Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for NKX1-2_RAX
Z-value: 0.77
Transcription factors associated with NKX1-2_RAX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
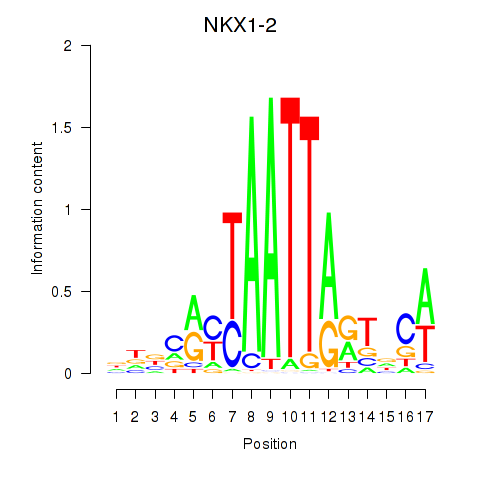
NKX1-2
|
ENSG00000229544.6 | NK1 homeobox 2 |
|
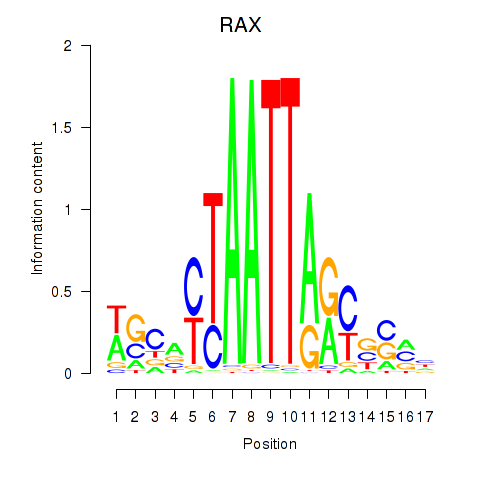
RAX
|
ENSG00000134438.9 | retina and anterior neural fold homeobox |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr10_126137746_126137897 | NKX1-2 | 729 | 0.569900 | 0.19 | 6.2e-01 | Click! |
| chr10_126137495_126137646 | NKX1-2 | 980 | 0.420831 | 0.18 | 6.4e-01 | Click! |
| chr10_126138132_126138283 | NKX1-2 | 343 | 0.841994 | -0.11 | 7.7e-01 | Click! |
| chr10_126138368_126138519 | NKX1-2 | 107 | 0.957241 | -0.06 | 8.9e-01 | Click! |
| chr18_56939565_56939853 | RAX | 908 | 0.623630 | 0.89 | 1.2e-03 | Click! |
| chr18_56936630_56936781 | RAX | 3912 | 0.253613 | 0.52 | 1.5e-01 | Click! |
| chr18_56932228_56932379 | RAX | 8314 | 0.215386 | 0.50 | 1.7e-01 | Click! |
| chr18_56939924_56940075 | RAX | 618 | 0.760815 | -0.20 | 6.1e-01 | Click! |
| chr18_56936216_56936367 | RAX | 4326 | 0.245541 | -0.13 | 7.3e-01 | Click! |
Activity of the NKX1-2_RAX motif across conditions
Conditions sorted by the z-value of the NKX1-2_RAX motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
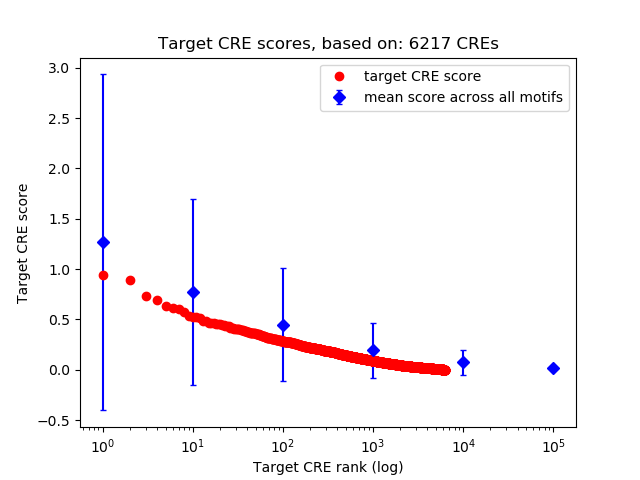
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_87799796_87800719 | 0.94 |
LMO4 |
LIM domain only 4 |
2906 |
0.4 |
| chr4_157891142_157892336 | 0.89 |
PDGFC |
platelet derived growth factor C |
316 |
0.91 |
| chr2_207998546_207999225 | 0.74 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
24 |
0.98 |
| chr1_68697735_68698156 | 0.69 |
WLS |
wntless Wnt ligand secretion mediator |
18 |
0.98 |
| chr1_196577911_196578299 | 0.63 |
KCNT2 |
potassium channel, subfamily T, member 2 |
250 |
0.94 |
| chr8_89311514_89311910 | 0.62 |
RP11-586K2.1 |
|
27353 |
0.2 |
| chr9_13278507_13279169 | 0.60 |
RP11-272P10.2 |
|
348 |
0.67 |
| chr8_116675050_116675578 | 0.57 |
TRPS1 |
trichorhinophalangeal syndrome I |
1409 |
0.6 |
| chr6_163817634_163818149 | 0.53 |
QKI |
QKI, KH domain containing, RNA binding |
17784 |
0.29 |
| chr1_225842736_225843127 | 0.53 |
ENAH |
enabled homolog (Drosophila) |
2087 |
0.34 |
| chr12_54418548_54418910 | 0.52 |
HOXC6 |
homeobox C6 |
3413 |
0.09 |
| chr3_112356169_112356393 | 0.51 |
CCDC80 |
coiled-coil domain containing 80 |
663 |
0.77 |
| chr4_15471989_15472140 | 0.49 |
CC2D2A |
coiled-coil and C2 domain containing 2A |
427 |
0.85 |
| chr8_37350646_37351319 | 0.48 |
RP11-150O12.6 |
|
23557 |
0.24 |
| chr5_82767889_82768074 | 0.47 |
VCAN |
versican |
237 |
0.96 |
| chr3_33318777_33319866 | 0.46 |
FBXL2 |
F-box and leucine-rich repeat protein 2 |
353 |
0.91 |
| chr4_77610213_77610516 | 0.46 |
AC107072.2 |
|
51588 |
0.12 |
| chr12_89033220_89033386 | 0.46 |
ENSG00000252850 |
. |
57887 |
0.14 |
| chr3_87038506_87038677 | 0.46 |
VGLL3 |
vestigial like 3 (Drosophila) |
1261 |
0.64 |
| chr15_99297407_99297593 | 0.45 |
ENSG00000264480 |
. |
30155 |
0.19 |
| chr8_56805751_56805987 | 0.45 |
RP11-318K15.2 |
|
285 |
0.88 |
| chr9_96819103_96819334 | 0.45 |
PTPDC1 |
protein tyrosine phosphatase domain containing 1 |
26142 |
0.21 |
| chr2_128492018_128492224 | 0.43 |
SFT2D3 |
SFT2 domain containing 3 |
33524 |
0.13 |
| chr6_148663224_148663375 | 0.43 |
SASH1 |
SAM and SH3 domain containing 1 |
430 |
0.89 |
| chr5_51565820_51566012 | 0.43 |
CTD-2203A3.1 |
Uncharacterized protein |
260267 |
0.02 |
| chr10_119303944_119304573 | 0.42 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
321 |
0.88 |
| chr22_36232810_36232961 | 0.42 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
3380 |
0.35 |
| chr9_16828464_16828715 | 0.41 |
BNC2 |
basonuclin 2 |
3697 |
0.36 |
| chr10_24754741_24755189 | 0.41 |
KIAA1217 |
KIAA1217 |
495 |
0.84 |
| chr2_101438663_101438947 | 0.41 |
AC092168.2 |
|
318 |
0.86 |
| chr1_182932139_182932350 | 0.40 |
ENSG00000264768 |
. |
3534 |
0.19 |
| chr15_96884556_96884898 | 0.40 |
ENSG00000222651 |
. |
8237 |
0.16 |
| chr20_10502111_10502382 | 0.40 |
SLX4IP |
SLX4 interacting protein |
86295 |
0.09 |
| chr12_52347769_52348164 | 0.40 |
ACVR1B |
activin A receptor, type IB |
802 |
0.57 |
| chr11_36252522_36252860 | 0.39 |
COMMD9 |
COMM domain containing 9 |
58283 |
0.13 |
| chr8_16622129_16622507 | 0.39 |
ENSG00000264092 |
. |
98687 |
0.08 |
| chr9_22008020_22008450 | 0.38 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
717 |
0.61 |
| chr18_53089837_53090372 | 0.38 |
TCF4 |
transcription factor 4 |
361 |
0.89 |
| chrX_119691520_119691671 | 0.38 |
CUL4B |
cullin 4B |
2203 |
0.3 |
| chr9_4137636_4137913 | 0.38 |
GLIS3 |
GLIS family zinc finger 3 |
7419 |
0.28 |
| chr6_28616143_28617086 | 0.38 |
ENSG00000272278 |
. |
572 |
0.79 |
| chr6_132437274_132437503 | 0.37 |
ENSG00000265669 |
. |
985 |
0.69 |
| chr10_4284322_4284712 | 0.37 |
ENSG00000207124 |
. |
272627 |
0.02 |
| chr3_134050593_134051228 | 0.37 |
AMOTL2 |
angiomotin like 2 |
39844 |
0.16 |
| chr2_188418561_188419245 | 0.37 |
AC007319.1 |
|
27 |
0.69 |
| chr1_79043181_79043547 | 0.37 |
IFI44L |
interferon-induced protein 44-like |
42243 |
0.17 |
| chr12_19358284_19358561 | 0.37 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
163 |
0.96 |
| chr10_12746098_12746480 | 0.36 |
ENSG00000221331 |
. |
21063 |
0.25 |
| chr3_79066759_79066910 | 0.36 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
860 |
0.75 |
| chr17_21415164_21415422 | 0.35 |
RP11-822E23.2 |
|
20942 |
0.2 |
| chr10_114552786_114553214 | 0.35 |
RP11-57H14.4 |
|
30249 |
0.21 |
| chr3_112362286_112362437 | 0.35 |
CCDC80 |
coiled-coil domain containing 80 |
2214 |
0.38 |
| chr7_55166552_55166703 | 0.35 |
EGFR |
epidermal growth factor receptor |
10789 |
0.29 |
| chr1_246198663_246198916 | 0.35 |
RP11-83A16.1 |
|
1936 |
0.45 |
| chr20_43965593_43965922 | 0.35 |
SDC4 |
syndecan 4 |
11307 |
0.11 |
| chr19_13127591_13127981 | 0.34 |
CTC-239J10.1 |
|
2461 |
0.15 |
| chr10_34149976_34150160 | 0.34 |
ENSG00000199200 |
. |
340934 |
0.01 |
| chr10_115032925_115033406 | 0.34 |
ENSG00000238380 |
. |
80019 |
0.11 |
| chr16_62068318_62068835 | 0.34 |
CDH8 |
cadherin 8, type 2 |
462 |
0.9 |
| chr1_177953163_177953341 | 0.34 |
SEC16B |
SEC16 homolog B (S. cerevisiae) |
13904 |
0.3 |
| chr5_154033985_154034136 | 0.33 |
ENSG00000221552 |
. |
31276 |
0.14 |
| chr9_94178893_94179044 | 0.33 |
NFIL3 |
nuclear factor, interleukin 3 regulated |
7176 |
0.29 |
| chr5_35851300_35851523 | 0.33 |
IL7R |
interleukin 7 receptor |
1386 |
0.45 |
| chr20_36040358_36040543 | 0.33 |
SRC |
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
27897 |
0.21 |
| chr2_151340168_151340387 | 0.32 |
RND3 |
Rho family GTPase 3 |
1619 |
0.57 |
| chr20_56706361_56706780 | 0.32 |
C20orf85 |
chromosome 20 open reading frame 85 |
19390 |
0.21 |
| chr8_58653688_58653839 | 0.32 |
ENSG00000252057 |
. |
184619 |
0.03 |
| chr7_33746403_33746586 | 0.32 |
RP11-89N17.1 |
HCG1643653; Uncharacterized protein |
19099 |
0.23 |
| chr18_3791000_3791154 | 0.32 |
RP11-874J12.3 |
|
19647 |
0.17 |
| chr1_10511084_10511235 | 0.32 |
CORT |
cortistatin |
1188 |
0.31 |
| chr12_115135696_115136278 | 0.32 |
TBX3 |
T-box 3 |
14018 |
0.21 |
| chr6_106474855_106475295 | 0.32 |
PRDM1 |
PR domain containing 1, with ZNF domain |
59120 |
0.13 |
| chr5_14153831_14154379 | 0.31 |
TRIO |
trio Rho guanine nucleotide exchange factor |
10276 |
0.32 |
| chr6_27515697_27516098 | 0.31 |
ENSG00000206671 |
. |
48294 |
0.13 |
| chr17_66700653_66700804 | 0.31 |
ENSG00000263690 |
. |
61972 |
0.13 |
| chr5_148786412_148786563 | 0.31 |
ENSG00000208035 |
. |
21994 |
0.11 |
| chr19_39149984_39150227 | 0.31 |
ACTN4 |
actinin, alpha 4 |
11727 |
0.12 |
| chr13_102107295_102107446 | 0.31 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
2352 |
0.4 |
| chr5_97790329_97790704 | 0.31 |
ENSG00000223053 |
. |
184419 |
0.03 |
| chr14_54816147_54816359 | 0.31 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
47420 |
0.17 |
| chr3_32001454_32001950 | 0.31 |
OSBPL10 |
oxysterol binding protein-like 10 |
21090 |
0.19 |
| chr5_131750481_131750657 | 0.30 |
C5orf56 |
chromosome 5 open reading frame 56 |
3888 |
0.16 |
| chr3_114169976_114170457 | 0.30 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
3314 |
0.33 |
| chr6_6005898_6006049 | 0.30 |
NRN1 |
neuritin 1 |
1227 |
0.56 |
| chr4_169769029_169769180 | 0.30 |
RP11-635L1.3 |
|
15016 |
0.19 |
| chr14_74191037_74191188 | 0.30 |
PNMA1 |
paraneoplastic Ma antigen 1 |
9984 |
0.11 |
| chr5_64438498_64438671 | 0.29 |
ENSG00000207439 |
. |
19388 |
0.29 |
| chr7_79766032_79766269 | 0.29 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
1079 |
0.66 |
| chr3_116583114_116583333 | 0.29 |
ENSG00000265433 |
. |
14009 |
0.25 |
| chr7_116312981_116313724 | 0.29 |
MET |
met proto-oncogene |
893 |
0.68 |
| chr2_213403391_213403972 | 0.29 |
ERBB4 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
116 |
0.98 |
| chr1_198873285_198873436 | 0.29 |
ENSG00000207759 |
. |
45078 |
0.17 |
| chr9_104248314_104249564 | 0.29 |
TMEM246 |
transmembrane protein 246 |
460 |
0.79 |
| chr12_94580963_94581249 | 0.29 |
RP11-74K11.2 |
|
1286 |
0.5 |
| chr8_127837007_127837644 | 0.29 |
ENSG00000212451 |
. |
153558 |
0.04 |
| chr14_101867418_101867569 | 0.29 |
ENSG00000258498 |
. |
159266 |
0.02 |
| chr7_80545378_80545529 | 0.29 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
3046 |
0.41 |
| chr4_134070427_134071051 | 0.29 |
PCDH10 |
protocadherin 10 |
269 |
0.96 |
| chr4_20258902_20259191 | 0.29 |
SLIT2 |
slit homolog 2 (Drosophila) |
2503 |
0.44 |
| chrX_34671991_34672311 | 0.28 |
TMEM47 |
transmembrane protein 47 |
3254 |
0.41 |
| chr7_128171824_128173030 | 0.28 |
RP11-274B21.1 |
|
1279 |
0.43 |
| chr2_63926212_63926391 | 0.28 |
ENSG00000221085 |
. |
3632 |
0.34 |
| chr5_92943123_92943644 | 0.28 |
ENSG00000251725 |
. |
13111 |
0.2 |
| chr12_66286178_66286489 | 0.28 |
RP11-366L20.2 |
Uncharacterized protein |
10386 |
0.18 |
| chr3_88191159_88191414 | 0.28 |
ZNF654 |
zinc finger protein 654 |
3032 |
0.24 |
| chr7_92327766_92328157 | 0.28 |
ENSG00000206763 |
. |
3167 |
0.33 |
| chr22_46476561_46476775 | 0.28 |
FLJ27365 |
hsa-mir-4763 |
476 |
0.66 |
| chr4_54063057_54063462 | 0.28 |
ENSG00000207385 |
. |
68475 |
0.13 |
| chr8_79468522_79468695 | 0.28 |
PKIA |
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
34858 |
0.19 |
| chr11_95646755_95646928 | 0.28 |
MTMR2 |
myotubularin related protein 2 |
600 |
0.82 |
| chr5_174178214_174178699 | 0.28 |
ENSG00000266890 |
. |
281 |
0.94 |
| chr1_150135684_150135897 | 0.28 |
PLEKHO1 |
pleckstrin homology domain containing, family O member 1 |
6667 |
0.15 |
| chr3_154798803_154798954 | 0.28 |
MME |
membrane metallo-endopeptidase |
125 |
0.98 |
| chr9_89738235_89738413 | 0.28 |
C9orf170 |
chromosome 9 open reading frame 170 |
25235 |
0.26 |
| chr17_42214986_42215221 | 0.27 |
ENSG00000212446 |
. |
1746 |
0.18 |
| chr3_114477857_114478359 | 0.27 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
10 |
0.99 |
| chr2_153349112_153349263 | 0.27 |
FMNL2 |
formin-like 2 |
126905 |
0.06 |
| chr5_3597777_3597928 | 0.27 |
IRX1 |
iroquois homeobox 1 |
1684 |
0.41 |
| chr1_186072218_186072510 | 0.27 |
HMCN1 |
hemicentin 1 |
68155 |
0.12 |
| chr3_59101471_59101696 | 0.27 |
C3orf67 |
chromosome 3 open reading frame 67 |
65773 |
0.15 |
| chr15_37179413_37179956 | 0.27 |
ENSG00000212511 |
. |
34843 |
0.22 |
| chr22_36232318_36232521 | 0.27 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
3846 |
0.33 |
| chr20_48319819_48319972 | 0.27 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
10520 |
0.19 |
| chr14_81484576_81484774 | 0.27 |
CEP128 |
centrosomal protein 128kDa |
58827 |
0.14 |
| chr3_87036535_87037613 | 0.27 |
VGLL3 |
vestigial like 3 (Drosophila) |
2778 |
0.42 |
| chr7_116142190_116142403 | 0.27 |
CAV2 |
caveolin 2 |
2462 |
0.22 |
| chr7_70201396_70202343 | 0.27 |
AUTS2 |
autism susceptibility candidate 2 |
7744 |
0.34 |
| chrX_73069883_73070423 | 0.27 |
RP13-216E22.5 |
|
119167 |
0.06 |
| chr15_96903910_96904291 | 0.27 |
AC087477.1 |
Uncharacterized protein |
387 |
0.85 |
| chr2_33662989_33663140 | 0.27 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
1673 |
0.49 |
| chr4_121990444_121990801 | 0.27 |
NDNF |
neuron-derived neurotrophic factor |
2534 |
0.31 |
| chr3_193856720_193856967 | 0.26 |
HES1 |
hes family bHLH transcription factor 1 |
2909 |
0.25 |
| chr2_216795737_216796089 | 0.26 |
ENSG00000212055 |
. |
52271 |
0.15 |
| chr1_88319546_88319697 | 0.26 |
ENSG00000199318 |
. |
400565 |
0.01 |
| chr9_97563812_97563963 | 0.26 |
C9orf3 |
chromosome 9 open reading frame 3 |
1429 |
0.41 |
| chr5_57751342_57751690 | 0.26 |
PLK2 |
polo-like kinase 2 |
4571 |
0.26 |
| chr8_8468799_8468974 | 0.26 |
ENSG00000264445 |
. |
22088 |
0.2 |
| chr18_52973612_52973763 | 0.26 |
TCF4 |
transcription factor 4 |
3830 |
0.35 |
| chr1_245461989_245462162 | 0.26 |
RP11-62I21.1 |
|
64272 |
0.12 |
| chr4_141013667_141013926 | 0.26 |
RP11-392B6.1 |
|
35373 |
0.19 |
| chr12_80794329_80794553 | 0.26 |
PTPRQ |
protein tyrosine phosphatase, receptor type, Q |
5333 |
0.26 |
| chr4_54187854_54188041 | 0.26 |
SCFD2 |
sec1 family domain containing 2 |
44281 |
0.15 |
| chr13_31443551_31443843 | 0.26 |
MEDAG |
mesenteric estrogen-dependent adipogenesis |
36631 |
0.17 |
| chr5_97726813_97726964 | 0.25 |
ENSG00000223053 |
. |
248047 |
0.02 |
| chr3_174159101_174159924 | 0.25 |
NAALADL2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
735 |
0.81 |
| chr8_80696427_80696819 | 0.25 |
ENSG00000238884 |
. |
15807 |
0.22 |
| chr3_14211699_14211850 | 0.25 |
LSM3 |
LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
8084 |
0.16 |
| chr9_15735359_15735549 | 0.25 |
CCDC171 |
coiled-coil domain containing 171 |
9049 |
0.26 |
| chr11_92117878_92118174 | 0.25 |
RP11-675M1.2 |
|
23316 |
0.23 |
| chr2_121334386_121334749 | 0.25 |
ENSG00000201006 |
. |
74282 |
0.11 |
| chr10_30744125_30744276 | 0.25 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
16449 |
0.22 |
| chr14_53509566_53509717 | 0.25 |
RP11-368P15.3 |
|
6168 |
0.27 |
| chr7_87974817_87974968 | 0.25 |
STEAP4 |
STEAP family member 4 |
38686 |
0.17 |
| chr6_5471426_5471612 | 0.25 |
RP1-232P20.1 |
|
13211 |
0.27 |
| chr4_71997150_71997347 | 0.24 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
55755 |
0.16 |
| chr9_110818837_110819103 | 0.24 |
ENSG00000222459 |
. |
137711 |
0.05 |
| chr8_32085203_32085460 | 0.24 |
NRG1-IT2 |
NRG1 intronic transcript 2 (non-protein coding) |
6887 |
0.22 |
| chr14_75896980_75897330 | 0.24 |
JDP2 |
Jun dimerization protein 2 |
1682 |
0.35 |
| chr8_57086542_57086822 | 0.24 |
PLAG1 |
pleiomorphic adenoma gene 1 |
37156 |
0.12 |
| chr6_85470567_85470756 | 0.24 |
TBX18 |
T-box 18 |
2412 |
0.42 |
| chr14_31887812_31887963 | 0.24 |
CTD-2213F21.2 |
|
1605 |
0.27 |
| chr11_697541_698020 | 0.24 |
TMEM80 |
transmembrane protein 80 |
1975 |
0.14 |
| chr21_17909068_17909746 | 0.24 |
ENSG00000207638 |
. |
2002 |
0.33 |
| chr11_15135960_15136328 | 0.24 |
INSC |
inscuteable homolog (Drosophila) |
340 |
0.92 |
| chr3_120215962_120216316 | 0.24 |
FSTL1 |
follistatin-like 1 |
46039 |
0.16 |
| chr9_77757026_77757296 | 0.24 |
ENSG00000200041 |
. |
39376 |
0.14 |
| chr6_3473109_3473260 | 0.24 |
SLC22A23 |
solute carrier family 22, member 23 |
15928 |
0.27 |
| chr2_66663569_66663965 | 0.24 |
MEIS1 |
Meis homeobox 1 |
758 |
0.57 |
| chr6_136570446_136570737 | 0.24 |
MTFR2 |
mitochondrial fission regulator 2 |
841 |
0.63 |
| chr1_178065779_178065930 | 0.24 |
RASAL2 |
RAS protein activator like 2 |
2578 |
0.43 |
| chr12_50748152_50748528 | 0.24 |
FAM186A |
family with sequence similarity 186, member A |
3672 |
0.22 |
| chr2_223916824_223917105 | 0.24 |
KCNE4 |
potassium voltage-gated channel, Isk-related family, member 4 |
102 |
0.98 |
| chr3_106060605_106060757 | 0.24 |
ENSG00000200610 |
. |
174063 |
0.04 |
| chr7_7545818_7546068 | 0.24 |
COL28A1 |
collagen, type XXVIII, alpha 1 |
29541 |
0.21 |
| chr15_96886136_96886425 | 0.23 |
ENSG00000222651 |
. |
9790 |
0.16 |
| chr9_81871858_81872685 | 0.23 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
314417 |
0.01 |
| chr15_42749459_42749764 | 0.23 |
ZNF106 |
zinc finger protein 106 |
100 |
0.96 |
| chr4_120446718_120446900 | 0.23 |
PDE5A |
phosphodiesterase 5A, cGMP-specific |
6555 |
0.24 |
| chr17_10039283_10039434 | 0.23 |
AC000003.2 |
CDNA FLJ25865 fis, clone CBR01927 |
9473 |
0.21 |
| chr12_47470653_47470804 | 0.23 |
PCED1B |
PC-esterase domain containing 1B |
2658 |
0.29 |
| chr12_53995183_53995708 | 0.23 |
ATF7 |
activating transcription factor 7 |
640 |
0.67 |
| chr21_35678304_35678597 | 0.23 |
ENSG00000266007 |
. |
1020 |
0.57 |
| chr6_45545466_45545617 | 0.23 |
ENSG00000252738 |
. |
68300 |
0.13 |
| chr12_26277242_26278053 | 0.23 |
BHLHE41 |
basic helix-loop-helix family, member e41 |
413 |
0.76 |
| chr17_57703305_57703456 | 0.23 |
CLTC |
clathrin, heavy chain (Hc) |
6087 |
0.21 |
| chr9_126331371_126331654 | 0.23 |
ENSG00000222722 |
. |
16224 |
0.24 |
| chr15_35950174_35950564 | 0.23 |
DPH6 |
diphthamine biosynthesis 6 |
111976 |
0.07 |
| chr11_46237993_46238166 | 0.23 |
CTD-2589M5.4 |
|
58019 |
0.1 |
| chr16_83583521_83583672 | 0.23 |
ENSG00000263785 |
. |
41645 |
0.18 |
| chr7_82070146_82070432 | 0.23 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
2742 |
0.43 |
| chr18_6730820_6730971 | 0.23 |
ARHGAP28 |
Rho GTPase activating protein 28 |
853 |
0.47 |
| chr5_172665530_172665713 | 0.23 |
NKX2-5 |
NK2 homeobox 5 |
3261 |
0.23 |
| chr11_27861058_27861209 | 0.22 |
RP11-587D21.4 |
|
38062 |
0.2 |
| chr3_152019172_152019323 | 0.22 |
MBNL1 |
muscleblind-like splicing regulator 1 |
1260 |
0.51 |
| chr14_88921501_88921652 | 0.22 |
PTPN21 |
protein tyrosine phosphatase, non-receptor type 21 |
14384 |
0.17 |
| chr12_50635075_50635226 | 0.22 |
ENSG00000221604 |
. |
7155 |
0.13 |
| chr13_52360273_52360424 | 0.22 |
DHRS12 |
dehydrogenase/reductase (SDR family) member 12 |
17886 |
0.19 |
| chr10_29010987_29011704 | 0.22 |
ENSG00000201001 |
. |
38387 |
0.14 |
| chr18_52495652_52496658 | 0.22 |
RAB27B |
RAB27B, member RAS oncogene family |
725 |
0.77 |
| chr6_116834038_116834189 | 0.22 |
TRAPPC3L |
trafficking protein particle complex 3-like |
607 |
0.55 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.4 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.1 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.2 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.2 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.2 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.2 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.3 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 1.0 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.2 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.2 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.0 | GO:0060027 | convergent extension involved in gastrulation(GO:0060027) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.2 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.5 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.2 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0061430 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.8 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.0 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.8 | GO:0048813 | dendrite morphogenesis(GO:0048813) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.9 | GO:0001843 | neural tube closure(GO:0001843) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.6 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.0 | GO:0090109 | regulation of focal adhesion assembly(GO:0051893) negative regulation of focal adhesion assembly(GO:0051895) regulation of cell-substrate junction assembly(GO:0090109) regulation of cell junction assembly(GO:1901888) negative regulation of cell junction assembly(GO:1901889) regulation of adherens junction organization(GO:1903391) negative regulation of adherens junction organization(GO:1903392) |
| 0.0 | 0.0 | GO:0030202 | heparin metabolic process(GO:0030202) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.0 | GO:0042663 | endodermal cell fate specification(GO:0001714) regulation of endodermal cell fate specification(GO:0042663) regulation of endodermal cell differentiation(GO:1903224) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.4 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.2 | GO:0007184 | SMAD protein import into nucleus(GO:0007184) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.2 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0010667 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.0 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0003307 | regulation of Wnt signaling pathway involved in heart development(GO:0003307) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) |
| 0.0 | 0.0 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.0 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.0 | GO:0060913 | cardiac cell fate determination(GO:0060913) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.0 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.0 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0061036 | positive regulation of cartilage development(GO:0061036) |
| 0.0 | 0.0 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0031529 | ruffle organization(GO:0031529) |
| 0.0 | 0.0 | GO:0016093 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.0 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0007320 | insemination(GO:0007320) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.0 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0001725 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.0 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.0 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.4 | GO:0036064 | ciliary basal body(GO:0036064) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 0.9 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.1 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.6 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.1 | 0.6 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 0.8 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.3 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.2 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.3 | GO:0030553 | cGMP binding(GO:0030553) |
| 0.0 | 0.1 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.1 | GO:0004656 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0000404 | heteroduplex DNA loop binding(GO:0000404) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.2 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.1 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0034483 | heparan sulfate sulfotransferase activity(GO:0034483) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.8 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.4 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.1 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.0 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.6 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.3 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.0 | REACTOME SIGNALING BY SCF KIT | Genes involved in Signaling by SCF-KIT |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |