334 | 0.19 |
PTPDC1 |
protein tyrosine phosphatase domain containing 1 |
26142 |
0.21 |
| chr11_36641126_36641415 | 0.19 |
RAG2 |
recombination activating gene 2 |
21484 |
0.19 |
| chrX_12929174_12929325 | 0.19 |
TLR8-AS1 |
TLR8 antisense RNA 1 |
2797 |
0.26 |
| chr5_150181351_150181502 | 0.19 |
AC010441.1 |
|
23559 |
0.14 |
| chr3_8152824_8153002 | 0.19 |
LMCD1-AS1 |
LMCD1 antisense RNA 1 (head to head) |
94919 |
0.09 |
| chr13_115028728_115028879 | 0.19 |
ENSG00000265450 |
. |
10500 |
0.15 |
| chr5_52106930_52107081 | 0.19 |
CTD-2288O8.1 |
|
23145 |
0.18 |
| chr21_34392151_34392837 | 0.19 |
OLIG2 |
oligodendrocyte lineage transcription factor 2 |
5659 |
0.22 |
| chr1_84107245_84107582 | 0.19 |
ENSG00000223231 |
. |
152147 |
0.04 |
| chr6_112555463_112555614 | 0.19 |
RP11-506B6.6 |
|
1756 |
0.35 |
| chr18_56029142_56029527 | 0.19 |
RP11-845C23.2 |
|
9464 |
0.19 |
| chr6_6546483_6547412 | 0.19 |
LY86-AS1 |
LY86 antisense RNA 1 |
40479 |
0.18 |
| chr15_39882698_39882873 | 0.19 |
CTD-2033D15.1 |
|
3647 |
0.23 |
| chr7_6292086_6292237 | 0.19 |
CYTH3 |
cytohesin 3 |
20114 |
0.16 |
| chr11_12964003_12964154 | 0.19 |
RP11-47J17.2 |
|
19606 |
0.17 |
| chr9_95525836_95526977 | 0.19 |
BICD2 |
bicaudal D homolog 2 (Drosophila) |
688 |
0.71 |
| chr21_40051384_40051625 | 0.19 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
17800 |
0.28 |
| chr1_59594796_59594947 | 0.19 |
FGGY |
FGGY carbohydrate kinase domain containing |
167439 |
0.04 |
| chr7_7856053_7856204 | 0.19 |
RPA3-AS1 |
RPA3 antisense RNA 1 |
44136 |
0.16 |
| chr4_134075824_134075975 | 0.19 |
PCDH10 |
protocadherin 10 |
5429 |
0.36 |
| chr12_121340609_121341729 | 0.19 |
SPPL3 |
signal peptide peptidase like 3 |
4 |
0.98 |
| chr10_102087281_102087432 | 0.19 |
PKD2L1 |
polycystic kidney disease 2-like 1 |
2373 |
0.21 |
| chr6_138195405_138195556 | 0.19 |
RP11-356I2.4 |
|
6110 |
0.23 |
| chr2_216546553_216546831 | 0.19 |
ENSG00000212055 |
. |
196950 |
0.03 |
| chr13_100647875_100648026 | 0.19 |
LINC00554 |
long intergenic non-protein coding RNA 554 |
2213 |
0.3 |
| chr7_19034879_19035030 | 0.19 |
AC004744.3 |
|
95898 |
0.07 |
| chr2_240302618_240303103 | 0.19 |
HDAC4 |
histone deacetylase 4 |
19783 |
0.17 |
| chr5_17282488_17282863 | 0.19 |
ENSG00000252908 |
. |
41955 |
0.15 |
| chr2_68599888_68600039 | 0.18 |
AC015969.3 |
|
7247 |
0.18 |
| chr17_13689465_13689699 | 0.18 |
ENSG00000236088 |
. |
6285 |
0.33 |
| chr16_87904899_87905050 | 0.18 |
SLC7A5 |
solute carrier family 7 (amino acid transporter light chain, L system), member 5 |
1880 |
0.31 |
| chr18_43588722_43588960 | 0.18 |
ENSG00000222179 |
. |
18930 |
0.17 |
| chrX_49801111_49801262 | 0.18 |
ENSG00000272080 |
. |
21980 |
0.08 |
| chr4_40240364_40240515 | 0.18 |
RHOH |
ras homolog family member H |
38475 |
0.16 |
| chr1_59221360_59221511 | 0.18 |
ENSG00000264081 |
. |
9172 |
0.2 |
| chr2_204394542_204394693 | 0.18 |
RAPH1 |
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
3539 |
0.35 |
| chr7_38347403_38347554 | 0.18 |
STARD3NL |
STARD3 N-terminal like |
129481 |
0.05 |
| chr3_161632795_161632946 | 0.18 |
OTOL1 |
otolin 1 |
418274 |
0.01 |
| chr4_108745368_108745519 | 0.18 |
SGMS2 |
sphingomyelin synthase 2 |
276 |
0.93 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.5 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.1 | 0.3 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.1 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.4 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.1 | 0.2 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.1 | 0.2 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.1 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.1 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.1 | 0.2 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.2 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.1 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.1 | 0.2 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.1 | 0.3 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.1 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.2 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.1 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.1 | 0.2 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.2 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.1 | 0.2 | GO:0055064 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0010715 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.2 | GO:2001259 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of calcium ion transmembrane transporter activity(GO:1901021) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.0 | GO:0060686 | negative regulation of prostatic bud formation(GO:0060686) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0090322 | regulation of superoxide metabolic process(GO:0090322) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0048245 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0051665 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) membrane raft localization(GO:0051665) |
| 0.0 | 0.3 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.2 | GO:0051459 | corticotropin secretion(GO:0051458) regulation of corticotropin secretion(GO:0051459) positive regulation of corticotropin secretion(GO:0051461) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.2 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.1 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0035930 | corticosteroid hormone secretion(GO:0035930) regulation of steroid hormone secretion(GO:2000831) regulation of corticosteroid hormone secretion(GO:2000846) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.0 | GO:0061316 | negative regulation of heart induction by canonical Wnt signaling pathway(GO:0003136) negative regulation of cardioblast cell fate specification(GO:0009997) cardioblast cell fate commitment(GO:0042684) cardioblast cell fate specification(GO:0042685) regulation of cardioblast cell fate specification(GO:0042686) negative regulation of cardioblast differentiation(GO:0051892) cardiac cell fate specification(GO:0060912) canonical Wnt signaling pathway involved in heart development(GO:0061316) negative regulation of heart induction(GO:1901320) negative regulation of cardiocyte differentiation(GO:1905208) regulation of cardiac cell fate specification(GO:2000043) negative regulation of cardiac cell fate specification(GO:2000044) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.2 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.2 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.2 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.3 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.0 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.1 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.1 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.2 | GO:2000178 | negative regulation of neuroblast proliferation(GO:0007406) negative regulation of neural precursor cell proliferation(GO:2000178) |
| 0.0 | 0.1 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0046184 | aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 0.1 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.3 | GO:0036296 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0043267 | negative regulation of potassium ion transport(GO:0043267) |
| 0.0 | 0.1 | GO:0052169 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.0 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.1 | GO:0050686 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.1 | GO:0045409 | negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.1 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0071371 | cellular response to gonadotropin stimulus(GO:0071371) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0034393 | positive regulation of muscle cell apoptotic process(GO:0010661) positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0021781 | glial cell fate commitment(GO:0021781) |
| 0.0 | 0.1 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0031558 | obsolete induction of apoptosis in response to chemical stimulus(GO:0031558) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.1 | GO:0010664 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.2 | GO:0046697 | decidualization(GO:0046697) |
| 0.0 | 0.0 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0009106 | lipoate metabolic process(GO:0009106) |
| 0.0 | 0.3 | GO:0007628 | adult walking behavior(GO:0007628) walking behavior(GO:0090659) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0061213 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.1 | GO:1903077 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.2 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.1 | GO:1901985 | positive regulation of histone acetylation(GO:0035066) positive regulation of protein acetylation(GO:1901985) positive regulation of peptidyl-lysine acetylation(GO:2000758) |
| 0.0 | 0.5 | GO:0035249 | synaptic transmission, glutamatergic(GO:0035249) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:1901881 | positive regulation of protein depolymerization(GO:1901881) |
| 0.0 | 0.1 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.1 | GO:0045348 | positive regulation of MHC class II biosynthetic process(GO:0045348) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0060789 | hair follicle placode formation(GO:0060789) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.1 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:0034285 | response to sucrose(GO:0009744) response to disaccharide(GO:0034285) |
| 0.0 | 0.0 | GO:1903224 | endodermal cell fate commitment(GO:0001711) endodermal cell fate specification(GO:0001714) endodermal cell differentiation(GO:0035987) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.0 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.1 | GO:0048268 | clathrin coat assembly(GO:0048268) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0090177 | establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.0 | 0.0 | GO:1901798 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.0 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) negative regulation of lipase activity(GO:0060192) |
| 0.0 | 0.1 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0050955 | sensory perception of temperature stimulus(GO:0050951) thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0000101 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.1 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.0 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.0 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.0 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.0 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.0 | GO:0090103 | cochlea morphogenesis(GO:0090103) |
| 0.0 | 0.0 | GO:0052572 | response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0010822 | positive regulation of mitochondrion organization(GO:0010822) positive regulation of release of cytochrome c from mitochondria(GO:0090200) positive regulation of apoptotic signaling pathway(GO:2001235) |
| 0.0 | 0.0 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.0 | GO:0048343 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0010744 | positive regulation of macrophage derived foam cell differentiation(GO:0010744) |
| 0.0 | 0.0 | GO:0003207 | cardiac chamber formation(GO:0003207) cardiac ventricle formation(GO:0003211) |
| 0.0 | 0.0 | GO:0031062 | positive regulation of histone methylation(GO:0031062) |
| 0.0 | 0.2 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.0 | GO:0048570 | notochord morphogenesis(GO:0048570) |
| 0.0 | 0.2 | GO:1901663 | quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.0 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.3 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.2 | GO:0042401 | cellular biogenic amine biosynthetic process(GO:0042401) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.0 | GO:0097384 | cellular lipid biosynthetic process(GO:0097384) |
| 0.0 | 0.0 | GO:0070168 | negative regulation of biomineral tissue development(GO:0070168) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.0 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.0 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0045416 | positive regulation of interleukin-8 biosynthetic process(GO:0045416) |
| 0.0 | 0.0 | GO:0042228 | interleukin-8 biosynthetic process(GO:0042228) |
| 0.0 | 0.1 | GO:0032689 | negative regulation of interferon-gamma production(GO:0032689) |
| 0.0 | 0.3 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.0 | GO:0070391 | response to lipoteichoic acid(GO:0070391) cellular response to lipoteichoic acid(GO:0071223) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.0 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.4 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.2 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0048668 | collateral sprouting(GO:0048668) |
| 0.0 | 0.0 | GO:0072698 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.0 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0042554 | superoxide anion generation(GO:0042554) |
| 0.0 | 0.1 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.0 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.1 | GO:0019682 | pentose-phosphate shunt(GO:0006098) glyceraldehyde-3-phosphate metabolic process(GO:0019682) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.0 | GO:0032717 | negative regulation of interleukin-8 production(GO:0032717) |
| 0.0 | 0.0 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:0051938 | L-glutamate import(GO:0051938) |
| 0.0 | 0.2 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.0 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.0 | GO:0043474 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.0 | GO:0042181 | ketone biosynthetic process(GO:0042181) |
| 0.0 | 0.0 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.0 | GO:0045830 | positive regulation of isotype switching(GO:0045830) |
| 0.0 | 0.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.0 | GO:0060706 | cell differentiation involved in embryonic placenta development(GO:0060706) |
| 0.0 | 0.1 | GO:0032461 | positive regulation of protein oligomerization(GO:0032461) |
| 0.0 | 0.0 | GO:0034698 | response to gonadotropin(GO:0034698) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0032964 | collagen biosynthetic process(GO:0032964) |
| 0.0 | 0.0 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0006069 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0032905 | transforming growth factor beta1 production(GO:0032905) regulation of transforming growth factor beta1 production(GO:0032908) |
| 0.0 | 0.0 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0070244 | negative regulation of thymocyte apoptotic process(GO:0070244) |
| 0.0 | 0.0 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.2 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0042597 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.8 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0099738 | cell cortex region(GO:0099738) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0005964 | phosphorylase kinase complex(GO:0005964) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.1 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.0 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.0 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.1 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.1 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.2 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.2 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.1 | 0.2 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.1 | 0.3 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.1 | 0.3 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.1 | 0.2 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.1 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.2 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.3 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.3 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.0 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.0 | 0.1 | GO:0047555 | 3',5'-cyclic-GMP phosphodiesterase activity(GO:0047555) |
| 0.0 | 0.1 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0004689 | phosphorylase kinase activity(GO:0004689) |
| 0.0 | 0.2 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.3 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.6 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0004445 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.1 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.1 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.2 | GO:0070001 | aspartic-type endopeptidase activity(GO:0004190) aspartic-type peptidase activity(GO:0070001) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0000976 | transcription regulatory region sequence-specific DNA binding(GO:0000976) |
| 0.0 | 0.1 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.0 | 0.2 | GO:0030291 | protein serine/threonine kinase inhibitor activity(GO:0030291) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.0 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.0 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.1 | GO:0016653 | oxidoreductase activity, acting on NAD(P)H, heme protein as acceptor(GO:0016653) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0051636 | obsolete Gram-negative bacterial cell surface binding(GO:0051636) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.0 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.0 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
| 0.1 | 0.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.2 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.0 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.4 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.2 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | PID PTP1B PATHWAY | Signaling events mediated by PTP1B |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID RHOA PATHWAY | RhoA signaling pathway |
| 0.0 | 0.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.5 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.4 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.1 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.1 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.1 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME TRIGLYCERIDE BIOSYNTHESIS | Genes involved in Triglyceride Biosynthesis |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.4 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.4 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.0 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.4 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.5 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.3 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE KERATIN METABOLISM | Genes involved in Keratan sulfate/keratin metabolism |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.4 | REACTOME INWARDLY RECTIFYING K CHANNELS | Genes involved in Inwardly rectifying K+ channels |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.0 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.0 | REACTOME BILE ACID AND BILE SALT METABOLISM | Genes involved in Bile acid and bile salt metabolism |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |