Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
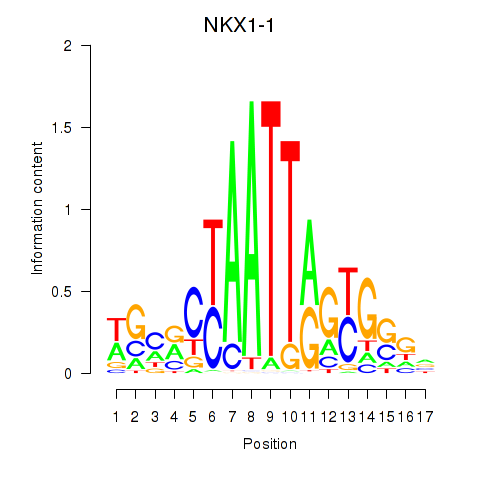
Results for NKX1-1
Z-value: 0.59
Transcription factors associated with NKX1-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX1-1
|
ENSG00000235608.1 | NK1 homeobox 1 |
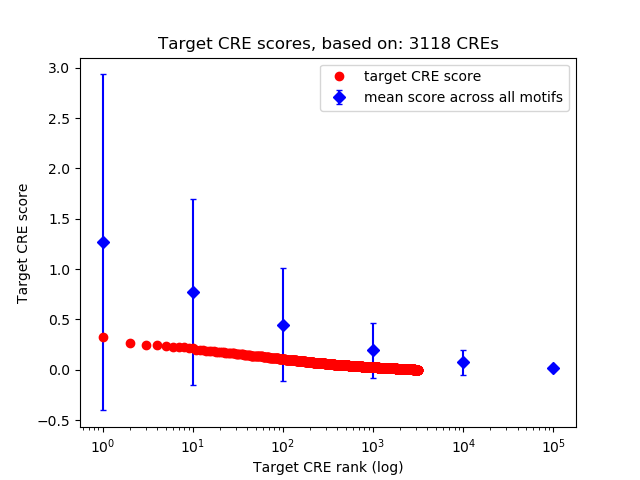
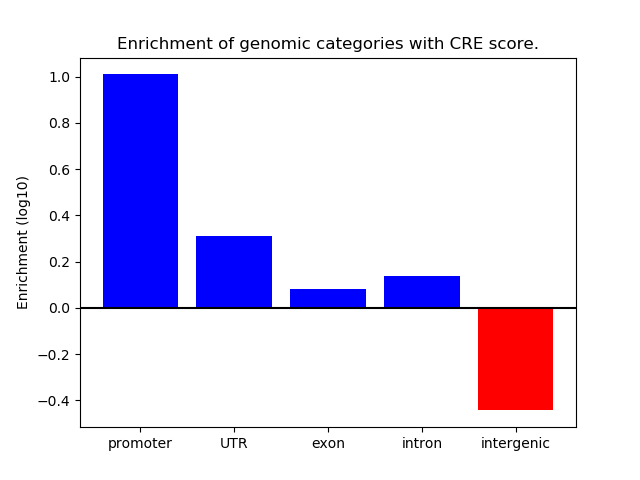
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_1397780_1397931 | NKX1-1 | 2264 | 0.241866 | 0.62 | 7.7e-02 | Click! |
| chr4_1514830_1514981 | NKX1-1 | 114786 | 0.046413 | 0.51 | 1.6e-01 | Click! |
| chr4_1514451_1514602 | NKX1-1 | 114407 | 0.046649 | 0.48 | 1.9e-01 | Click! |
| chr4_1523235_1523394 | NKX1-1 | 123195 | 0.041550 | 0.34 | 3.7e-01 | Click! |
| chr4_1513980_1514175 | NKX1-1 | 113958 | 0.046929 | -0.16 | 6.7e-01 | Click! |
Activity of the NKX1-1 motif across conditions
Conditions sorted by the z-value of the NKX1-1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
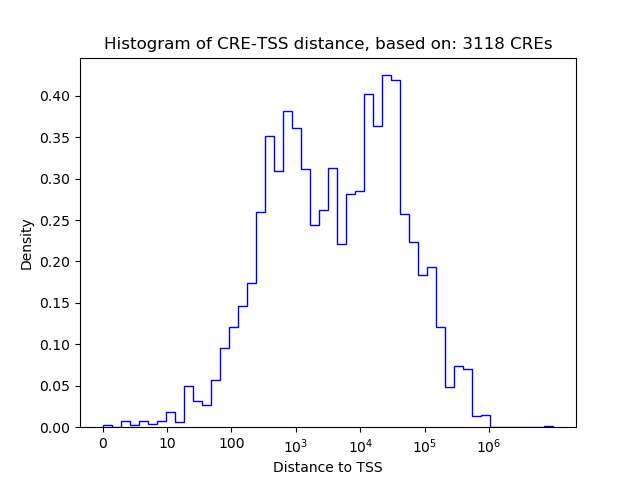
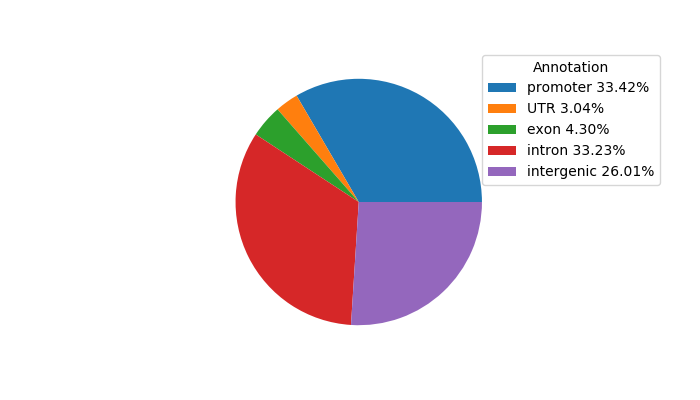
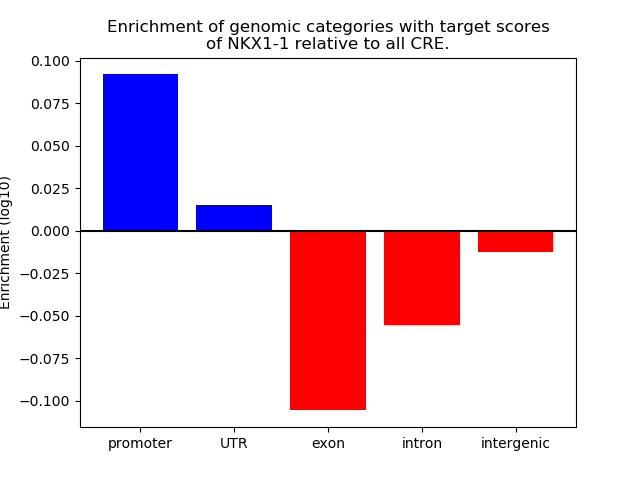
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_196577911_196578299 | 0.33 |
KCNT2 |
potassium channel, subfamily T, member 2 |
250 |
0.94 |
| chr13_68202936_68203087 | 0.27 |
PCDH9 |
protocadherin 9 |
398543 |
0.01 |
| chr12_28124078_28124290 | 0.25 |
PTHLH |
parathyroid hormone-like hormone |
719 |
0.71 |
| chr2_101620301_101620452 | 0.24 |
RPL31 |
ribosomal protein L31 |
1175 |
0.38 |
| chr4_83810146_83810528 | 0.23 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
1834 |
0.28 |
| chr6_117804579_117805184 | 0.23 |
DCBLD1 |
discoidin, CUB and LCCL domain containing 1 |
1056 |
0.49 |
| chr1_23494434_23494599 | 0.23 |
LUZP1 |
leucine zipper protein 1 |
794 |
0.54 |
| chr9_13278507_13279169 | 0.22 |
RP11-272P10.2 |
|
348 |
0.67 |
| chr4_89522534_89522685 | 0.22 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
4336 |
0.21 |
| chr4_157891142_157892336 | 0.21 |
PDGFC |
platelet derived growth factor C |
316 |
0.91 |
| chr6_27263961_27264170 | 0.20 |
POM121L2 |
POM121 transmembrane nucleoporin-like 2 |
15026 |
0.2 |
| chr5_57752049_57752200 | 0.20 |
PLK2 |
polo-like kinase 2 |
3963 |
0.27 |
| chr16_13401641_13401792 | 0.19 |
AC003009.1 |
|
24724 |
0.26 |
| chr7_80513051_80513202 | 0.19 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
35373 |
0.24 |
| chr21_16435398_16436179 | 0.18 |
NRIP1 |
nuclear receptor interacting protein 1 |
1338 |
0.57 |
| chr22_46476561_46476775 | 0.18 |
FLJ27365 |
hsa-mir-4763 |
476 |
0.66 |
| chr17_35289223_35289419 | 0.18 |
RP11-445F12.1 |
|
4600 |
0.15 |
| chr5_180529470_180529621 | 0.18 |
OR2V1 |
olfactory receptor, family 2, subfamily V, member 1 |
22759 |
0.13 |
| chr6_136888235_136888386 | 0.18 |
MAP7 |
microtubule-associated protein 7 |
16353 |
0.15 |
| chr20_43965593_43965922 | 0.18 |
SDC4 |
syndecan 4 |
11307 |
0.11 |
| chr10_102760785_102760994 | 0.17 |
LZTS2 |
leucine zipper, putative tumor suppressor 2 |
1279 |
0.27 |
| chrX_25020829_25021251 | 0.17 |
ARX |
aristaless related homeobox |
13025 |
0.26 |
| chr9_72745702_72745853 | 0.17 |
MAMDC2-AS1 |
MAMDC2 antisense RNA 1 |
17001 |
0.22 |
| chr15_70384069_70384220 | 0.17 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
2980 |
0.31 |
| chr11_122049732_122050124 | 0.17 |
ENSG00000207994 |
. |
26912 |
0.16 |
| chr18_20612011_20612377 | 0.17 |
ENSG00000223023 |
. |
7635 |
0.17 |
| chr21_27778313_27779012 | 0.17 |
AP001597.1 |
|
400 |
0.89 |
| chr8_94507892_94508285 | 0.16 |
ENSG00000222416 |
. |
54583 |
0.14 |
| chr5_180523773_180523999 | 0.16 |
OR2V1 |
olfactory receptor, family 2, subfamily V, member 1 |
28418 |
0.12 |
| chr12_115120620_115120912 | 0.16 |
TBX3 |
T-box 3 |
629 |
0.77 |
| chr2_163171316_163171467 | 0.16 |
IFIH1 |
interferon induced with helicase C domain 1 |
3803 |
0.25 |
| chr12_53896533_53896684 | 0.16 |
TARBP2 |
TAR (HIV-1) RNA binding protein 2 |
773 |
0.43 |
| chr2_163157606_163157781 | 0.16 |
IFIH1 |
interferon induced with helicase C domain 1 |
17501 |
0.19 |
| chr14_62032902_62033075 | 0.16 |
RP11-47I22.3 |
Uncharacterized protein |
4326 |
0.24 |
| chr1_109741279_109741478 | 0.16 |
ENSG00000238310 |
. |
9092 |
0.15 |
| chr10_131555251_131555402 | 0.15 |
RP11-109A6.2 |
|
55505 |
0.15 |
| chr18_59619337_59619488 | 0.15 |
RNF152 |
ring finger protein 152 |
57948 |
0.16 |
| chr6_27263399_27263763 | 0.15 |
POM121L2 |
POM121 transmembrane nucleoporin-like 2 |
15510 |
0.2 |
| chr6_49794571_49794884 | 0.15 |
RP3-417L20.4 |
|
9584 |
0.19 |
| chr17_41668751_41668949 | 0.15 |
ETV4 |
ets variant 4 |
11862 |
0.16 |
| chr4_186768313_186768795 | 0.14 |
SORBS2 |
sorbin and SH3 domain containing 2 |
34276 |
0.19 |
| chr14_103058041_103058305 | 0.14 |
RCOR1 |
REST corepressor 1 |
825 |
0.6 |
| chr15_69367071_69367289 | 0.14 |
RP11-809H16.4 |
|
31275 |
0.17 |
| chr4_75550401_75550552 | 0.14 |
AC142293.3 |
|
35812 |
0.18 |
| chr10_30347491_30347726 | 0.14 |
KIAA1462 |
KIAA1462 |
845 |
0.76 |
| chr13_114599987_114600138 | 0.14 |
GAS6 |
growth arrest-specific 6 |
33016 |
0.2 |
| chr6_37210883_37211034 | 0.14 |
ENSG00000238375 |
. |
8124 |
0.16 |
| chr11_12716213_12716424 | 0.14 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
19268 |
0.26 |
| chr1_45826795_45827032 | 0.14 |
ENSG00000238945 |
. |
2493 |
0.23 |
| chr8_334889_335040 | 0.14 |
FBXO25 |
F-box protein 25 |
21464 |
0.22 |
| chr6_41472777_41473161 | 0.14 |
RP11-328M4.2 |
|
40843 |
0.13 |
| chr4_31656800_31656951 | 0.14 |
RP11-619J20.1 |
|
861283 |
0.0 |
| chr15_67334428_67334662 | 0.14 |
SMAD3 |
SMAD family member 3 |
21556 |
0.25 |
| chr5_172665530_172665713 | 0.14 |
NKX2-5 |
NK2 homeobox 5 |
3261 |
0.23 |
| chr4_1005974_1006310 | 0.13 |
FGFRL1 |
fibroblast growth factor receptor-like 1 |
97 |
0.95 |
| chr4_163496269_163496420 | 0.13 |
FSTL5 |
follistatin-like 5 |
411157 |
0.01 |
| chr9_133711223_133711921 | 0.13 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
1119 |
0.56 |
| chr3_79417981_79418132 | 0.13 |
ENSG00000265193 |
. |
138981 |
0.05 |
| chr16_19606642_19606793 | 0.13 |
C16orf62 |
chromosome 16 open reading frame 62 |
12381 |
0.18 |
| chr11_94661378_94661529 | 0.13 |
RP11-856F16.2 |
|
2767 |
0.3 |
| chr19_4402510_4402661 | 0.13 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
74 |
0.93 |
| chr2_63815441_63815904 | 0.13 |
WDPCP |
WD repeat containing planar cell polarity effector |
20 |
0.54 |
| chr5_127874204_127874722 | 0.13 |
FBN2 |
fibrillin 2 |
547 |
0.58 |
| chr14_93552039_93552233 | 0.13 |
ITPK1-AS1 |
ITPK1 antisense RNA 1 |
18339 |
0.18 |
| chr19_2962281_2962480 | 0.13 |
TLE6 |
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
15064 |
0.12 |
| chr8_29200137_29200318 | 0.13 |
DUSP4 |
dual specificity phosphatase 4 |
6095 |
0.17 |
| chr2_188418561_188419245 | 0.13 |
AC007319.1 |
|
27 |
0.69 |
| chr2_242256486_242256668 | 0.12 |
HDLBP |
high density lipoprotein binding protein |
101 |
0.93 |
| chr8_98880755_98880906 | 0.12 |
MATN2 |
matrilin 2 |
238 |
0.95 |
| chr8_124284035_124284186 | 0.12 |
ZHX1 |
zinc fingers and homeoboxes 1 |
2437 |
0.16 |
| chr16_85202371_85202571 | 0.12 |
CTC-786C10.1 |
|
2411 |
0.35 |
| chr11_95646755_95646928 | 0.12 |
MTMR2 |
myotubularin related protein 2 |
600 |
0.82 |
| chr6_62996046_62996197 | 0.12 |
KHDRBS2 |
KH domain containing, RNA binding, signal transduction associated 2 |
11 |
0.99 |
| chr7_63385679_63385830 | 0.12 |
ENSG00000265017 |
. |
1460 |
0.47 |
| chr8_126323291_126323442 | 0.12 |
RP11-550A5.2 |
|
40292 |
0.16 |
| chr1_206593163_206593314 | 0.12 |
SRGAP2 |
SLIT-ROBO Rho GTPase activating protein 2 |
13502 |
0.18 |
| chr6_165341311_165341481 | 0.12 |
C6orf118 |
chromosome 6 open reading frame 118 |
380946 |
0.01 |
| chr13_26043049_26043230 | 0.12 |
ATP8A2 |
ATPase, aminophospholipid transporter, class I, type 8A, member 2 |
26 |
0.98 |
| chr12_42875750_42875901 | 0.12 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
1049 |
0.48 |
| chr9_110400275_110400607 | 0.12 |
ENSG00000202308 |
. |
25086 |
0.24 |
| chr14_91091040_91091191 | 0.12 |
TTC7B |
tetratricopeptide repeat domain 7B |
6748 |
0.2 |
| chr8_124195558_124195709 | 0.11 |
RP11-539E17.5 |
|
605 |
0.42 |
| chr1_115258427_115259019 | 0.11 |
NRAS |
neuroblastoma RAS viral (v-ras) oncogene homolog |
792 |
0.54 |
| chr5_174178214_174178699 | 0.11 |
ENSG00000266890 |
. |
281 |
0.94 |
| chr8_36807823_36808005 | 0.11 |
KCNU1 |
potassium channel, subfamily U, member 1 |
21491 |
0.29 |
| chr3_154797621_154798368 | 0.11 |
MME |
membrane metallo-endopeptidase |
94 |
0.98 |
| chr17_47547430_47548013 | 0.11 |
NGFR |
nerve growth factor receptor |
24934 |
0.13 |
| chr3_159483781_159484285 | 0.11 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
1141 |
0.44 |
| chr9_82795299_82795450 | 0.11 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
473693 |
0.01 |
| chr2_5837836_5837987 | 0.11 |
SOX11 |
SRY (sex determining region Y)-box 11 |
5112 |
0.22 |
| chr4_66536403_66536611 | 0.11 |
EPHA5 |
EPH receptor A5 |
294 |
0.95 |
| chr13_106426830_106427002 | 0.11 |
ENSG00000252550 |
. |
123076 |
0.06 |
| chr4_75858398_75858601 | 0.11 |
PARM1 |
prostate androgen-regulated mucin-like protein 1 |
173 |
0.97 |
| chr6_47666951_47667231 | 0.11 |
GPR115 |
G protein-coupled receptor 115 |
802 |
0.64 |
| chr10_105559478_105559629 | 0.11 |
SH3PXD2A |
SH3 and PX domains 2A |
1502 |
0.47 |
| chr18_77662426_77662577 | 0.11 |
KCNG2 |
potassium voltage-gated channel, subfamily G, member 2 |
38833 |
0.15 |
| chr15_33323538_33323689 | 0.11 |
FMN1 |
formin 1 |
36472 |
0.19 |
| chr10_119302142_119302346 | 0.11 |
EMX2 |
empty spiracles homeobox 2 |
265 |
0.64 |
| chr11_2834821_2835116 | 0.11 |
KCNQ1-AS1 |
KCNQ1 antisense RNA 1 |
47830 |
0.11 |
| chr6_7312252_7312483 | 0.11 |
SSR1 |
signal sequence receptor, alpha |
1040 |
0.57 |
| chr12_59314342_59314656 | 0.11 |
RP11-150C16.1 |
|
79 |
0.67 |
| chr13_53192037_53192369 | 0.10 |
HNRNPA1L2 |
heterogeneous nuclear ribonucleoprotein A1-like 2 |
598 |
0.79 |
| chr13_107160462_107160937 | 0.10 |
EFNB2 |
ephrin-B2 |
26763 |
0.24 |
| chr2_44935412_44935824 | 0.10 |
ENSG00000252896 |
. |
73651 |
0.12 |
| chr14_76604893_76605206 | 0.10 |
GPATCH2L |
G patch domain containing 2-like |
13213 |
0.26 |
| chr5_102897969_102898137 | 0.10 |
NUDT12 |
nudix (nucleoside diphosphate linked moiety X)-type motif 12 |
437 |
0.91 |
| chr3_16024121_16024347 | 0.10 |
ENSG00000207815 |
. |
108956 |
0.06 |
| chr10_6763782_6764322 | 0.10 |
PRKCQ |
protein kinase C, theta |
141789 |
0.05 |
| chr2_8135764_8135915 | 0.10 |
ENSG00000221255 |
. |
418867 |
0.01 |
| chr5_36951473_36951624 | 0.10 |
NIPBL |
Nipped-B homolog (Drosophila) |
74656 |
0.12 |
| chr7_132088071_132088231 | 0.10 |
AC011625.1 |
|
51058 |
0.17 |
| chr11_105892671_105892946 | 0.10 |
MSANTD4 |
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
146 |
0.96 |
| chr1_146641853_146642189 | 0.10 |
PRKAB2 |
protein kinase, AMP-activated, beta 2 non-catalytic subunit |
2025 |
0.25 |
| chr6_136887685_136887836 | 0.10 |
MAP7 |
microtubule-associated protein 7 |
15803 |
0.16 |
| chr6_26312991_26313348 | 0.10 |
HIST1H4H |
histone cluster 1, H4h |
27407 |
0.07 |
| chr2_5619821_5619972 | 0.10 |
AC108025.2 |
|
211282 |
0.02 |
| chr11_16809886_16810037 | 0.10 |
PLEKHA7 |
pleckstrin homology domain containing, family A member 7 |
24069 |
0.19 |
| chr11_117778647_117778798 | 0.10 |
TMPRSS13 |
transmembrane protease, serine 13 |
21358 |
0.14 |
| chr1_2461773_2461924 | 0.10 |
HES5 |
hes family bHLH transcription factor 5 |
164 |
0.9 |
| chr1_226109753_226109904 | 0.10 |
PYCR2 |
pyrroline-5-carboxylate reductase family, member 2 |
2131 |
0.19 |
| chr7_32929941_32930760 | 0.10 |
KBTBD2 |
kelch repeat and BTB (POZ) domain containing 2 |
415 |
0.87 |
| chr10_17547257_17547425 | 0.10 |
ST8SIA6 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 |
51012 |
0.13 |
| chr11_109294220_109294371 | 0.10 |
C11orf87 |
chromosome 11 open reading frame 87 |
1449 |
0.58 |
| chr1_209975427_209975724 | 0.10 |
IRF6 |
interferon regulatory factor 6 |
64 |
0.96 |
| chr9_13278250_13278433 | 0.10 |
RP11-272P10.2 |
|
845 |
0.51 |
| chr14_42609890_42610041 | 0.10 |
ENSG00000222084 |
. |
159989 |
0.04 |
| chr19_4343203_4343398 | 0.10 |
MPND |
MPN domain containing |
224 |
0.83 |
| chr20_10502111_10502382 | 0.09 |
SLX4IP |
SLX4 interacting protein |
86295 |
0.09 |
| chr8_80696427_80696819 | 0.09 |
ENSG00000238884 |
. |
15807 |
0.22 |
| chr15_41274840_41275069 | 0.09 |
RP11-540O11.6 |
|
16518 |
0.12 |
| chr17_40574158_40574426 | 0.09 |
PTRF |
polymerase I and transcript release factor |
1243 |
0.31 |
| chr12_17033936_17034087 | 0.09 |
ENSG00000212358 |
. |
190428 |
0.03 |
| chr10_12746098_12746480 | 0.09 |
ENSG00000221331 |
. |
21063 |
0.25 |
| chr8_31771478_31771629 | 0.09 |
NRG1-IT1 |
NRG1 intronic transcript 1 (non-protein coding) |
112182 |
0.07 |
| chr15_31690130_31690281 | 0.09 |
KLF13 |
Kruppel-like factor 13 |
31848 |
0.24 |
| chr15_96870341_96870527 | 0.09 |
NR2F2-AS1 |
NR2F2 antisense RNA 1 |
89 |
0.95 |
| chr3_170527527_170527678 | 0.09 |
ENSG00000222411 |
. |
32359 |
0.19 |
| chr5_36873918_36874176 | 0.09 |
NIPBL |
Nipped-B homolog (Drosophila) |
2814 |
0.4 |
| chr10_44277299_44277450 | 0.09 |
ZNF32 |
zinc finger protein 32 |
133070 |
0.05 |
| chr3_171848594_171848745 | 0.09 |
FNDC3B |
fibronectin type III domain containing 3B |
3905 |
0.33 |
| chr2_216876893_216877044 | 0.09 |
MREG |
melanoregulin |
1378 |
0.5 |
| chr5_36842382_36842533 | 0.09 |
NIPBL |
Nipped-B homolog (Drosophila) |
34404 |
0.22 |
| chr13_37006714_37007207 | 0.09 |
CCNA1 |
cyclin A1 |
465 |
0.85 |
| chr5_82767889_82768074 | 0.09 |
VCAN |
versican |
237 |
0.96 |
| chr15_43576768_43576919 | 0.09 |
RP11-402F9.3 |
|
17451 |
0.1 |
| chr7_29603770_29603921 | 0.09 |
PRR15 |
proline rich 15 |
418 |
0.58 |
| chrX_45482024_45482567 | 0.09 |
ENSG00000207870 |
. |
123399 |
0.06 |
| chr7_70596624_70596819 | 0.09 |
WBSCR17 |
Williams-Beuren syndrome chromosome region 17 |
434 |
0.91 |
| chr8_37915323_37915474 | 0.09 |
KB-1836B5.1 |
|
5001 |
0.15 |
| chr19_37743129_37743340 | 0.09 |
ZNF383 |
zinc finger protein 383 |
24782 |
0.13 |
| chr12_70381241_70381594 | 0.09 |
RP11-611E13.3 |
|
40556 |
0.18 |
| chr10_114552786_114553214 | 0.09 |
RP11-57H14.4 |
|
30249 |
0.21 |
| chr7_70060715_70061143 | 0.09 |
AUTS2 |
autism susceptibility candidate 2 |
133196 |
0.06 |
| chr6_165341863_165342014 | 0.09 |
C6orf118 |
chromosome 6 open reading frame 118 |
380404 |
0.01 |
| chr4_160357796_160357947 | 0.09 |
ENSG00000251979 |
. |
70309 |
0.12 |
| chr12_6495821_6496120 | 0.09 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
1685 |
0.24 |
| chr10_83897233_83897384 | 0.09 |
ENSG00000221378 |
. |
87290 |
0.09 |
| chr22_46929020_46929201 | 0.09 |
CELSR1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
2081 |
0.33 |
| chrX_24168685_24169495 | 0.09 |
ZFX |
zinc finger protein, X-linked |
789 |
0.52 |
| chr12_117627018_117627293 | 0.09 |
FBXO21 |
F-box protein 21 |
936 |
0.7 |
| chr4_169361832_169361983 | 0.09 |
DDX60L |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 60-like |
15303 |
0.2 |
| chr2_100591103_100591254 | 0.09 |
AFF3 |
AF4/FMR2 family, member 3 |
129814 |
0.05 |
| chr16_77619617_77619768 | 0.09 |
ENSG00000221226 |
. |
124506 |
0.05 |
| chr5_115909245_115909579 | 0.09 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
679 |
0.82 |
| chr11_13690121_13690272 | 0.09 |
FAR1 |
fatty acyl CoA reductase 1 |
21 |
0.94 |
| chr14_78078422_78078640 | 0.08 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
4585 |
0.22 |
| chr3_54674397_54674548 | 0.08 |
ESRG |
embryonic stem cell related (non-protein coding) |
588 |
0.86 |
| chr11_66383426_66383834 | 0.08 |
RBM14 |
RNA binding motif protein 14 |
423 |
0.31 |
| chr7_4046197_4046504 | 0.08 |
SDK1 |
sidekick cell adhesion molecule 1 |
55798 |
0.18 |
| chr3_57578310_57578700 | 0.08 |
ARF4 |
ADP-ribosylation factor 4 |
4547 |
0.12 |
| chr4_74571443_74571594 | 0.08 |
IL8 |
interleukin 8 |
34705 |
0.18 |
| chr9_24036289_24036440 | 0.08 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
210029 |
0.03 |
| chr14_89017580_89017758 | 0.08 |
PTPN21 |
protein tyrosine phosphatase, non-receptor type 21 |
135 |
0.68 |
| chr19_57350948_57351186 | 0.08 |
PEG3 |
paternally expressed 3 |
997 |
0.38 |
| chr19_13127591_13127981 | 0.08 |
CTC-239J10.1 |
|
2461 |
0.15 |
| chr2_17997369_17997520 | 0.08 |
MSGN1 |
mesogenin 1 |
319 |
0.9 |
| chr8_110619590_110619741 | 0.08 |
SYBU |
syntabulin (syntaxin-interacting) |
639 |
0.72 |
| chr20_52211254_52212184 | 0.08 |
ZNF217 |
zinc finger protein 217 |
1341 |
0.45 |
| chr4_152337049_152337200 | 0.08 |
FAM160A1 |
family with sequence similarity 160, member A1 |
6620 |
0.28 |
| chr6_131948545_131949340 | 0.08 |
MED23 |
mediator complex subunit 23 |
258 |
0.77 |
| chr17_4403572_4403723 | 0.08 |
AC118754.4 |
|
678 |
0.53 |
| chr1_171220672_171220826 | 0.08 |
RP1-45C12.1 |
|
184 |
0.91 |
| chr1_162532826_162532977 | 0.08 |
UAP1 |
UDP-N-acteylglucosamine pyrophosphorylase 1 |
1578 |
0.3 |
| chr14_70317063_70317281 | 0.08 |
SMOC1 |
SPARC related modular calcium binding 1 |
28961 |
0.17 |
| chr3_158519414_158519896 | 0.08 |
MFSD1 |
major facilitator superfamily domain containing 1 |
1 |
0.54 |
| chr6_26553804_26554291 | 0.08 |
HMGN4 |
high mobility group nucleosomal binding domain 4 |
15414 |
0.14 |
| chr3_116583114_116583333 | 0.08 |
ENSG00000265433 |
. |
14009 |
0.25 |
| chr9_126770623_126770924 | 0.08 |
RP11-85O21.2 |
|
902 |
0.48 |
| chrX_44401210_44401465 | 0.08 |
FUNDC1 |
FUN14 domain containing 1 |
910 |
0.68 |
| chr7_99699979_99700130 | 0.08 |
AP4M1 |
adaptor-related protein complex 4, mu 1 subunit |
370 |
0.46 |
| chr5_114631582_114631761 | 0.08 |
CCDC112 |
coiled-coil domain containing 112 |
91 |
0.97 |
| chr10_118534538_118534689 | 0.08 |
HSPA12A |
heat shock 70kDa protein 12A |
32528 |
0.17 |
| chr2_62081759_62082202 | 0.08 |
FAM161A |
family with sequence similarity 161, member A |
702 |
0.69 |
| chr12_30906652_30906803 | 0.08 |
CAPRIN2 |
caprin family member 2 |
518 |
0.86 |
| chr1_244174827_244174978 | 0.08 |
ZBTB18 |
zinc finger and BTB domain containing 18 |
39683 |
0.17 |
| chr12_54120989_54121200 | 0.08 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
118 |
0.96 |
| chr1_51796370_51796715 | 0.08 |
TTC39A |
tetratricopeptide repeat domain 39A |
77 |
0.95 |
| chr4_146101952_146102435 | 0.08 |
OTUD4 |
OTU domain containing 4 |
880 |
0.65 |
| chr2_178261408_178261672 | 0.08 |
AGPS |
alkylglycerone phosphate synthase |
4044 |
0.17 |
| chr5_1887316_1887467 | 0.08 |
IRX4 |
iroquois homeobox 4 |
41 |
0.51 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0051973 | positive regulation of telomerase activity(GO:0051973) |
| 0.0 | 0.1 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.1 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0060766 | negative regulation of androgen receptor signaling pathway(GO:0060766) |
| 0.0 | 0.0 | GO:0003171 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.0 | GO:0060197 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.1 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0046504 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.0 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.3 | GO:0071805 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.0 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.0 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:1904358 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0031332 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.2 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.0 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.0 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.0 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME AUTODEGRADATION OF THE E3 UBIQUITIN LIGASE COP1 | Genes involved in Autodegradation of the E3 ubiquitin ligase COP1 |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.0 | REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | Genes involved in NGF signalling via TRKA from the plasma membrane |