Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
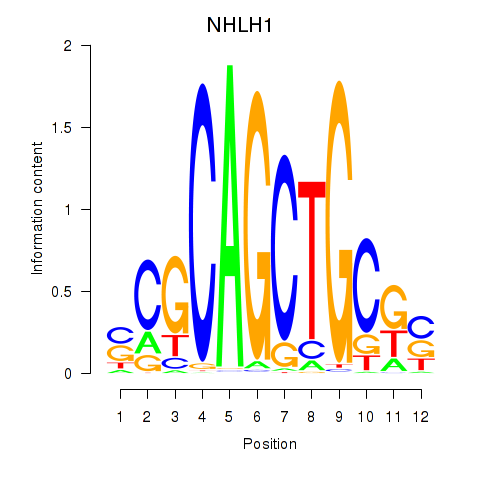
Results for NHLH1
Z-value: 1.21
Transcription factors associated with NHLH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NHLH1
|
ENSG00000171786.5 | nescient helix-loop-helix 1 |
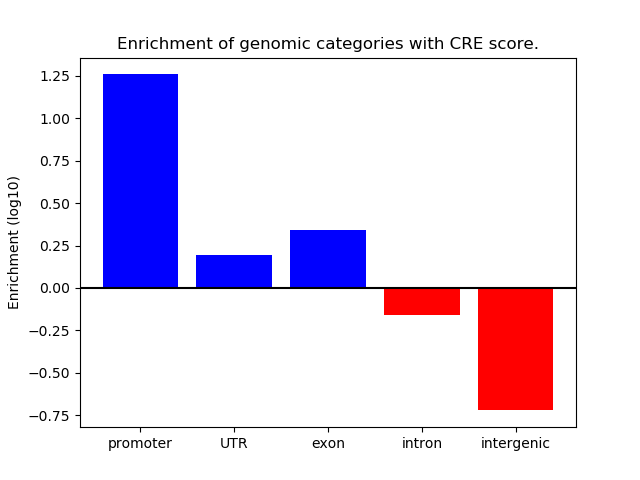
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_160346259_160346410 | NHLH1 | 9477 | 0.114862 | -0.69 | 4.1e-02 | Click! |
| chr1_160346041_160346210 | NHLH1 | 9268 | 0.115183 | -0.40 | 2.9e-01 | Click! |
| chr1_160345564_160345744 | NHLH1 | 8797 | 0.115952 | 0.08 | 8.4e-01 | Click! |
Activity of the NHLH1 motif across conditions
Conditions sorted by the z-value of the NHLH1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
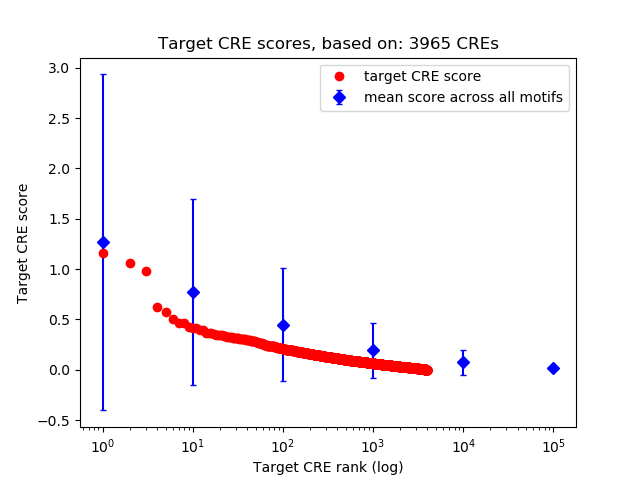
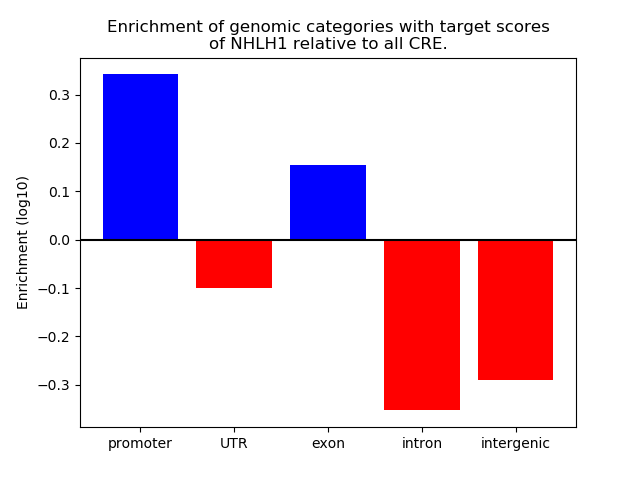
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_1092852_1093142 | 1.16 |
IDI1 |
isopentenyl-diphosphate delta isomerase 1 |
1340 |
0.3 |
| chr10_1092625_1092849 | 1.06 |
IDI1 |
isopentenyl-diphosphate delta isomerase 1 |
1600 |
0.26 |
| chr10_1092407_1092589 | 0.98 |
IDI1 |
isopentenyl-diphosphate delta isomerase 1 |
1839 |
0.23 |
| chr18_11688870_11689119 | 0.63 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
39 |
0.98 |
| chr13_26624895_26625081 | 0.58 |
SHISA2 |
shisa family member 2 |
181 |
0.97 |
| chr11_65374818_65375338 | 0.51 |
MAP3K11 |
mitogen-activated protein kinase kinase kinase 11 |
581 |
0.5 |
| chr22_31644425_31644631 | 0.47 |
LIMK2 |
LIM domain kinase 2 |
55 |
0.96 |
| chr9_117880836_117880987 | 0.46 |
TNC |
tenascin C |
375 |
0.9 |
| chr19_3061066_3061410 | 0.43 |
AES |
amino-terminal enhancer of split |
164 |
0.92 |
| chr12_115120954_115121265 | 0.42 |
TBX3 |
T-box 3 |
286 |
0.93 |
| chr11_64012846_64013110 | 0.42 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
310 |
0.51 |
| chrX_51637827_51638413 | 0.40 |
MAGED1 |
melanoma antigen family D, 1 |
1385 |
0.5 |
| chr7_55087325_55087551 | 0.40 |
EGFR |
epidermal growth factor receptor |
627 |
0.83 |
| chr3_128720113_128720357 | 0.37 |
EFCC1 |
EF-hand and coiled-coil domain containing 1 |
237 |
0.9 |
| chr1_27692801_27692961 | 0.36 |
MAP3K6 |
mitogen-activated protein kinase kinase kinase 6 |
468 |
0.71 |
| chr8_141599151_141599550 | 0.36 |
AGO2 |
argonaute RISC catalytic component 2 |
16637 |
0.22 |
| chr14_103523327_103523546 | 0.35 |
CDC42BPB |
CDC42 binding protein kinase beta (DMPK-like) |
363 |
0.87 |
| chr15_83953612_83953905 | 0.35 |
BNC1 |
basonuclin 1 |
292 |
0.93 |
| chr19_13616677_13616828 | 0.35 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
286 |
0.93 |
| chr7_157483101_157483509 | 0.35 |
AC006003.3 |
|
49843 |
0.15 |
| chr11_6341290_6341712 | 0.34 |
PRKCDBP |
protein kinase C, delta binding protein |
261 |
0.9 |
| chr1_19401262_19401413 | 0.34 |
UBR4 |
ubiquitin protein ligase E3 component n-recognin 4 |
7397 |
0.2 |
| chr6_99796840_99797257 | 0.33 |
FAXC |
failed axon connections homolog (Drosophila) |
483 |
0.83 |
| chr14_105399017_105399245 | 0.33 |
PLD4 |
phospholipase D family, member 4 |
634 |
0.69 |
| chr1_2125005_2125156 | 0.33 |
C1orf86 |
chromosome 1 open reading frame 86 |
1100 |
0.36 |
| chr3_64008917_64009121 | 0.33 |
PSMD6 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
113 |
0.94 |
| chr17_46620793_46621086 | 0.32 |
HOXB-AS1 |
HOXB cluster antisense RNA 1 |
26 |
0.94 |
| chr17_7493173_7493324 | 0.32 |
SOX15 |
SRY (sex determining region Y)-box 15 |
142 |
0.84 |
| chr4_120549971_120550122 | 0.31 |
PDE5A |
phosphodiesterase 5A, cGMP-specific |
100 |
0.98 |
| chr10_93392557_93392792 | 0.31 |
PPP1R3C |
protein phosphatase 1, regulatory subunit 3C |
137 |
0.98 |
| chr10_88728773_88729195 | 0.31 |
ENSG00000272734 |
. |
49 |
0.52 |
| chr5_112630315_112630670 | 0.31 |
MCC |
mutated in colorectal cancers |
154 |
0.97 |
| chr9_102058986_102059155 | 0.31 |
ENSG00000222337 |
. |
12766 |
0.24 |
| chr17_1618285_1618721 | 0.30 |
ENSG00000186594 |
. |
1218 |
0.24 |
| chr10_118500836_118501537 | 0.30 |
HSPA12A |
heat shock 70kDa protein 12A |
899 |
0.64 |
| chr5_72922368_72922731 | 0.30 |
ARHGEF28 |
Rho guanine nucleotide exchange factor (GEF) 28 |
528 |
0.79 |
| chr2_220306830_220307128 | 0.30 |
SPEG |
SPEG complex locus |
206 |
0.88 |
| chr11_804070_804221 | 0.30 |
PIDD |
p53-induced death domain protein |
324 |
0.68 |
| chr2_196521999_196522280 | 0.30 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
34 |
0.98 |
| chr4_144435743_144436361 | 0.30 |
SMARCA5-AS1 |
SMARCA5 antisense RNA 1 |
264 |
0.9 |
| chr13_114566489_114566848 | 0.29 |
GAS6 |
growth arrest-specific 6 |
372 |
0.9 |
| chr1_2461361_2461622 | 0.29 |
HES5 |
hes family bHLH transcription factor 5 |
193 |
0.88 |
| chr17_62051691_62051850 | 0.29 |
SCN4A |
sodium channel, voltage-gated, type IV, alpha subunit |
1492 |
0.3 |
| chr20_50073707_50073858 | 0.29 |
ENSG00000266761 |
. |
4268 |
0.31 |
| chr11_64950203_64950490 | 0.29 |
CAPN1 |
calpain 1, (mu/I) large subunit |
413 |
0.62 |
| chr10_134001657_134001828 | 0.29 |
DPYSL4 |
dihydropyrimidinase-like 4 |
1338 |
0.42 |
| chrX_1572414_1572620 | 0.29 |
ASMTL |
acetylserotonin O-methyltransferase-like |
138 |
0.96 |
| chr14_105767785_105767936 | 0.29 |
BRF1 |
BRF1, RNA polymerase III transcription initiation factor 90 kDa subunit |
245 |
0.85 |
| chr7_7812087_7812527 | 0.28 |
RPA3-AS1 |
RPA3 antisense RNA 1 |
315 |
0.93 |
| chrY_1522413_1522621 | 0.28 |
NA |
NA |
> 106 |
NA |
| chr11_96040354_96040983 | 0.27 |
ENSG00000266192 |
. |
33934 |
0.17 |
| chr4_52917326_52917683 | 0.27 |
SPATA18 |
spermatogenesis associated 18 |
7 |
0.98 |
| chr1_110230464_110231016 | 0.27 |
GSTM1 |
glutathione S-transferase mu 1 |
260 |
0.84 |
| chr18_11908463_11908810 | 0.27 |
MPPE1 |
metallophosphoesterase 1 |
5 |
0.55 |
| chr11_133939469_133939706 | 0.27 |
JAM3 |
junctional adhesion molecule 3 |
621 |
0.79 |
| chr13_114908669_114908820 | 0.27 |
RASA3 |
RAS p21 protein activator 3 |
10658 |
0.22 |
| chr11_789656_789830 | 0.27 |
CEND1 |
cell cycle exit and neuronal differentiation 1 |
380 |
0.63 |
| chr1_227505826_227506086 | 0.26 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
130 |
0.98 |
| chr6_126111801_126112009 | 0.26 |
NCOA7 |
nuclear receptor coactivator 7 |
96 |
0.97 |
| chr5_1295340_1295548 | 0.25 |
TERT |
telomerase reverse transcriptase |
282 |
0.9 |
| chrX_2418812_2418977 | 0.25 |
ZBED1 |
zinc finger, BED-type containing 1 |
42 |
0.6 |
| chr15_67356472_67356683 | 0.25 |
SMAD3 |
SMAD family member 3 |
476 |
0.88 |
| chr16_29910277_29910428 | 0.25 |
SEZ6L2 |
seizure related 6 homolog (mouse)-like 2 |
33 |
0.93 |
| chr8_141598821_141598972 | 0.25 |
AGO2 |
argonaute RISC catalytic component 2 |
17091 |
0.22 |
| chr2_217235630_217236249 | 0.25 |
MARCH4 |
membrane-associated ring finger (C3HC4) 4, E3 ubiquitin protein ligase |
811 |
0.59 |
| chrY_2368812_2368979 | 0.24 |
ENSG00000251841 |
. |
283895 |
0.01 |
| chr1_152080352_152080527 | 0.24 |
TCHH |
trichohyalin |
6117 |
0.14 |
| chr2_131797674_131798057 | 0.24 |
ARHGEF4 |
Rho guanine nucleotide exchange factor (GEF) 4 |
985 |
0.6 |
| chr10_103347501_103347768 | 0.24 |
POLL |
polymerase (DNA directed), lambda |
249 |
0.65 |
| chr15_27212862_27213085 | 0.24 |
GABRG3 |
gamma-aminobutyric acid (GABA) A receptor, gamma 3 |
3456 |
0.3 |
| chr16_56660545_56660940 | 0.24 |
MT1E |
metallothionein 1E |
1032 |
0.26 |
| chr3_73673648_73674178 | 0.24 |
PDZRN3 |
PDZ domain containing ring finger 3 |
78 |
0.96 |
| chr14_104902871_104903022 | 0.24 |
ENSG00000222761 |
. |
35561 |
0.19 |
| chr5_177541312_177541594 | 0.24 |
N4BP3 |
NEDD4 binding protein 3 |
1009 |
0.51 |
| chr15_83952244_83952401 | 0.24 |
BNC1 |
basonuclin 1 |
251 |
0.94 |
| chr16_67241052_67241203 | 0.23 |
LRRC29 |
leucine rich repeat containing 29 |
3861 |
0.08 |
| chr16_88701814_88701965 | 0.23 |
IL17C |
interleukin 17C |
3112 |
0.13 |
| chr1_114695259_114695512 | 0.23 |
SYT6 |
synaptotagmin VI |
154 |
0.97 |
| chr12_103352417_103352568 | 0.23 |
PAH |
phenylalanine hydroxylase |
348 |
0.8 |
| chr20_3730561_3730712 | 0.23 |
C20orf27 |
chromosome 20 open reading frame 27 |
8689 |
0.12 |
| chr6_84761983_84762373 | 0.23 |
MRAP2 |
melanocortin 2 receptor accessory protein 2 |
18703 |
0.28 |
| chr12_123459866_123460094 | 0.23 |
OGFOD2 |
2-oxoglutarate and iron-dependent oxygenase domain containing 2 |
194 |
0.53 |
| chr20_24615638_24615855 | 0.22 |
SYNDIG1 |
synapse differentiation inducing 1 |
165911 |
0.04 |
| chr1_1474981_1475180 | 0.22 |
TMEM240 |
transmembrane protein 240 |
657 |
0.54 |
| chr6_167413267_167413526 | 0.22 |
FGFR1OP |
FGFR1 oncogene partner |
500 |
0.57 |
| chrX_100334236_100334411 | 0.22 |
TMEM35 |
transmembrane protein 35 |
614 |
0.68 |
| chr20_62959121_62959318 | 0.22 |
PCMTD2 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
62354 |
0.13 |
| chr6_170598495_170598646 | 0.22 |
DLL1 |
delta-like 1 (Drosophila) |
991 |
0.41 |
| chr13_114524953_114525104 | 0.22 |
GAS6-AS1 |
GAS6 antisense RNA 1 |
6425 |
0.22 |
| chr2_192110869_192111020 | 0.22 |
MYO1B |
myosin IB |
65 |
0.98 |
| chr10_135106416_135106777 | 0.22 |
TUBGCP2 |
tubulin, gamma complex associated protein 2 |
2058 |
0.16 |
| chr2_63279236_63279503 | 0.22 |
OTX1 |
orthodenticle homeobox 1 |
1432 |
0.46 |
| chr11_72532728_72533160 | 0.21 |
ATG16L2 |
autophagy related 16-like 2 (S. cerevisiae) |
423 |
0.77 |
| chr14_23308247_23308521 | 0.21 |
MMP14 |
matrix metallopeptidase 14 (membrane-inserted) |
1863 |
0.16 |
| chr1_155293290_155293460 | 0.21 |
RUSC1 |
RUN and SH3 domain containing 1 |
353 |
0.56 |
| chr16_9058022_9058398 | 0.21 |
USP7 |
ubiquitin specific peptidase 7 (herpes virus-associated) |
161 |
0.96 |
| chr17_18266986_18267178 | 0.21 |
SHMT1 |
serine hydroxymethyltransferase 1 (soluble) |
226 |
0.89 |
| chr6_26614195_26614680 | 0.21 |
ABT1 |
activator of basal transcription 1 |
17257 |
0.17 |
| chr18_60277973_60278124 | 0.21 |
ENSG00000243549 |
. |
39786 |
0.17 |
| chr16_56677548_56677767 | 0.21 |
MT1DP |
metallothionein 1D, pseudogene |
20 |
0.93 |
| chr20_13202499_13202944 | 0.21 |
ISM1 |
isthmin 1, angiogenesis inhibitor |
303 |
0.91 |
| chr6_116381607_116382020 | 0.21 |
FRK |
fyn-related kinase |
108 |
0.97 |
| chr1_31886215_31886421 | 0.21 |
SERINC2 |
serine incorporator 2 |
342 |
0.86 |
| chrX_107068956_107069440 | 0.20 |
MID2 |
midline 2 |
89 |
0.97 |
| chr11_119186999_119187624 | 0.20 |
MCAM |
melanoma cell adhesion molecule |
515 |
0.61 |
| chr8_145909540_145910009 | 0.20 |
ARHGAP39 |
Rho GTPase activating protein 39 |
1420 |
0.34 |
| chr17_1011223_1011414 | 0.20 |
ABR |
active BCR-related |
1022 |
0.58 |
| chr4_62067754_62067905 | 0.20 |
LPHN3 |
latrophilin 3 |
31 |
0.99 |
| chr18_23806471_23806681 | 0.20 |
TAF4B |
TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa |
126 |
0.97 |
| chrX_139586225_139586732 | 0.20 |
SOX3 |
SRY (sex determining region Y)-box 3 |
747 |
0.78 |
| chr9_135038389_135038692 | 0.20 |
NTNG2 |
netrin G2 |
1206 |
0.56 |
| chr22_24383455_24383606 | 0.20 |
GSTT1 |
glutathione S-transferase theta 1 |
712 |
0.53 |
| chr10_118608958_118609220 | 0.20 |
ENO4 |
enolase family member 4 |
5 |
0.62 |
| chr11_86666090_86666398 | 0.20 |
FZD4 |
frizzled family receptor 4 |
189 |
0.96 |
| chr12_58120609_58120962 | 0.20 |
AGAP2-AS1 |
AGAP2 antisense RNA 1 |
731 |
0.39 |
| chr1_205417300_205417451 | 0.20 |
ENSG00000199059 |
. |
151 |
0.94 |
| chr4_77870925_77871335 | 0.20 |
SEPT11 |
septin 11 |
115 |
0.97 |
| chr19_45281152_45281359 | 0.20 |
CBLC |
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
129 |
0.93 |
| chr12_90102796_90103077 | 0.20 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
141 |
0.96 |
| chrX_140270541_140271303 | 0.20 |
LDOC1 |
leucine zipper, down-regulated in cancer 1 |
388 |
0.86 |
| chr6_72596608_72597181 | 0.19 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
167 |
0.98 |
| chr1_20811158_20811808 | 0.19 |
CAMK2N1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
1230 |
0.48 |
| chr19_59070035_59070244 | 0.19 |
UBE2M |
ubiquitin-conjugating enzyme E2M |
182 |
0.66 |
| chr11_70245277_70245996 | 0.19 |
CTTN |
cortactin |
989 |
0.33 |
| chr14_62583839_62584088 | 0.19 |
RP11-355I22.7 |
|
6370 |
0.33 |
| chrX_40944624_40944775 | 0.19 |
USP9X |
ubiquitin specific peptidase 9, X-linked |
189 |
0.97 |
| chr22_39686630_39686781 | 0.19 |
RP3-333H23.8 |
|
2288 |
0.19 |
| chr16_15083543_15083773 | 0.19 |
PDXDC1 |
pyridoxal-dependent decarboxylase domain containing 1 |
13038 |
0.11 |
| chr16_89988377_89988901 | 0.19 |
TUBB3 |
Tubulin beta-3 chain |
315 |
0.55 |
| chr22_24353470_24353828 | 0.19 |
AP000351.8 |
|
1568 |
0.24 |
| chr22_20906440_20906695 | 0.19 |
MED15 |
mediator complex subunit 15 |
990 |
0.46 |
| chr6_1637512_1637663 | 0.19 |
FOXC1 |
forkhead box C1 |
26906 |
0.26 |
| chr19_35600814_35600965 | 0.19 |
HPN-AS1 |
HPN antisense RNA 1 |
3681 |
0.11 |
| chr6_170501628_170501779 | 0.19 |
RP11-302L19.1 |
|
23962 |
0.21 |
| chr6_153303975_153304186 | 0.19 |
FBXO5 |
F-box protein 5 |
73 |
0.93 |
| chr9_116356768_116357019 | 0.19 |
RGS3 |
regulator of G-protein signaling 3 |
1127 |
0.54 |
| chr1_17914943_17915229 | 0.19 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
175 |
0.97 |
| chr7_44298813_44298964 | 0.19 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
16983 |
0.18 |
| chr2_242499136_242499442 | 0.18 |
BOK-AS1 |
BOK antisense RNA 1 |
897 |
0.38 |
| chr14_90863482_90863819 | 0.18 |
CALM1 |
calmodulin 1 (phosphorylase kinase, delta) |
250 |
0.92 |
| chr13_96056493_96056697 | 0.18 |
CLDN10 |
claudin 10 |
29263 |
0.19 |
| chr6_41438506_41438660 | 0.18 |
RP11-328M4.2 |
|
75229 |
0.08 |
| chr9_98224571_98225205 | 0.18 |
PTCH1 |
patched 1 |
17879 |
0.18 |
| chr11_128646889_128647040 | 0.18 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
12279 |
0.22 |
| chr1_193154702_193155811 | 0.18 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
528 |
0.8 |
| chr2_75426626_75426804 | 0.18 |
TACR1 |
tachykinin receptor 1 |
111 |
0.98 |
| chr9_90328572_90328919 | 0.18 |
CTSL |
cathepsin L |
11689 |
0.2 |
| chr17_80832203_80832354 | 0.18 |
TBCD |
tubulin folding cofactor D |
9750 |
0.18 |
| chr19_35521533_35521780 | 0.18 |
SCN1B |
sodium channel, voltage-gated, type I, beta subunit |
19 |
0.95 |
| chr7_150498130_150498281 | 0.18 |
TMEM176B |
transmembrane protein 176B |
243 |
0.66 |
| chr3_57996038_57996378 | 0.18 |
FLNB |
filamin B, beta |
2081 |
0.42 |
| chr2_73613395_73613732 | 0.18 |
ALMS1 |
Alstrom syndrome 1 |
677 |
0.73 |
| chr19_6532612_6532841 | 0.18 |
TNFSF9 |
tumor necrosis factor (ligand) superfamily, member 9 |
1716 |
0.23 |
| chr8_92082532_92082845 | 0.18 |
OTUD6B |
OTU domain containing 6B |
135 |
0.67 |
| chr8_22443451_22443742 | 0.18 |
AC037459.4 |
Uncharacterized protein |
3191 |
0.12 |
| chr12_47164319_47164534 | 0.18 |
SLC38A4 |
solute carrier family 38, member 4 |
1339 |
0.62 |
| chr13_111040147_111040457 | 0.18 |
ENSG00000238629 |
. |
26250 |
0.19 |
| chr10_104210626_104210853 | 0.17 |
ENSG00000269609 |
. |
351 |
0.57 |
| chr8_65494091_65494563 | 0.17 |
RP11-21C4.1 |
|
118 |
0.96 |
| chr6_28554604_28555086 | 0.17 |
SCAND3 |
SCAN domain containing 3 |
267 |
0.92 |
| chr20_60925711_60925862 | 0.17 |
RP11-157P1.5 |
|
2265 |
0.19 |
| chr8_29522987_29523177 | 0.17 |
ENSG00000221003 |
. |
263039 |
0.01 |
| chr7_65822632_65822783 | 0.17 |
ENSG00000252126 |
. |
13416 |
0.2 |
| chr5_134944090_134944241 | 0.17 |
CXCL14 |
chemokine (C-X-C motif) ligand 14 |
29196 |
0.16 |
| chr17_47653239_47653774 | 0.17 |
NXPH3 |
neurexophilin 3 |
22 |
0.95 |
| chr10_27531146_27531464 | 0.17 |
ACBD5 |
acyl-CoA binding domain containing 5 |
246 |
0.89 |
| chr19_17185147_17185420 | 0.17 |
HAUS8 |
HAUS augmin-like complex, subunit 8 |
727 |
0.47 |
| chr12_72666876_72667203 | 0.17 |
TRHDE |
thyrotropin-releasing hormone degrading enzyme |
224 |
0.85 |
| chr4_170037457_170037608 | 0.17 |
RP11-327O17.2 |
|
85413 |
0.08 |
| chr9_96589949_96590137 | 0.17 |
ENSG00000265347 |
. |
8404 |
0.29 |
| chr19_10531284_10531435 | 0.17 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
28 |
0.9 |
| chr6_100051015_100051219 | 0.17 |
PRDM13 |
PR domain containing 13 |
3489 |
0.22 |
| chr17_78833031_78833182 | 0.17 |
RP11-28G8.1 |
|
53674 |
0.1 |
| chr6_150390611_150390768 | 0.17 |
ULBP3 |
UL16 binding protein 3 |
458 |
0.82 |
| chrX_82766329_82766480 | 0.17 |
RP3-326L13.2 |
|
2182 |
0.36 |
| chr19_38146354_38146681 | 0.17 |
ZFP30 |
ZFP30 zinc finger protein |
149 |
0.95 |
| chrX_69654840_69655096 | 0.17 |
DLG3 |
discs, large homolog 3 (Drosophila) |
9743 |
0.14 |
| chr12_50015766_50015944 | 0.17 |
PRPF40B |
PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) |
1364 |
0.35 |
| chr19_5904904_5905447 | 0.16 |
VMAC |
vimentin-type intermediate filament associated coiled-coil protein |
306 |
0.68 |
| chr19_4304677_4304951 | 0.16 |
FSD1 |
fibronectin type III and SPRY domain containing 1 |
128 |
0.91 |
| chr14_23447268_23447766 | 0.16 |
AJUBA |
ajuba LIM protein |
571 |
0.5 |
| chr19_21666073_21666487 | 0.16 |
ZNF429 |
zinc finger protein 429 |
22086 |
0.22 |
| chr20_62185445_62185854 | 0.16 |
C20orf195 |
chromosome 20 open reading frame 195 |
153 |
0.91 |
| chr4_176593006_176593157 | 0.16 |
GPM6A |
glycoprotein M6A |
115457 |
0.06 |
| chr6_153451642_153452011 | 0.16 |
RGS17 |
regulator of G-protein signaling 17 |
558 |
0.79 |
| chr17_19771318_19771635 | 0.16 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
227 |
0.94 |
| chr16_81349076_81349337 | 0.16 |
GAN |
gigaxonin |
649 |
0.67 |
| chr22_46469619_46469920 | 0.16 |
RP6-109B7.4 |
|
3998 |
0.12 |
| chr6_3455456_3455921 | 0.16 |
SLC22A23 |
solute carrier family 22, member 23 |
589 |
0.84 |
| chr17_6459033_6459314 | 0.16 |
PITPNM3 |
PITPNM family member 3 |
607 |
0.7 |
| chr15_95869891_95870263 | 0.16 |
ENSG00000222076 |
. |
418956 |
0.01 |
| chrX_48930077_48930476 | 0.16 |
PRAF2 |
PRA1 domain family, member 2 |
1372 |
0.22 |
| chr10_17272457_17272817 | 0.16 |
VIM |
vimentin |
29 |
0.92 |
| chr11_120206324_120206586 | 0.16 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
1332 |
0.42 |
| chr11_62211663_62212209 | 0.16 |
CTD-2531D15.4 |
|
17740 |
0.11 |
| chr15_99645907_99646604 | 0.16 |
RP11-654A16.3 |
|
225 |
0.85 |
| chr7_141400668_141400819 | 0.16 |
KIAA1147 |
KIAA1147 |
672 |
0.56 |
| chr2_106012197_106012348 | 0.16 |
FHL2 |
four and a half LIM domains 2 |
970 |
0.58 |
| chr4_99404298_99404452 | 0.16 |
RP11-724M22.1 |
|
13140 |
0.27 |
| chr5_175560458_175560649 | 0.16 |
FAM153B |
family with sequence similarity 153, member B |
48644 |
0.12 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0016114 | terpenoid biosynthetic process(GO:0016114) |
| 0.1 | 0.4 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.4 | GO:0032351 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.1 | 0.5 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.3 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.1 | 0.1 | GO:0045631 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.1 | 0.3 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.1 | 0.6 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.1 | 0.3 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.1 | 0.3 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.1 | 0.2 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.1 | 0.2 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.2 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.1 | 0.2 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.3 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.1 | 0.2 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.1 | 0.3 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.1 | 0.2 | GO:1900121 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.1 | 0.4 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.1 | 0.2 | GO:0010193 | response to ozone(GO:0010193) |
| 0.1 | 0.5 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.1 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.3 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.0 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.2 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0022009 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.1 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.2 | GO:0006545 | glycine biosynthetic process(GO:0006545) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0061140 | lung secretory cell differentiation(GO:0061140) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.6 | GO:0032091 | negative regulation of protein binding(GO:0032091) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0061162 | establishment of apical/basal cell polarity(GO:0035089) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0061099 | negative regulation of protein tyrosine kinase activity(GO:0061099) |
| 0.0 | 0.1 | GO:0051608 | histamine transport(GO:0051608) |
| 0.0 | 0.6 | GO:0031532 | actin cytoskeleton reorganization(GO:0031532) |
| 0.0 | 0.1 | GO:0071072 | negative regulation of phospholipid biosynthetic process(GO:0071072) |
| 0.0 | 0.3 | GO:0060324 | face development(GO:0060324) |
| 0.0 | 0.1 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.2 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.2 | GO:0033008 | positive regulation of mast cell activation involved in immune response(GO:0033008) positive regulation of leukocyte degranulation(GO:0043302) positive regulation of mast cell degranulation(GO:0043306) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.1 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.1 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.3 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0060120 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.0 | 0.1 | GO:1901889 | negative regulation of focal adhesion assembly(GO:0051895) negative regulation of cell junction assembly(GO:1901889) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0022011 | myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.0 | GO:0090085 | regulation of protein deubiquitination(GO:0090085) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0071637 | monocyte chemotactic protein-1 production(GO:0071605) regulation of monocyte chemotactic protein-1 production(GO:0071637) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0060487 | lung epithelial cell differentiation(GO:0060487) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.3 | GO:0043403 | skeletal muscle tissue regeneration(GO:0043403) |
| 0.0 | 0.1 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0048712 | negative regulation of astrocyte differentiation(GO:0048712) |
| 0.0 | 0.1 | GO:0048147 | negative regulation of fibroblast proliferation(GO:0048147) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.1 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.2 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.2 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.4 | GO:0007257 | activation of JUN kinase activity(GO:0007257) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0043949 | regulation of cAMP-mediated signaling(GO:0043949) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.2 | GO:0002566 | somatic diversification of immune receptors via somatic mutation(GO:0002566) somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.3 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.0 | GO:2000192 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0090177 | establishment of planar polarity involved in neural tube closure(GO:0090177) |
| 0.0 | 0.2 | GO:0032400 | melanosome localization(GO:0032400) |
| 0.0 | 0.0 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0045006 | DNA deamination(GO:0045006) |
| 0.0 | 0.2 | GO:0016998 | cell wall macromolecule catabolic process(GO:0016998) |
| 0.0 | 0.1 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.2 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.0 | GO:0043501 | skeletal muscle adaptation(GO:0043501) |
| 0.0 | 0.0 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0010614 | negative regulation of cardiac muscle hypertrophy(GO:0010614) negative regulation of muscle hypertrophy(GO:0014741) |
| 0.0 | 0.1 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.1 | GO:0001573 | ganglioside metabolic process(GO:0001573) |
| 0.0 | 0.1 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0045351 | type I interferon biosynthetic process(GO:0045351) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.0 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.0 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.1 | GO:0015811 | sulfur amino acid transport(GO:0000101) L-cystine transport(GO:0015811) |
| 0.0 | 0.0 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0007252 | I-kappaB phosphorylation(GO:0007252) |
| 0.0 | 0.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.0 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0090026 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0030643 | cellular phosphate ion homeostasis(GO:0030643) cellular divalent inorganic anion homeostasis(GO:0072501) cellular trivalent inorganic anion homeostasis(GO:0072502) |
| 0.0 | 0.1 | GO:0042354 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0048636 | positive regulation of striated muscle tissue development(GO:0045844) positive regulation of muscle organ development(GO:0048636) positive regulation of muscle tissue development(GO:1901863) |
| 0.0 | 0.0 | GO:0019322 | pentose biosynthetic process(GO:0019322) |
| 0.0 | 0.0 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.1 | GO:0014003 | oligodendrocyte development(GO:0014003) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.0 | GO:0070272 | proton-transporting ATP synthase complex assembly(GO:0043461) proton-transporting ATP synthase complex biogenesis(GO:0070272) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.1 | GO:0070265 | necrotic cell death(GO:0070265) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.1 | GO:0060907 | positive regulation of macrophage cytokine production(GO:0060907) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0032648 | regulation of interferon-beta production(GO:0032648) |
| 0.0 | 0.1 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.0 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.1 | GO:0009209 | GTP biosynthetic process(GO:0006183) UTP biosynthetic process(GO:0006228) CTP biosynthetic process(GO:0006241) pyrimidine ribonucleoside triphosphate metabolic process(GO:0009208) pyrimidine ribonucleoside triphosphate biosynthetic process(GO:0009209) CTP metabolic process(GO:0046036) UTP metabolic process(GO:0046051) |
| 0.0 | 0.6 | GO:0006200 | obsolete ATP catabolic process(GO:0006200) |
| 0.0 | 0.0 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.0 | GO:0051770 | positive regulation of nitric-oxide synthase biosynthetic process(GO:0051770) |
| 0.0 | 0.1 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.0 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.0 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.0 | GO:0032647 | interferon-alpha production(GO:0032607) regulation of interferon-alpha production(GO:0032647) positive regulation of interferon-alpha production(GO:0032727) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.1 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.2 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0090576 | RNA polymerase III transcription factor complex(GO:0090576) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.3 | GO:0000783 | telomere cap complex(GO:0000782) nuclear telomere cap complex(GO:0000783) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.3 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.5 | GO:0043204 | perikaryon(GO:0043204) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.9 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.5 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 1.2 | GO:0042579 | peroxisome(GO:0005777) microbody(GO:0042579) |
| 0.0 | 0.4 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.3 | GO:0005637 | nuclear inner membrane(GO:0005637) |
| 0.0 | 0.1 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.0 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.1 | 0.4 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.1 | 0.4 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.3 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.1 | 1.2 | GO:0016863 | intramolecular oxidoreductase activity, transposing C=C bonds(GO:0016863) |
| 0.1 | 0.3 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.3 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.2 | GO:0003720 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.1 | 0.4 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.4 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0046980 | tapasin binding(GO:0046980) |
| 0.0 | 0.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.5 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.3 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.3 | GO:0003709 | obsolete RNA polymerase III transcription factor activity(GO:0003709) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.6 | GO:0004114 | 3',5'-cyclic-nucleotide phosphodiesterase activity(GO:0004114) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0099604 | calcium-release channel activity(GO:0015278) ligand-gated calcium channel activity(GO:0099604) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.2 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.2 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.4 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0051117 | ATPase binding(GO:0051117) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.2 | GO:0004709 | MAP kinase kinase kinase activity(GO:0004709) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.2 | GO:0008417 | fucosyltransferase activity(GO:0008417) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0008649 | rRNA methyltransferase activity(GO:0008649) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0016453 | C-acetyltransferase activity(GO:0016453) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0042301 | phosphate ion binding(GO:0042301) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.0 | GO:0099602 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.2 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.2 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.4 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0035254 | glutamate receptor binding(GO:0035254) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.0 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.0 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.0 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.0 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.0 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.0 | GO:0019238 | cyclohydrolase activity(GO:0019238) |
| 0.0 | 0.0 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0016861 | intramolecular oxidoreductase activity, interconverting aldoses and ketoses(GO:0016861) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | PID PI3K PLC TRK PATHWAY | Trk receptor signaling mediated by PI3K and PLC-gamma |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.1 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 1.2 | PID CDC42 PATHWAY | CDC42 signaling events |
| 0.0 | 0.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.7 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.3 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.2 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.2 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.2 | PID DNA PK PATHWAY | DNA-PK pathway in nonhomologous end joining |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.3 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.4 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.3 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.0 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.1 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION INITIATION FROM TYPE 2 PROMOTER | Genes involved in RNA Polymerase III Transcription Initiation From Type 2 Promoter |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.9 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.0 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |