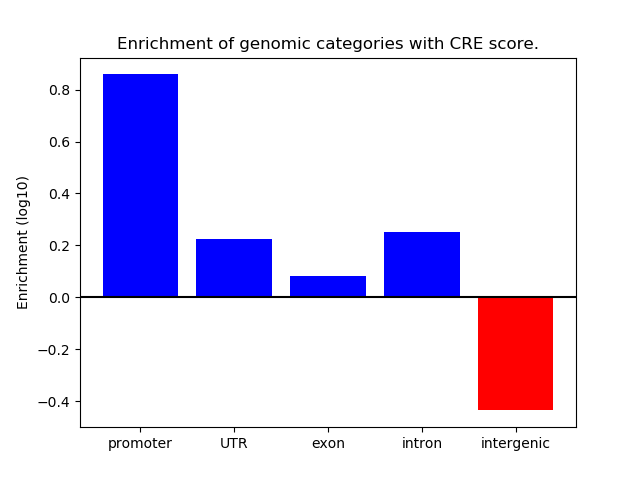
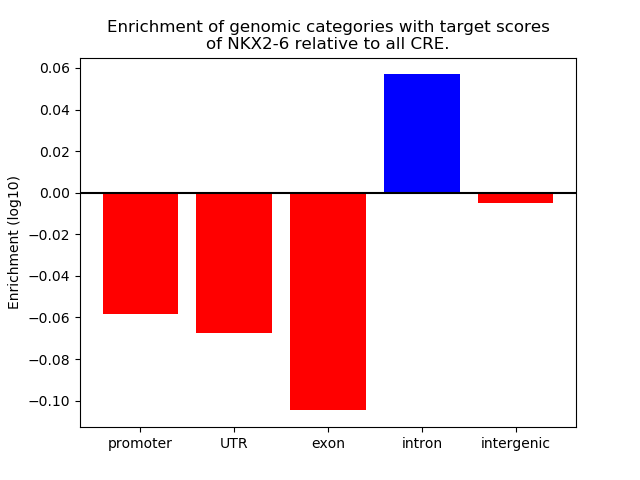
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
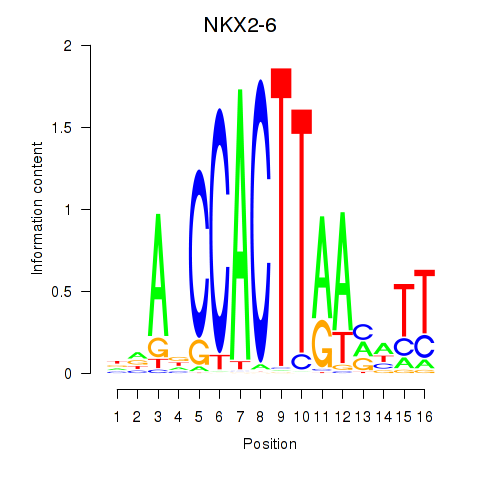
Results for NKX2-6
Z-value: 0.51
Transcription factors associated with NKX2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-6
|
ENSG00000180053.6 | NK2 homeobox 6 |
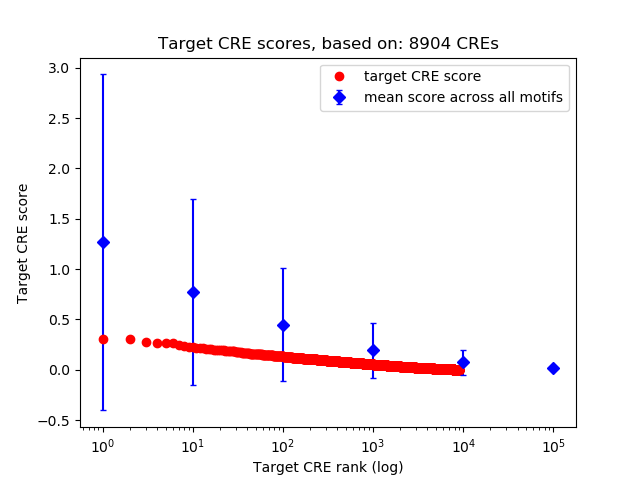
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_23562532_23562683 | NKX2-6 | 1315 | 0.359028 | 0.74 | 2.2e-02 | Click! |
| chr8_23563794_23563945 | NKX2-6 | 53 | 0.935410 | -0.46 | 2.2e-01 | Click! |
| chr8_23562338_23562489 | NKX2-6 | 1509 | 0.323056 | 0.24 | 5.4e-01 | Click! |
| chr8_23563950_23564574 | NKX2-6 | 151 | 0.764890 | -0.03 | 9.4e-01 | Click! |
Activity of the NKX2-6 motif across conditions
Conditions sorted by the z-value of the NKX2-6 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
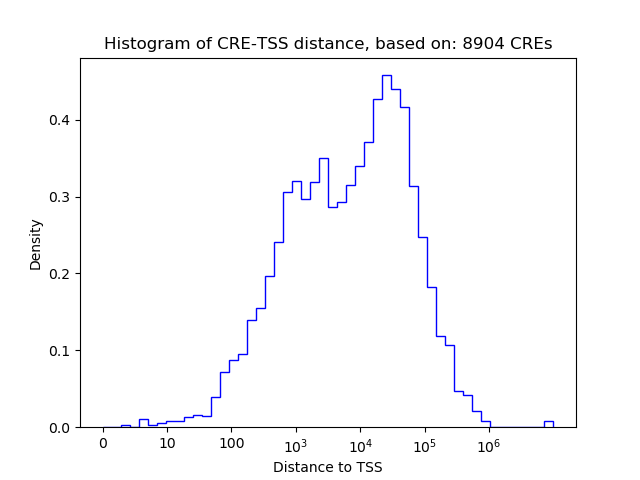
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr3_132315920_132316071 | 0.31 |
ACKR4 |
atypical chemokine receptor 4 |
86 |
0.98 |
| chr2_175197281_175197503 | 0.30 |
SP9 |
Sp9 transcription factor |
2282 |
0.26 |
| chr8_106743100_106743756 | 0.28 |
RP11-642D21.2 |
|
45435 |
0.16 |
| chr22_36310772_36312004 | 0.27 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
23557 |
0.25 |
| chr12_20523872_20524102 | 0.26 |
RP11-284H19.1 |
|
791 |
0.64 |
| chr3_127540841_127541975 | 0.26 |
MGLL |
monoglyceride lipase |
214 |
0.96 |
| chr7_119912758_119913222 | 0.25 |
KCND2 |
potassium voltage-gated channel, Shal-related subfamily, member 2 |
732 |
0.81 |
| chr11_121968562_121968780 | 0.23 |
ENSG00000207971 |
. |
1881 |
0.27 |
| chr6_28175291_28175856 | 0.22 |
ZSCAN9 |
zinc finger and SCAN domain containing 9 |
17091 |
0.12 |
| chr2_159610172_159610621 | 0.22 |
AC005042.4 |
|
18882 |
0.21 |
| chr7_116142833_116142984 | 0.22 |
CAV2 |
caveolin 2 |
3074 |
0.2 |
| chr6_72129322_72130506 | 0.21 |
ENSG00000207827 |
. |
16590 |
0.22 |
| chr17_57229359_57229924 | 0.21 |
SKA2 |
spindle and kinetochore associated complex subunit 2 |
479 |
0.57 |
| chr6_56614643_56614951 | 0.21 |
DST |
dystonin |
35880 |
0.21 |
| chr11_69458516_69459047 | 0.21 |
CCND1 |
cyclin D1 |
2807 |
0.29 |
| chr7_80547175_80548098 | 0.20 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
863 |
0.76 |
| chr6_86114788_86115158 | 0.20 |
NT5E |
5'-nucleotidase, ecto (CD73) |
44836 |
0.19 |
| chr7_15725092_15725754 | 0.20 |
MEOX2 |
mesenchyme homeobox 2 |
1014 |
0.63 |
| chr4_169799680_169799831 | 0.20 |
ENSG00000201176 |
. |
6751 |
0.22 |
| chr8_494094_494251 | 0.19 |
TDRP |
testis development related protein |
665 |
0.79 |
| chr18_61554936_61555215 | 0.19 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
82 |
0.97 |
| chr12_13350846_13351208 | 0.19 |
EMP1 |
epithelial membrane protein 1 |
1307 |
0.55 |
| chr1_158969828_158970040 | 0.19 |
IFI16 |
interferon, gamma-inducible protein 16 |
176 |
0.96 |
| chr20_19916034_19916185 | 0.19 |
RIN2 |
Ras and Rab interactor 2 |
45899 |
0.15 |
| chr21_31311288_31312406 | 0.19 |
GRIK1 |
glutamate receptor, ionotropic, kainate 1 |
3 |
0.99 |
| chr20_6749993_6750220 | 0.19 |
BMP2 |
bone morphogenetic protein 2 |
1795 |
0.51 |
| chr10_4719640_4719886 | 0.18 |
AKR1E2 |
aldo-keto reductase family 1, member E2 |
109058 |
0.07 |
| chr4_74975461_74975696 | 0.18 |
CXCL2 |
chemokine (C-X-C motif) ligand 2 |
10568 |
0.16 |
| chr20_13976647_13977145 | 0.18 |
SEL1L2 |
sel-1 suppressor of lin-12-like 2 (C. elegans) |
193 |
0.78 |
| chr7_13959407_13959706 | 0.18 |
ETV1 |
ets variant 1 |
66510 |
0.14 |
| chr9_86916681_86916945 | 0.18 |
RP11-380F14.2 |
|
23679 |
0.22 |
| chr8_8154312_8154463 | 0.17 |
ALG1L13P |
asparagine-linked glycosylation 1-like 13, pseudogene |
55454 |
0.11 |
| chr4_180980346_180980880 | 0.17 |
NA |
NA |
> 106 |
NA |
| chr4_23890131_23890306 | 0.17 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
1440 |
0.57 |
| chr2_226896266_226896823 | 0.17 |
NYAP2 |
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
622947 |
0.0 |
| chr1_13910795_13911459 | 0.17 |
PDPN |
podoplanin |
362 |
0.87 |
| chr1_167721376_167721626 | 0.17 |
MPZL1 |
myelin protein zero-like 1 |
13306 |
0.23 |
| chr8_6691754_6692464 | 0.17 |
DEFB1 |
defensin, beta 1 |
43435 |
0.14 |
| chr11_16947220_16947414 | 0.17 |
PLEKHA7 |
pleckstrin homology domain containing, family A member 7 |
44412 |
0.12 |
| chr2_216297601_216297993 | 0.16 |
FN1 |
fibronectin 1 |
2993 |
0.27 |
| chrX_3264180_3264467 | 0.16 |
MXRA5 |
matrix-remodelling associated 5 |
359 |
0.92 |
| chr8_37350646_37351319 | 0.16 |
RP11-150O12.6 |
|
23557 |
0.24 |
| chr13_52165265_52165666 | 0.16 |
ENSG00000242893 |
. |
2886 |
0.18 |
| chr10_16909529_16909705 | 0.16 |
RSU1 |
Ras suppressor protein 1 |
50090 |
0.17 |
| chr3_49447935_49448923 | 0.16 |
RHOA |
ras homolog family member A |
932 |
0.32 |
| chr8_38857241_38857490 | 0.16 |
ADAM9 |
ADAM metallopeptidase domain 9 |
2860 |
0.19 |
| chr1_162534114_162534517 | 0.16 |
UAP1 |
UDP-N-acteylglucosamine pyrophosphorylase 1 |
1512 |
0.34 |
| chr8_87934784_87934935 | 0.16 |
CNBD1 |
cyclic nucleotide binding domain containing 1 |
56186 |
0.17 |
| chr15_83951840_83952048 | 0.16 |
BNC1 |
basonuclin 1 |
127 |
0.97 |
| chr12_124990844_124991534 | 0.16 |
NCOR2 |
nuclear receptor corepressor 2 |
11391 |
0.3 |
| chr14_71838527_71838748 | 0.16 |
ENSG00000207444 |
. |
26417 |
0.18 |
| chr13_110957949_110958238 | 0.16 |
COL4A2 |
collagen, type IV, alpha 2 |
66 |
0.97 |
| chr4_80572253_80572434 | 0.15 |
OR7E94P |
olfactory receptor, family 7, subfamily E, member 94 pseudogene |
63059 |
0.15 |
| chr1_179110770_179111258 | 0.15 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
1165 |
0.52 |
| chr10_95227944_95228273 | 0.15 |
MYOF |
myoferlin |
13843 |
0.19 |
| chr8_91782901_91783052 | 0.15 |
NECAB1 |
N-terminal EF-hand calcium binding protein 1 |
20802 |
0.19 |
| chr8_30668680_30668831 | 0.15 |
PPP2CB |
protein phosphatase 2, catalytic subunit, beta isozyme |
963 |
0.63 |
| chr4_129998585_129998736 | 0.15 |
C4orf33 |
chromosome 4 open reading frame 33 |
15812 |
0.24 |
| chr8_104090727_104091208 | 0.15 |
ENSG00000265667 |
. |
13982 |
0.15 |
| chr7_91923432_91923918 | 0.15 |
ANKIB1 |
ankyrin repeat and IBR domain containing 1 |
101 |
0.97 |
| chr1_214723042_214723197 | 0.15 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
1447 |
0.54 |
| chr5_158611818_158612659 | 0.15 |
RNF145 |
ring finger protein 145 |
22404 |
0.16 |
| chr7_78398482_78399243 | 0.15 |
MAGI2 |
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
1534 |
0.56 |
| chr5_107041117_107041487 | 0.15 |
ENSG00000239708 |
. |
29465 |
0.21 |
| chrX_19716439_19716715 | 0.15 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
27461 |
0.25 |
| chr9_97811337_97811671 | 0.15 |
ENSG00000207563 |
. |
35986 |
0.11 |
| chr5_158126537_158126688 | 0.15 |
CTD-2363C16.1 |
|
283402 |
0.01 |
| chr3_29327656_29328192 | 0.15 |
RBMS3-AS3 |
RBMS3 antisense RNA 3 |
4293 |
0.26 |
| chr13_24121051_24121276 | 0.15 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
23346 |
0.25 |
| chr3_99766860_99767059 | 0.15 |
FILIP1L |
filamin A interacting protein 1-like |
66398 |
0.1 |
| chr4_157891142_157892336 | 0.15 |
PDGFC |
platelet derived growth factor C |
316 |
0.91 |
| chr6_56406612_56407191 | 0.14 |
DST |
dystonin |
68113 |
0.13 |
| chr7_12663902_12664053 | 0.14 |
SCIN |
scinderin |
19839 |
0.21 |
| chr4_85886471_85886934 | 0.14 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
801 |
0.75 |
| chr10_102921756_102921944 | 0.14 |
RP11-31L23.3 |
|
22382 |
0.12 |
| chr7_27274920_27275071 | 0.14 |
EVX1 |
even-skipped homeobox 1 |
7169 |
0.1 |
| chr10_51574745_51574998 | 0.14 |
NCOA4 |
nuclear receptor coactivator 4 |
1414 |
0.4 |
| chr1_16466376_16466578 | 0.14 |
RP11-276H7.2 |
|
15229 |
0.11 |
| chr15_59598169_59598320 | 0.14 |
RP11-429D19.1 |
|
34883 |
0.11 |
| chrX_10602528_10602751 | 0.14 |
MID1 |
midline 1 (Opitz/BBB syndrome) |
13965 |
0.27 |
| chr22_43017042_43017193 | 0.14 |
ENSG00000270022 |
. |
5867 |
0.12 |
| chr12_3034547_3034860 | 0.14 |
ENSG00000238689 |
. |
3428 |
0.16 |
| chr7_82071524_82071810 | 0.14 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
1364 |
0.61 |
| chr14_59949074_59949274 | 0.14 |
L3HYPDH |
L-3-hydroxyproline dehydratase (trans-) |
1229 |
0.4 |
| chr11_121972599_121972750 | 0.14 |
RP11-166D19.1 |
|
200 |
0.92 |
| chr5_171269419_171269604 | 0.14 |
SMIM23 |
small integral membrane protein 23 |
56621 |
0.13 |
| chr1_206687074_206687508 | 0.14 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
6412 |
0.15 |
| chr2_192710069_192710427 | 0.14 |
AC098617.1 |
|
1017 |
0.51 |
| chr19_6103641_6103897 | 0.14 |
CTB-66B24.1 |
|
5564 |
0.15 |
| chr6_18053801_18053981 | 0.14 |
KIF13A |
kinesin family member 13A |
66037 |
0.11 |
| chr8_37555995_37556200 | 0.14 |
ZNF703 |
zinc finger protein 703 |
2828 |
0.18 |
| chr12_59309288_59309823 | 0.14 |
LRIG3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
1191 |
0.55 |
| chr1_200710499_200710966 | 0.14 |
CAMSAP2 |
calmodulin regulated spectrin-associated protein family, member 2 |
1825 |
0.44 |
| chr15_68573802_68573977 | 0.14 |
FEM1B |
fem-1 homolog b (C. elegans) |
1592 |
0.39 |
| chr15_72529055_72529849 | 0.14 |
PKM |
pyruvate kinase, muscle |
5288 |
0.17 |
| chr3_49169788_49170138 | 0.14 |
LAMB2 |
laminin, beta 2 (laminin S) |
490 |
0.65 |
| chr20_47299051_47299422 | 0.13 |
ENSG00000251876 |
. |
56749 |
0.16 |
| chr7_102990167_102990648 | 0.13 |
PSMC2 |
proteasome (prosome, macropain) 26S subunit, ATPase, 2 |
2297 |
0.25 |
| chr3_61560941_61561100 | 0.13 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
13435 |
0.31 |
| chr1_101359369_101359680 | 0.13 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
696 |
0.48 |
| chr1_70688549_70688700 | 0.13 |
SRSF11 |
serine/arginine-rich splicing factor 11 |
1479 |
0.33 |
| chr5_16934966_16935117 | 0.13 |
MYO10 |
myosin X |
1006 |
0.59 |
| chr9_20511940_20512213 | 0.13 |
ENSG00000264941 |
. |
9736 |
0.23 |
| chr14_65928823_65928974 | 0.13 |
ENSG00000207781 |
. |
8922 |
0.25 |
| chr12_56513564_56513771 | 0.13 |
RP11-603J24.6 |
|
56 |
0.9 |
| chr5_141278602_141278753 | 0.13 |
PCDH1 |
protocadherin 1 |
19866 |
0.14 |
| chr6_155005040_155005564 | 0.13 |
SCAF8 |
SR-related CTD-associated factor 8 |
49157 |
0.17 |
| chr1_109824775_109824999 | 0.13 |
PSRC1 |
proline/serine-rich coiled-coil 1 |
586 |
0.7 |
| chr3_11551023_11551396 | 0.13 |
VGLL4 |
vestigial like 4 (Drosophila) |
59189 |
0.14 |
| chr12_105651079_105651410 | 0.13 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
21228 |
0.19 |
| chr6_119667569_119668515 | 0.13 |
MAN1A1 |
mannosidase, alpha, class 1A, member 1 |
2859 |
0.34 |
| chr4_114214241_114214392 | 0.13 |
ANK2 |
ankyrin 2, neuronal |
178 |
0.97 |
| chr7_27205863_27206297 | 0.13 |
HOXA9 |
homeobox A9 |
931 |
0.25 |
| chr2_127822596_127822803 | 0.13 |
BIN1 |
bridging integrator 1 |
41878 |
0.16 |
| chr7_17030060_17030455 | 0.13 |
AGR3 |
anterior gradient 3 |
108646 |
0.07 |
| chr9_21555046_21555197 | 0.13 |
MIR31HG |
MIR31 host gene (non-protein coding) |
4547 |
0.21 |
| chr19_16403865_16404198 | 0.13 |
CTD-2562J15.6 |
|
355 |
0.85 |
| chr2_17722476_17722627 | 0.13 |
VSNL1 |
visinin-like 1 |
124 |
0.98 |
| chr1_150481112_150481644 | 0.12 |
ECM1 |
extracellular matrix protein 1 |
795 |
0.48 |
| chr12_56916774_56917003 | 0.12 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
1105 |
0.41 |
| chr2_31360151_31360442 | 0.12 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
591 |
0.81 |
| chr9_97811962_97812255 | 0.12 |
ENSG00000207563 |
. |
35382 |
0.12 |
| chr5_151065179_151065606 | 0.12 |
SPARC |
secreted protein, acidic, cysteine-rich (osteonectin) |
601 |
0.67 |
| chr12_6494464_6494615 | 0.12 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
254 |
0.87 |
| chr22_43386709_43386953 | 0.12 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
11615 |
0.21 |
| chr11_114861879_114862030 | 0.12 |
ENSG00000252870 |
. |
137139 |
0.05 |
| chr9_97768076_97768588 | 0.12 |
C9orf3 |
chromosome 9 open reading frame 3 |
1054 |
0.55 |
| chr5_82767889_82768074 | 0.12 |
VCAN |
versican |
237 |
0.96 |
| chr5_54916685_54916836 | 0.12 |
SLC38A9 |
solute carrier family 38, member 9 |
71688 |
0.1 |
| chr10_35102956_35103650 | 0.12 |
PARD3 |
par-3 family cell polarity regulator |
946 |
0.5 |
| chr13_113689501_113689756 | 0.12 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
9348 |
0.15 |
| chr2_1734119_1734469 | 0.12 |
PXDN |
peroxidasin homolog (Drosophila) |
13920 |
0.26 |
| chr17_4762663_4762814 | 0.12 |
ENSG00000263599 |
. |
16183 |
0.08 |
| chr7_101596567_101596967 | 0.12 |
CTB-181H17.1 |
|
6629 |
0.27 |
| chr1_170630999_170631537 | 0.12 |
PRRX1 |
paired related homeobox 1 |
1007 |
0.68 |
| chr10_62541267_62541606 | 0.12 |
CDK1 |
cyclin-dependent kinase 1 |
1525 |
0.51 |
| chr11_61456591_61456863 | 0.12 |
DAGLA |
diacylglycerol lipase, alpha |
8822 |
0.18 |
| chr6_16339042_16339210 | 0.12 |
ENSG00000265642 |
. |
89628 |
0.08 |
| chr12_89846997_89847148 | 0.12 |
POC1B |
POC1 centriolar protein B |
43962 |
0.13 |
| chr2_46546253_46546404 | 0.12 |
EPAS1 |
endothelial PAS domain protein 1 |
21787 |
0.24 |
| chr3_11755297_11755448 | 0.12 |
VGLL4 |
vestigial like 4 (Drosophila) |
3361 |
0.34 |
| chr14_102446484_102446635 | 0.12 |
DYNC1H1 |
dynein, cytoplasmic 1, heavy chain 1 |
15694 |
0.19 |
| chr13_43842946_43843190 | 0.12 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
92145 |
0.1 |
| chr7_107221551_107221702 | 0.12 |
BCAP29 |
B-cell receptor-associated protein 29 |
422 |
0.69 |
| chr7_100464814_100466140 | 0.12 |
TRIP6 |
thyroid hormone receptor interactor 6 |
717 |
0.47 |
| chr1_98514169_98514606 | 0.12 |
ENSG00000225206 |
. |
2660 |
0.42 |
| chr6_18389672_18389881 | 0.12 |
RNF144B |
ring finger protein 144B |
2195 |
0.34 |
| chr14_92997841_92998087 | 0.12 |
RIN3 |
Ras and Rab interactor 3 |
17816 |
0.26 |
| chr5_149462966_149463221 | 0.12 |
CSF1R |
colony stimulating factor 1 receptor |
2897 |
0.21 |
| chr5_78204137_78204521 | 0.12 |
ARSB |
arylsulfatase B |
77279 |
0.11 |
| chr2_47527627_47527778 | 0.12 |
EPCAM |
epithelial cell adhesion molecule |
44595 |
0.13 |
| chr4_141018823_141018974 | 0.12 |
RP11-392B6.1 |
|
30271 |
0.2 |
| chr1_212812342_212812619 | 0.12 |
RP11-338C15.5 |
|
12367 |
0.14 |
| chr14_68613241_68613392 | 0.12 |
ENSG00000244677 |
. |
3309 |
0.29 |
| chr3_4624343_4624494 | 0.12 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
66242 |
0.12 |
| chr15_92438505_92438656 | 0.12 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
41229 |
0.19 |
| chrX_31284819_31285011 | 0.11 |
DMD |
dystrophin |
59 |
0.99 |
| chr2_159992813_159993143 | 0.11 |
ENSG00000202029 |
. |
109324 |
0.06 |
| chr1_178269315_178269642 | 0.11 |
RASAL2 |
RAS protein activator like 2 |
41128 |
0.2 |
| chr4_140793537_140793902 | 0.11 |
MAML3 |
mastermind-like 3 (Drosophila) |
17487 |
0.25 |
| chr8_38585652_38586394 | 0.11 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
82 |
0.97 |
| chr5_61681454_61681605 | 0.11 |
DIMT1 |
DIM1 dimethyladenosine transferase 1 homolog (S. cerevisiae) |
18196 |
0.17 |
| chr1_57043984_57044408 | 0.11 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
1045 |
0.66 |
| chr12_110720958_110721647 | 0.11 |
ATP2A2 |
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 |
1816 |
0.41 |
| chr2_153382153_153382407 | 0.11 |
FMNL2 |
formin-like 2 |
93812 |
0.09 |
| chr6_108486676_108487297 | 0.11 |
OSTM1 |
osteopetrosis associated transmembrane protein 1 |
58 |
0.78 |
| chr7_27236053_27236457 | 0.11 |
HOTTIP |
HOXA distal transcript antisense RNA |
1939 |
0.13 |
| chr3_177978802_177979100 | 0.11 |
KCNMB2 |
potassium large conductance calcium-activated channel, subfamily M, beta member 2 |
11769 |
0.3 |
| chr3_99360396_99360669 | 0.11 |
COL8A1 |
collagen, type VIII, alpha 1 |
3078 |
0.35 |
| chr20_6764834_6765429 | 0.11 |
BMP2 |
bone morphogenetic protein 2 |
16820 |
0.28 |
| chr19_10736478_10736985 | 0.11 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
530 |
0.65 |
| chr4_113803321_113803472 | 0.11 |
RP11-119H12.6 |
|
90 |
0.98 |
| chr16_54464970_54465224 | 0.11 |
ENSG00000264079 |
. |
118211 |
0.06 |
| chr4_20195598_20195749 | 0.11 |
SLIT2 |
slit homolog 2 (Drosophila) |
59210 |
0.17 |
| chr2_97593411_97593614 | 0.11 |
ENSG00000252845 |
. |
29040 |
0.12 |
| chr9_117439582_117439814 | 0.11 |
ENSG00000272241 |
. |
60871 |
0.1 |
| chr10_17241830_17242242 | 0.11 |
TRDMT1 |
tRNA aspartic acid methyltransferase 1 |
1543 |
0.37 |
| chr5_139968434_139968585 | 0.11 |
ENSG00000200235 |
. |
9257 |
0.08 |
| chr12_118745072_118745584 | 0.11 |
TAOK3 |
TAO kinase 3 |
51464 |
0.15 |
| chr22_29106631_29107007 | 0.11 |
CHEK2 |
checkpoint kinase 2 |
1100 |
0.47 |
| chr3_188149558_188149830 | 0.11 |
LPP-AS1 |
LPP antisense RNA 1 |
136760 |
0.05 |
| chr12_54778714_54778870 | 0.11 |
ZNF385A |
zinc finger protein 385A |
256 |
0.84 |
| chr12_13408941_13409103 | 0.11 |
EMP1 |
epithelial membrane protein 1 |
44577 |
0.17 |
| chrX_107683395_107684019 | 0.11 |
COL4A5 |
collagen, type IV, alpha 5 |
595 |
0.63 |
| chr1_245331414_245331723 | 0.11 |
KIF26B |
kinesin family member 26B |
13281 |
0.2 |
| chr16_54321004_54321411 | 0.11 |
IRX3 |
iroquois homeobox 3 |
532 |
0.83 |
| chr7_114054549_114054700 | 0.11 |
FOXP2 |
forkhead box P2 |
295 |
0.93 |
| chr12_56075239_56075869 | 0.11 |
METTL7B |
methyltransferase like 7B |
224 |
0.86 |
| chr1_33455162_33455432 | 0.11 |
RP1-117O3.2 |
|
2621 |
0.21 |
| chr3_156468450_156468930 | 0.11 |
TIPARP |
TCDD-inducible poly(ADP-ribose) polymerase |
69735 |
0.11 |
| chr4_108817732_108818165 | 0.11 |
SGMS2 |
sphingomyelin synthase 2 |
2544 |
0.25 |
| chr5_98112819_98113202 | 0.11 |
RGMB |
repulsive guidance molecule family member b |
3671 |
0.26 |
| chr10_62724503_62724836 | 0.11 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
20664 |
0.25 |
| chr5_171380881_171381289 | 0.11 |
FBXW11 |
F-box and WD repeat domain containing 11 |
23675 |
0.23 |
| chr4_141013667_141013926 | 0.11 |
RP11-392B6.1 |
|
35373 |
0.19 |
| chr9_129467204_129467355 | 0.11 |
ENSG00000266403 |
. |
13333 |
0.21 |
| chr9_73539563_73539988 | 0.11 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
55801 |
0.16 |
| chr15_40395062_40395658 | 0.11 |
BMF |
Bcl2 modifying factor |
2927 |
0.21 |
| chr9_23850343_23850766 | 0.11 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
24219 |
0.29 |
| chr2_202015949_202016344 | 0.11 |
CFLAR-AS1 |
CFLAR antisense RNA 1 |
263 |
0.88 |
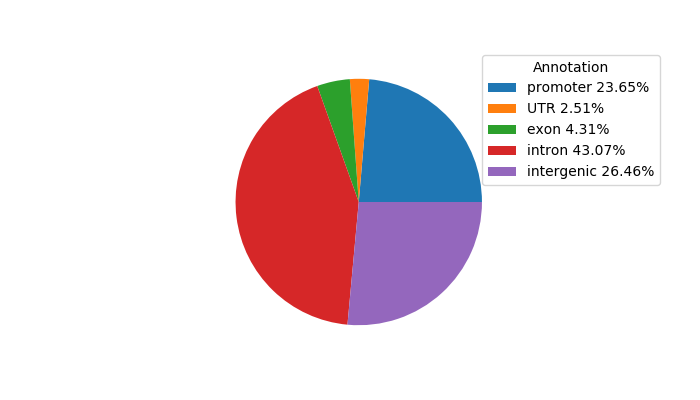
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.2 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.3 | GO:0036303 | lymphangiogenesis(GO:0001946) lymph vessel morphogenesis(GO:0036303) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.1 | GO:0060236 | regulation of mitotic spindle organization(GO:0060236) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.2 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.3 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.1 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.2 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:1901532 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0001547 | antral ovarian follicle growth(GO:0001547) |
| 0.0 | 0.2 | GO:0030224 | monocyte differentiation(GO:0030224) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0043268 | positive regulation of potassium ion transport(GO:0043268) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.0 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0042219 | cellular modified amino acid catabolic process(GO:0042219) |
| 0.0 | 0.2 | GO:0051058 | negative regulation of small GTPase mediated signal transduction(GO:0051058) |
| 0.0 | 0.0 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.0 | GO:0072193 | positive regulation of smooth muscle cell differentiation(GO:0051152) ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.0 | GO:0048875 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0039656 | modulation by virus of host gene expression(GO:0039656) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.0 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0035284 | central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.0 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.0 | 0.1 | GO:0008595 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.0 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0031392 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.0 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0019614 | phenol-containing compound catabolic process(GO:0019336) catechol-containing compound catabolic process(GO:0019614) catecholamine catabolic process(GO:0042424) |
| 0.0 | 0.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.0 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.0 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.2 | GO:0048146 | positive regulation of fibroblast proliferation(GO:0048146) |
| 0.0 | 0.0 | GO:0021898 | commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.0 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0031122 | cytoplasmic microtubule organization(GO:0031122) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.1 | 0.2 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.0 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0002142 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.2 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.0 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.3 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.1 | GO:0030544 | Hsp70 protein binding(GO:0030544) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.0 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.0 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0044620 | ACP phosphopantetheine attachment site binding involved in fatty acid biosynthetic process(GO:0000036) ACP phosphopantetheine attachment site binding(GO:0044620) prosthetic group binding(GO:0051192) |
| 0.0 | 0.2 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.0 | GO:0016901 | oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0000247 | C-8 sterol isomerase activity(GO:0000247) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 0.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.2 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.0 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.0 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.5 | NABA COLLAGENS | Genes encoding collagen proteins |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.0 | REACTOME P53 DEPENDENT G1 DNA DAMAGE RESPONSE | Genes involved in p53-Dependent G1 DNA Damage Response |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.0 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.6 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |