Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
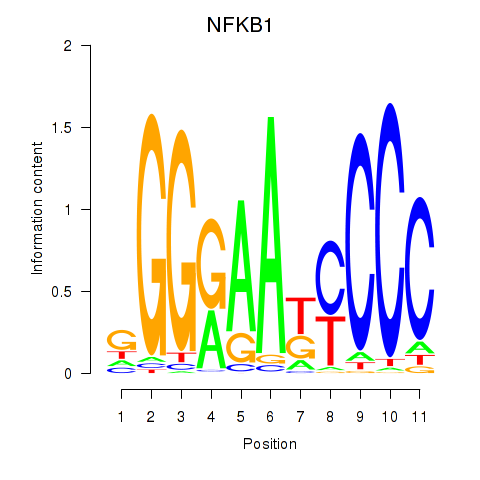
Results for NFKB1
Z-value: 1.34
Transcription factors associated with NFKB1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB1
|
ENSG00000109320.7 | nuclear factor kappa B subunit 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_103425319_103425470 | NFKB1 | 862 | 0.614179 | 0.96 | 2.6e-05 | Click! |
| chr4_103425152_103425303 | NFKB1 | 695 | 0.692990 | 0.96 | 3.2e-05 | Click! |
| chr4_103541720_103541875 | NFKB1 | 42776 | 0.157378 | -0.92 | 4.0e-04 | Click! |
| chr4_103534410_103534561 | NFKB1 | 35464 | 0.174211 | 0.90 | 1.1e-03 | Click! |
| chr4_103449015_103449166 | NFKB1 | 24558 | 0.176648 | 0.86 | 2.7e-03 | Click! |
Activity of the NFKB1 motif across conditions
Conditions sorted by the z-value of the NFKB1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr9_134143747_134144006 | 1.05 |
FAM78A |
family with sequence similarity 78, member A |
2004 |
0.31 |
| chr15_45409056_45409207 | 0.87 |
DUOX2 |
dual oxidase 2 |
2589 |
0.14 |
| chr4_100738641_100738957 | 0.86 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
796 |
0.72 |
| chr19_49839028_49839359 | 0.85 |
CD37 |
CD37 molecule |
472 |
0.63 |
| chr7_26333293_26333815 | 0.79 |
SNX10 |
sorting nexin 10 |
880 |
0.67 |
| chr8_126964027_126964178 | 0.71 |
ENSG00000206695 |
. |
50907 |
0.19 |
| chr7_30766706_30766866 | 0.62 |
INMT |
indolethylamine N-methyltransferase |
24965 |
0.16 |
| chr2_113932280_113932569 | 0.62 |
AC016683.5 |
|
490 |
0.59 |
| chr19_38908265_38908571 | 0.62 |
RASGRP4 |
RAS guanyl releasing protein 4 |
8384 |
0.09 |
| chr1_9688741_9688898 | 0.62 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
22971 |
0.15 |
| chr2_219744680_219744914 | 0.60 |
WNT10A |
wingless-type MMTV integration site family, member 10A |
288 |
0.85 |
| chr5_153582547_153582698 | 0.60 |
GALNT10 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) |
12288 |
0.25 |
| chr1_31227757_31227908 | 0.60 |
LAPTM5 |
lysosomal protein transmembrane 5 |
2835 |
0.23 |
| chr20_43597999_43598419 | 0.59 |
STK4 |
serine/threonine kinase 4 |
3042 |
0.19 |
| chr19_1074414_1074782 | 0.57 |
HMHA1 |
histocompatibility (minor) HA-1 |
2844 |
0.13 |
| chr10_121298283_121299012 | 0.57 |
RGS10 |
regulator of G-protein signaling 10 |
2602 |
0.34 |
| chr22_37545294_37545803 | 0.54 |
IL2RB |
interleukin 2 receptor, beta |
482 |
0.7 |
| chr9_134604865_134605016 | 0.54 |
ENSG00000240853 |
. |
4086 |
0.24 |
| chr7_37487305_37488000 | 0.54 |
ELMO1 |
engulfment and cell motility 1 |
901 |
0.61 |
| chrX_70837784_70838022 | 0.53 |
CXCR3 |
chemokine (C-X-C motif) receptor 3 |
457 |
0.81 |
| chr22_50232554_50232757 | 0.53 |
BRD1 |
bromodomain containing 1 |
11495 |
0.18 |
| chr15_41775317_41775468 | 0.52 |
RTF1 |
Rtf1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) |
8525 |
0.13 |
| chr17_80279192_80279518 | 0.52 |
CD7 |
CD7 molecule |
3877 |
0.13 |
| chr20_4795121_4795462 | 0.51 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
478 |
0.82 |
| chr19_54672117_54672268 | 0.50 |
TMC4 |
transmembrane channel-like 4 |
4255 |
0.08 |
| chr2_219259559_219259710 | 0.50 |
CTDSP1 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 1 |
3427 |
0.11 |
| chr12_111115977_111116128 | 0.50 |
HVCN1 |
hydrogen voltage-gated channel 1 |
10462 |
0.16 |
| chr22_18923115_18923266 | 0.50 |
PRODH |
proline dehydrogenase (oxidase) 1 |
465 |
0.79 |
| chr22_30475055_30475206 | 0.50 |
HORMAD2 |
HORMA domain containing 2 |
1033 |
0.38 |
| chr6_132113440_132113591 | 0.50 |
ENSG00000266807 |
. |
203 |
0.94 |
| chr17_38024211_38024362 | 0.50 |
ZPBP2 |
zona pellucida binding protein 2 |
131 |
0.95 |
| chr14_61788916_61789580 | 0.49 |
PRKCH |
protein kinase C, eta |
134 |
0.7 |
| chr1_8000859_8001010 | 0.49 |
TNFRSF9 |
tumor necrosis factor receptor superfamily, member 9 |
8 |
0.98 |
| chr1_92269793_92270018 | 0.49 |
ENSG00000239794 |
. |
25726 |
0.22 |
| chr17_75103749_75103900 | 0.48 |
ENSG00000234912 |
. |
18435 |
0.18 |
| chr3_71833354_71833608 | 0.48 |
PROK2 |
prokineticin 2 |
731 |
0.68 |
| chr5_176857238_176857389 | 0.47 |
GRK6 |
G protein-coupled receptor kinase 6 |
3458 |
0.12 |
| chr13_71865361_71865710 | 0.47 |
DACH1 |
dachshund homolog 1 (Drosophila) |
575372 |
0.0 |
| chr9_123688442_123688941 | 0.46 |
TRAF1 |
TNF receptor-associated factor 1 |
2356 |
0.33 |
| chr17_72755238_72755446 | 0.46 |
SLC9A3R1 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
2800 |
0.14 |
| chr16_29822482_29822845 | 0.46 |
PRRT2 |
proline-rich transmembrane protein 2 |
514 |
0.38 |
| chr16_29678066_29678245 | 0.45 |
QPRT |
quinolinate phosphoribosyltransferase |
3555 |
0.15 |
| chr20_35463892_35464165 | 0.45 |
SOGA1 |
suppressor of glucose, autophagy associated 1 |
19344 |
0.15 |
| chr1_26872928_26873171 | 0.45 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
706 |
0.6 |
| chr16_78776576_78776802 | 0.45 |
RP11-319G9.3 |
|
154373 |
0.04 |
| chr6_75458992_75459143 | 0.44 |
ENSG00000264884 |
. |
29508 |
0.26 |
| chr2_25585755_25585961 | 0.44 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
20399 |
0.2 |
| chrX_128915626_128915992 | 0.43 |
SASH3 |
SAM and SH3 domain containing 3 |
1849 |
0.35 |
| chr6_138028959_138029110 | 0.43 |
ENSG00000216097 |
. |
9051 |
0.27 |
| chr11_721054_721205 | 0.43 |
AP006621.9 |
|
5918 |
0.08 |
| chr20_62368373_62368670 | 0.42 |
LIME1 |
Lck interacting transmembrane adaptor 1 |
116 |
0.89 |
| chr12_7065402_7065553 | 0.42 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
4939 |
0.06 |
| chr5_134879321_134879990 | 0.42 |
NEUROG1 |
neurogenin 1 |
8016 |
0.18 |
| chr7_74179826_74180081 | 0.42 |
NCF1 |
neutrophil cytosolic factor 1 |
8356 |
0.2 |
| chr6_32159695_32160229 | 0.42 |
GPSM3 |
G-protein signaling modulator 3 |
683 |
0.42 |
| chr7_72626118_72626352 | 0.42 |
NCF1B |
neutrophil cytosolic factor 1B pseudogene |
8376 |
0.18 |
| chr16_17624024_17624268 | 0.41 |
XYLT1 |
xylosyltransferase I |
59408 |
0.17 |
| chr19_2083107_2083981 | 0.41 |
MOB3A |
MOB kinase activator 3A |
1847 |
0.21 |
| chr1_895544_895695 | 0.41 |
KLHL17 |
kelch-like family member 17 |
348 |
0.67 |
| chr7_74596048_74596381 | 0.41 |
GTF2IRD2B |
GTF2I repeat domain containing 2B |
39310 |
0.17 |
| chrX_30594869_30596024 | 0.41 |
CXorf21 |
chromosome X open reading frame 21 |
515 |
0.84 |
| chr6_106969857_106970345 | 0.40 |
AIM1 |
absent in melanoma 1 |
10371 |
0.2 |
| chr2_10548224_10548465 | 0.40 |
HPCAL1 |
hippocalcin-like 1 |
11803 |
0.19 |
| chr1_154300198_154300512 | 0.40 |
ATP8B2 |
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
79 |
0.94 |
| chr1_166459489_166459697 | 0.39 |
FAM78B |
family with sequence similarity 78, member B |
323387 |
0.01 |
| chr1_2455307_2455458 | 0.39 |
PANK4 |
pantothenate kinase 4 |
2653 |
0.14 |
| chr17_37932472_37932886 | 0.39 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
1799 |
0.27 |
| chr1_6526104_6526255 | 0.39 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
12 |
0.96 |
| chr19_40447803_40447954 | 0.39 |
FCGBP |
Fc fragment of IgG binding protein |
7345 |
0.17 |
| chr7_2445424_2445909 | 0.39 |
CHST12 |
carbohydrate (chondroitin 4) sulfotransferase 12 |
1932 |
0.35 |
| chr5_140997925_140998467 | 0.38 |
AC008781.7 |
|
215 |
0.56 |
| chr17_264289_264440 | 0.38 |
AC108004.3 |
|
550 |
0.69 |
| chr17_66286056_66286766 | 0.38 |
SLC16A6 |
solute carrier family 16, member 6 |
846 |
0.47 |
| chr17_43297952_43298150 | 0.38 |
CTD-2020K17.1 |
|
259 |
0.8 |
| chrX_48793999_48794379 | 0.38 |
PIM2 |
pim-2 oncogene |
17888 |
0.08 |
| chr11_69065156_69065401 | 0.38 |
MYEOV |
myeloma overexpressed |
3653 |
0.34 |
| chr20_52205540_52206428 | 0.37 |
ZNF217 |
zinc finger protein 217 |
4394 |
0.22 |
| chr2_10860609_10860760 | 0.36 |
ATP6V1C2 |
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2 |
1091 |
0.47 |
| chr12_122228420_122228575 | 0.36 |
RHOF |
ras homolog family member F (in filopodia) |
2769 |
0.23 |
| chr14_22994691_22994955 | 0.36 |
TRAJ15 |
T cell receptor alpha joining 15 |
3757 |
0.14 |
| chr19_5805561_5805712 | 0.36 |
DUS3L |
dihydrouridine synthase 3-like (S. cerevisiae) |
14387 |
0.08 |
| chr15_81586254_81586487 | 0.35 |
IL16 |
interleukin 16 |
2884 |
0.28 |
| chr10_124136372_124136523 | 0.35 |
PLEKHA1 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 1 |
2097 |
0.31 |
| chr10_135089818_135090113 | 0.35 |
ADAM8 |
ADAM metallopeptidase domain 8 |
389 |
0.74 |
| chr14_94425760_94425911 | 0.35 |
ASB2 |
ankyrin repeat and SOCS box containing 2 |
2068 |
0.26 |
| chr5_125937096_125937334 | 0.34 |
PHAX |
phosphorylated adaptor for RNA export |
1255 |
0.44 |
| chr1_207495095_207495855 | 0.34 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
339 |
0.92 |
| chr19_46283149_46283300 | 0.34 |
DMPK |
dystrophia myotonica-protein kinase |
575 |
0.52 |
| chr9_139927070_139927409 | 0.34 |
FUT7 |
fucosyltransferase 7 (alpha (1,3) fucosyltransferase) |
223 |
0.78 |
| chr6_11852612_11852763 | 0.34 |
ADTRP |
androgen-dependent TFPI-regulating protein |
45408 |
0.18 |
| chr8_59717634_59717850 | 0.34 |
ENSG00000201231 |
. |
9369 |
0.29 |
| chr22_37639776_37640249 | 0.33 |
RAC2 |
ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) |
276 |
0.88 |
| chr16_4256693_4256974 | 0.33 |
SRL |
sarcalumenin |
32911 |
0.15 |
| chr1_154474731_154474966 | 0.33 |
TDRD10 |
tudor domain containing 10 |
153 |
0.63 |
| chr19_48824738_48825145 | 0.33 |
EMP3 |
epithelial membrane protein 3 |
175 |
0.9 |
| chr10_111971435_111971586 | 0.33 |
MXI1 |
MAX interactor 1, dimerization protein |
1521 |
0.41 |
| chr6_18155802_18156067 | 0.33 |
KDM1B |
lysine (K)-specific demethylase 1B |
251 |
0.78 |
| chr3_112215334_112215665 | 0.33 |
BTLA |
B and T lymphocyte associated |
2706 |
0.33 |
| chr12_12870481_12870701 | 0.32 |
CDKN1B |
cyclin-dependent kinase inhibitor 1B (p27, Kip1) |
165 |
0.93 |
| chr10_99609976_99610583 | 0.32 |
GOLGA7B |
golgin A7 family, member B |
283 |
0.91 |
| chr15_31638589_31638828 | 0.32 |
KLF13 |
Kruppel-like factor 13 |
6572 |
0.32 |
| chr10_73869476_73869627 | 0.32 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
20761 |
0.19 |
| chr7_44790648_44790799 | 0.32 |
ZMIZ2 |
zinc finger, MIZ-type containing 2 |
2193 |
0.28 |
| chr20_48419063_48419214 | 0.31 |
ENSG00000252540 |
. |
7790 |
0.16 |
| chr17_74268969_74269120 | 0.31 |
UBALD2 |
UBA-like domain containing 2 |
7260 |
0.13 |
| chr1_208417858_208418217 | 0.31 |
PLXNA2 |
plexin A2 |
372 |
0.93 |
| chr2_240219542_240220118 | 0.31 |
ENSG00000265215 |
. |
7327 |
0.19 |
| chr3_142800787_142800938 | 0.31 |
CHST2 |
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
37311 |
0.15 |
| chr18_10451924_10452075 | 0.31 |
APCDD1 |
adenomatosis polyposis coli down-regulated 1 |
2626 |
0.36 |
| chr9_38042423_38042574 | 0.30 |
SHB |
Src homology 2 domain containing adaptor protein B |
26710 |
0.21 |
| chr19_1469984_1470717 | 0.30 |
ENSG00000267317 |
. |
7586 |
0.07 |
| chr1_16488827_16488993 | 0.30 |
RP11-276H7.3 |
|
3149 |
0.15 |
| chr7_134855680_134855894 | 0.30 |
C7orf49 |
chromosome 7 open reading frame 49 |
240 |
0.89 |
| chr1_226922169_226922483 | 0.30 |
ITPKB |
inositol-trisphosphate 3-kinase B |
2833 |
0.32 |
| chr15_79236726_79237037 | 0.30 |
CTSH |
cathepsin H |
552 |
0.76 |
| chr3_185647070_185647439 | 0.30 |
TRA2B |
transformer 2 beta homolog (Drosophila) |
5573 |
0.22 |
| chr22_31686910_31687128 | 0.30 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
1362 |
0.25 |
| chr17_73840046_73840332 | 0.30 |
UNC13D |
unc-13 homolog D (C. elegans) |
226 |
0.84 |
| chr9_123690633_123691433 | 0.30 |
TRAF1 |
TNF receptor-associated factor 1 |
14 |
0.98 |
| chr19_1068869_1069054 | 0.29 |
HMHA1 |
histocompatibility (minor) HA-1 |
1464 |
0.22 |
| chr3_119187850_119188681 | 0.29 |
POGLUT1 |
protein O-glucosyltransferase 1 |
455 |
0.54 |
| chr10_129861236_129861473 | 0.29 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
15520 |
0.26 |
| chr12_49391673_49391824 | 0.29 |
RP11-386G11.5 |
|
402 |
0.59 |
| chr7_128578118_128579002 | 0.29 |
IRF5 |
interferon regulatory factor 5 |
289 |
0.89 |
| chr4_90228457_90229530 | 0.29 |
GPRIN3 |
GPRIN family member 3 |
168 |
0.97 |
| chrX_53741093_53741244 | 0.29 |
HUWE1 |
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
27495 |
0.23 |
| chr1_226899370_226899800 | 0.29 |
ITPKB |
inositol-trisphosphate 3-kinase B |
25574 |
0.18 |
| chr1_247242173_247242440 | 0.29 |
ZNF670 |
zinc finger protein 670 |
249 |
0.91 |
| chr17_41738065_41738244 | 0.29 |
MEOX1 |
mesenchyme homeobox 1 |
777 |
0.64 |
| chr2_74153401_74153920 | 0.29 |
DGUOK |
deoxyguanosine kinase |
293 |
0.89 |
| chr22_39708334_39708626 | 0.29 |
ENSG00000209480 |
. |
1436 |
0.25 |
| chr17_47572677_47573321 | 0.28 |
NGFR |
nerve growth factor receptor |
344 |
0.86 |
| chr16_68321366_68321603 | 0.28 |
ENSG00000252026 |
. |
3263 |
0.1 |
| chr1_6520532_6520815 | 0.28 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
5017 |
0.12 |
| chr1_31229648_31230364 | 0.28 |
LAPTM5 |
lysosomal protein transmembrane 5 |
661 |
0.67 |
| chr19_11533070_11533503 | 0.28 |
RGL3 |
ral guanine nucleotide dissociation stimulator-like 3 |
3268 |
0.1 |
| chr19_50400390_50400724 | 0.28 |
IL4I1 |
interleukin 4 induced 1 |
345 |
0.67 |
| chr2_219745053_219745526 | 0.28 |
WNT10A |
wingless-type MMTV integration site family, member 10A |
204 |
0.9 |
| chr6_30798000_30798388 | 0.28 |
ENSG00000202241 |
. |
33833 |
0.07 |
| chr11_69457554_69457705 | 0.28 |
CCND1 |
cyclin D1 |
1655 |
0.41 |
| chr16_28996496_28996848 | 0.28 |
LAT |
linker for activation of T cells |
11 |
0.94 |
| chr5_150465316_150465727 | 0.28 |
TNIP1 |
TNFAIP3 interacting protein 1 |
1198 |
0.49 |
| chr1_55681102_55681610 | 0.28 |
USP24 |
ubiquitin specific peptidase 24 |
570 |
0.79 |
| chr16_80690406_80690960 | 0.28 |
ENSG00000265341 |
. |
8777 |
0.18 |
| chr16_3210084_3210235 | 0.28 |
CASP16 |
caspase 16, apoptosis-related cysteine peptidase (putative) |
15915 |
0.08 |
| chr1_16292676_16293171 | 0.28 |
ZBTB17 |
zinc finger and BTB domain containing 17 |
9651 |
0.13 |
| chr3_9690857_9691008 | 0.28 |
MTMR14 |
myotubularin related protein 14 |
185 |
0.94 |
| chr7_5183512_5184010 | 0.27 |
ZNF890P |
zinc finger protein 890, pseudogene |
16335 |
0.17 |
| chr9_139424881_139425156 | 0.27 |
ENSG00000263403 |
. |
10940 |
0.09 |
| chr11_118016214_118016693 | 0.27 |
SCN4B |
sodium channel, voltage-gated, type IV, beta subunit |
7082 |
0.15 |
| chr22_37703418_37703750 | 0.27 |
CYTH4 |
cytohesin 4 |
2425 |
0.26 |
| chr20_48329922_48330172 | 0.27 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
368 |
0.87 |
| chr8_28205045_28205788 | 0.27 |
ZNF395 |
zinc finger protein 395 |
3667 |
0.19 |
| chr19_661226_661492 | 0.27 |
RNF126 |
ring finger protein 126 |
43 |
0.95 |
| chrX_48794595_48794842 | 0.27 |
PIM2 |
pim-2 oncogene |
18417 |
0.08 |
| chr22_44392181_44392448 | 0.27 |
PARVB |
parvin, beta |
2777 |
0.29 |
| chr17_61778727_61779133 | 0.27 |
LIMD2 |
LIM domain containing 2 |
398 |
0.8 |
| chr19_46145825_46145976 | 0.26 |
EML2 |
echinoderm microtubule associated protein like 2 |
80 |
0.9 |
| chr8_21769341_21769492 | 0.26 |
DOK2 |
docking protein 2, 56kDa |
1758 |
0.36 |
| chr15_45003095_45003549 | 0.26 |
B2M |
beta-2-microglobulin |
353 |
0.83 |
| chr10_15211128_15211858 | 0.26 |
NMT2 |
N-myristoyltransferase 2 |
801 |
0.67 |
| chr20_4788995_4789146 | 0.26 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
6699 |
0.21 |
| chr19_49362125_49362344 | 0.26 |
PLEKHA4 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
397 |
0.71 |
| chr14_103285531_103285798 | 0.26 |
TRAF3 |
TNF receptor-associated factor 3 |
11027 |
0.17 |
| chr8_134069456_134069607 | 0.26 |
SLA |
Src-like-adaptor |
3072 |
0.32 |
| chr2_95691472_95692130 | 0.25 |
MAL |
mal, T-cell differentiation protein |
263 |
0.76 |
| chr15_41234314_41234975 | 0.25 |
CHAC1 |
ChaC, cation transport regulator homolog 1 (E. coli) |
10516 |
0.12 |
| chr19_18061359_18061510 | 0.25 |
KCNN1 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 |
668 |
0.61 |
| chr1_16490997_16491148 | 0.25 |
RP11-276H7.3 |
|
5311 |
0.12 |
| chr1_27113866_27114017 | 0.25 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
22 |
0.96 |
| chr13_25254086_25254684 | 0.25 |
ATP12A |
ATPase, H+/K+ transporting, nongastric, alpha polypeptide |
164 |
0.96 |
| chr6_43483991_43484670 | 0.25 |
YIPF3 |
Yip1 domain family, member 3 |
22 |
0.85 |
| chr15_86127194_86127729 | 0.25 |
RP11-815J21.2 |
|
4052 |
0.21 |
| chr2_26569363_26569689 | 0.25 |
GPR113 |
G protein-coupled receptor 113 |
85 |
0.88 |
| chr1_32739822_32739973 | 0.25 |
LCK |
lymphocyte-specific protein tyrosine kinase |
34 |
0.95 |
| chr17_38020283_38020945 | 0.25 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
173 |
0.93 |
| chr10_105437541_105438126 | 0.25 |
SH3PXD2A |
SH3 and PX domains 2A |
89 |
0.97 |
| chr7_158938159_158938310 | 0.25 |
VIPR2 |
vasoactive intestinal peptide receptor 2 |
585 |
0.86 |
| chr19_3786043_3786321 | 0.25 |
MATK |
megakaryocyte-associated tyrosine kinase |
98 |
0.94 |
| chr13_109148449_109148673 | 0.25 |
MYO16 |
myosin XVI |
99939 |
0.08 |
| chr7_44679217_44679482 | 0.25 |
OGDH |
oxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) |
15288 |
0.18 |
| chr17_56421831_56421982 | 0.24 |
SUPT4H1 |
suppressor of Ty 4 homolog 1 (S. cerevisiae) |
7116 |
0.11 |
| chr5_42924026_42924397 | 0.24 |
SEPP1 |
selenoprotein P, plasma, 1 |
36717 |
0.17 |
| chr18_13136915_13137798 | 0.24 |
RP11-794M8.1 |
|
78968 |
0.08 |
| chr10_114135452_114136486 | 0.24 |
ACSL5 |
acyl-CoA synthetase long-chain family member 5 |
12 |
0.98 |
| chr10_65021596_65021747 | 0.24 |
JMJD1C |
jumonji domain containing 1C |
7155 |
0.29 |
| chr10_75634514_75635168 | 0.24 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
498 |
0.7 |
| chr14_21571987_21572281 | 0.24 |
ZNF219 |
zinc finger protein 219 |
396 |
0.72 |
| chr19_39106930_39107372 | 0.24 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
1413 |
0.27 |
| chr9_125667781_125668094 | 0.24 |
RC3H2 |
ring finger and CCCH-type domains 2 |
317 |
0.82 |
| chr14_91879963_91880132 | 0.24 |
CCDC88C |
coiled-coil domain containing 88C |
3643 |
0.29 |
| chr1_154979027_154979178 | 0.24 |
ZBTB7B |
zinc finger and BTB domain containing 7B |
3807 |
0.09 |
| chr12_105063987_105064138 | 0.24 |
ENSG00000264295 |
. |
78651 |
0.1 |
| chr14_52313408_52314007 | 0.24 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
137 |
0.97 |
| chr22_50967987_50968138 | 0.24 |
TYMP |
thymidine phosphorylase |
357 |
0.66 |
| chr6_32158116_32158500 | 0.24 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
345 |
0.7 |
| chr19_8274217_8274814 | 0.24 |
CERS4 |
ceramide synthase 4 |
3 |
0.97 |
| chr16_57318716_57318953 | 0.24 |
PLLP |
plasmolipin |
235 |
0.9 |
| chr17_55338552_55338703 | 0.24 |
MSI2 |
musashi RNA-binding protein 2 |
4248 |
0.31 |
| chr2_25537366_25537517 | 0.24 |
ENSG00000221445 |
. |
14149 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.1 | 0.8 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.1 | 0.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.3 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.2 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.1 | 0.3 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.1 | 0.2 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.1 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.5 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.1 | 0.2 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.1 | 0.3 | GO:0032776 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.1 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.2 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.2 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.1 | 0.2 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.1 | 0.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.1 | 0.3 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 0.2 | GO:0010867 | positive regulation of triglyceride biosynthetic process(GO:0010867) |
| 0.1 | 0.2 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.1 | 0.1 | GO:0072110 | glomerular mesangial cell proliferation(GO:0072110) regulation of glomerular mesangial cell proliferation(GO:0072124) |
| 0.1 | 0.1 | GO:1903519 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) regulation of apoptotic process involved in morphogenesis(GO:1902337) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) regulation of apoptotic process involved in development(GO:1904748) |
| 0.1 | 0.3 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.1 | 0.2 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.1 | 0.3 | GO:0002475 | antigen processing and presentation via MHC class Ib(GO:0002475) |
| 0.1 | 0.3 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.1 | 0.3 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.3 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.2 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) |
| 0.0 | 0.5 | GO:0045730 | respiratory burst(GO:0045730) |
| 0.0 | 0.1 | GO:0002579 | positive regulation of antigen processing and presentation(GO:0002579) positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.2 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.2 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.2 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.2 | GO:0010457 | centriole-centriole cohesion(GO:0010457) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.2 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.2 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.2 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.2 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.3 | GO:0006000 | fructose metabolic process(GO:0006000) |
| 0.0 | 0.3 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.2 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.5 | GO:0035036 | binding of sperm to zona pellucida(GO:0007339) sperm-egg recognition(GO:0035036) |
| 0.0 | 0.2 | GO:0042117 | monocyte activation(GO:0042117) |
| 0.0 | 0.5 | GO:0002920 | regulation of humoral immune response(GO:0002920) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.3 | GO:1904377 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.6 | GO:0033209 | tumor necrosis factor-mediated signaling pathway(GO:0033209) |
| 0.0 | 0.4 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.0 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0031507 | heterochromatin assembly(GO:0031507) heterochromatin organization(GO:0070828) |
| 0.0 | 0.1 | GO:0034375 | high-density lipoprotein particle remodeling(GO:0034375) |
| 0.0 | 0.2 | GO:0006857 | oligopeptide transport(GO:0006857) |
| 0.0 | 0.3 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.3 | GO:0016601 | Rac protein signal transduction(GO:0016601) |
| 0.0 | 0.5 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.1 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0009313 | oligosaccharide catabolic process(GO:0009313) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.0 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.0 | GO:0035844 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.2 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.0 | GO:0048548 | regulation of pinocytosis(GO:0048548) |
| 0.0 | 0.3 | GO:0031648 | protein destabilization(GO:0031648) |
| 0.0 | 0.1 | GO:0060317 | cardiac epithelial to mesenchymal transition(GO:0060317) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.0 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.1 | GO:1902622 | regulation of neutrophil chemotaxis(GO:0090022) regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.0 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.1 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.0 | GO:0010954 | positive regulation of protein processing(GO:0010954) positive regulation of protein maturation(GO:1903319) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.0 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0045824 | negative regulation of innate immune response(GO:0045824) |
| 0.0 | 0.1 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0002467 | germinal center formation(GO:0002467) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.1 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.0 | 0.2 | GO:0043097 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.2 | GO:0015858 | nucleoside transport(GO:0015858) |
| 0.0 | 0.1 | GO:0008356 | asymmetric cell division(GO:0008356) |
| 0.0 | 0.1 | GO:0060122 | inner ear receptor stereocilium organization(GO:0060122) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.1 | GO:0034227 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0042789 | mRNA transcription from RNA polymerase II promoter(GO:0042789) |
| 0.0 | 0.1 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.0 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.1 | GO:0032329 | serine transport(GO:0032329) |
| 0.0 | 0.0 | GO:0071624 | positive regulation of granulocyte chemotaxis(GO:0071624) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0070189 | kynurenine metabolic process(GO:0070189) |
| 0.0 | 0.0 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.0 | GO:0000966 | RNA 5'-end processing(GO:0000966) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0006777 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.0 | GO:0032495 | response to muramyl dipeptide(GO:0032495) |
| 0.0 | 0.1 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.0 | GO:2000193 | positive regulation of fatty acid transport(GO:2000193) |
| 0.0 | 0.0 | GO:0003077 | obsolete negative regulation of diuresis(GO:0003077) |
| 0.0 | 0.1 | GO:0070301 | cellular response to hydrogen peroxide(GO:0070301) |
| 0.0 | 0.1 | GO:0009109 | coenzyme catabolic process(GO:0009109) |
| 0.0 | 0.1 | GO:0044110 | growth involved in symbiotic interaction(GO:0044110) growth of symbiont involved in interaction with host(GO:0044116) growth of symbiont in host(GO:0044117) regulation of growth of symbiont in host(GO:0044126) negative regulation of growth of symbiont in host(GO:0044130) modulation of growth of symbiont involved in interaction with host(GO:0044144) negative regulation of growth of symbiont involved in interaction with host(GO:0044146) |
| 0.0 | 0.2 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.0 | GO:0060413 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.0 | 0.1 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.0 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.0 | 0.1 | GO:0006878 | cellular copper ion homeostasis(GO:0006878) |
| 0.0 | 0.0 | GO:0032375 | negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) |
| 0.0 | 0.0 | GO:0007042 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.0 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.1 | GO:0008089 | anterograde axonal transport(GO:0008089) |
| 0.0 | 0.0 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.0 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0090398 | cellular senescence(GO:0090398) |
| 0.0 | 0.1 | GO:0035235 | ionotropic glutamate receptor signaling pathway(GO:0035235) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.0 | GO:0048752 | semicircular canal morphogenesis(GO:0048752) |
| 0.0 | 0.0 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.0 | GO:0048799 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.8 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.1 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.0 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0045945 | positive regulation of transcription from RNA polymerase III promoter(GO:0045945) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.6 | GO:0000288 | nuclear-transcribed mRNA catabolic process, deadenylation-dependent decay(GO:0000288) |
| 0.0 | 0.3 | GO:0015914 | phospholipid transport(GO:0015914) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.2 | GO:0009268 | response to pH(GO:0009268) |
| 0.0 | 0.0 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.0 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0015012 | heparan sulfate proteoglycan biosynthetic process(GO:0015012) |
| 0.0 | 0.4 | GO:0043966 | histone H3 acetylation(GO:0043966) |
| 0.0 | 0.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.0 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 0.0 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.4 | GO:0042552 | myelination(GO:0042552) |
| 0.0 | 0.0 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.2 | GO:0042992 | negative regulation of transcription factor import into nucleus(GO:0042992) |
| 0.0 | 0.0 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.0 | 0.2 | GO:0006342 | chromatin silencing(GO:0006342) |
| 0.0 | 0.1 | GO:0042401 | polyamine biosynthetic process(GO:0006596) cellular biogenic amine biosynthetic process(GO:0042401) |
| 0.0 | 0.0 | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.0 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.0 | GO:0010523 | negative regulation of calcium ion transport into cytosol(GO:0010523) |
| 0.0 | 0.2 | GO:0019835 | cytolysis(GO:0019835) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.2 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.0 | GO:0002823 | negative regulation of adaptive immune response(GO:0002820) negative regulation of adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains(GO:0002823) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.3 | GO:0070820 | tertiary granule(GO:0070820) |
| 0.1 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.1 | 0.3 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.1 | 0.2 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.1 | 0.2 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.2 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.2 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.3 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.3 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.4 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.3 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.2 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.0 | GO:0016600 | flotillin complex(GO:0016600) |
| 0.0 | 0.2 | GO:0042597 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0043230 | extracellular organelle(GO:0043230) |
| 0.0 | 0.0 | GO:0016580 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.7 | GO:0031902 | late endosome membrane(GO:0031902) |
| 0.0 | 0.1 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.0 | 0.2 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.2 | GO:0032420 | stereocilium(GO:0032420) |
| 0.0 | 0.1 | GO:0032040 | small-subunit processome(GO:0032040) |
| 0.0 | 0.0 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.2 | GO:0000307 | cyclin-dependent protein kinase holoenzyme complex(GO:0000307) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.5 | GO:0017053 | transcriptional repressor complex(GO:0017053) |
| 0.0 | 0.1 | GO:0000153 | cytoplasmic ubiquitin ligase complex(GO:0000153) |
| 0.0 | 0.2 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.1 | 0.3 | GO:0034648 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.3 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.2 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.1 | 0.5 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.1 | 0.3 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.1 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.1 | 0.3 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.1 | 0.7 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.1 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.2 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.1 | 0.2 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.1 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.6 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.1 | 0.3 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.3 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.5 | GO:0034979 | NAD-dependent protein deacetylase activity(GO:0034979) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.2 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.2 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.2 | GO:0050664 | oxidoreductase activity, acting on NAD(P)H, oxygen as acceptor(GO:0050664) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.3 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0046979 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.2 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.3 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.2 | GO:0046527 | glucosyltransferase activity(GO:0046527) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.4 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.0 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.2 | GO:0052866 | phosphatidylinositol phosphate phosphatase activity(GO:0052866) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.4 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.2 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.4 | GO:0016645 | oxidoreductase activity, acting on the CH-NH group of donors(GO:0016645) |
| 0.0 | 0.2 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.2 | GO:0008430 | selenium binding(GO:0008430) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0061659 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0005391 | sodium:potassium-exchanging ATPase activity(GO:0005391) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.2 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0015556 | C4-dicarboxylate transmembrane transporter activity(GO:0015556) |
| 0.0 | 0.6 | GO:0004722 | protein serine/threonine phosphatase activity(GO:0004722) |
| 0.0 | 0.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.2 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.2 | GO:0016986 | obsolete transcription initiation factor activity(GO:0016986) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0030250 | cyclase activator activity(GO:0010853) guanylate cyclase activator activity(GO:0030250) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.7 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.0 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.0 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.0 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0017150 | tRNA dihydrouridine synthase activity(GO:0017150) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.0 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
| 0.0 | 0.1 | GO:0070567 | cytidylyltransferase activity(GO:0070567) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 1.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 1.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.5 | PID KIT PATHWAY | Signaling events mediated by Stem cell factor receptor (c-Kit) |
| 0.0 | 0.9 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.6 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 1.0 | PID CD8 TCR DOWNSTREAM PATHWAY | Downstream signaling in naïve CD8+ T cells |
| 0.0 | 0.4 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.2 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | PID IFNG PATHWAY | IFN-gamma pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.2 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.1 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.1 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.3 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.8 | WNT SIGNALING | Genes related to Wnt-mediated signal transduction |
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.6 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.3 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.9 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.2 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.0 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.4 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.1 | REACTOME INTEGRATION OF ENERGY METABOLISM | Genes involved in Integration of energy metabolism |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.1 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME PIP3 ACTIVATES AKT SIGNALING | Genes involved in PIP3 activates AKT signaling |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.3 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.4 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | Genes involved in CREB phosphorylation through the activation of Ras |
| 0.0 | 0.7 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.3 | REACTOME INHIBITION OF THE PROTEOLYTIC ACTIVITY OF APC C REQUIRED FOR THE ONSET OF ANAPHASE BY MITOTIC SPINDLE CHECKPOINT COMPONENTS | Genes involved in Inhibition of the proteolytic activity of APC/C required for the onset of anaphase by mitotic spindle checkpoint components |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.1 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.5 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.1 | REACTOME IL 3 5 AND GM CSF SIGNALING | Genes involved in Interleukin-3, 5 and GM-CSF signaling |
| 0.0 | 0.2 | REACTOME PRE NOTCH EXPRESSION AND PROCESSING | Genes involved in Pre-NOTCH Expression and Processing |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.4 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.3 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.5 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |