Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
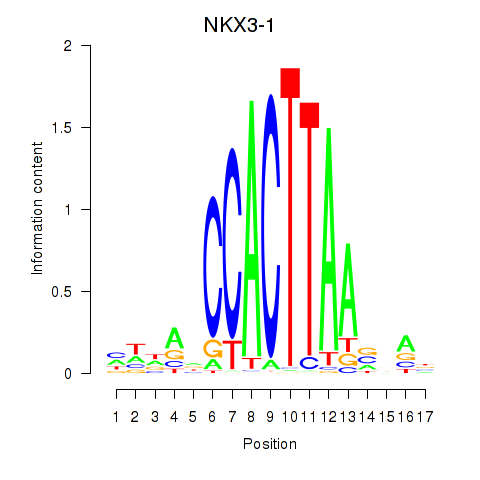
Results for NKX3-1
Z-value: 0.66
Transcription factors associated with NKX3-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX3-1
|
ENSG00000167034.9 | NK3 homeobox 1 |
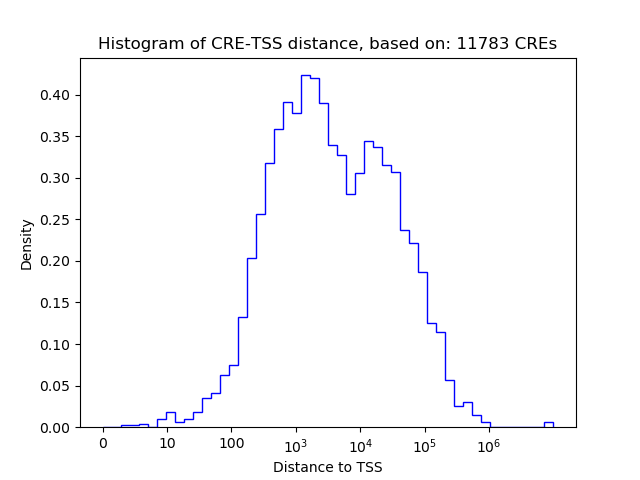
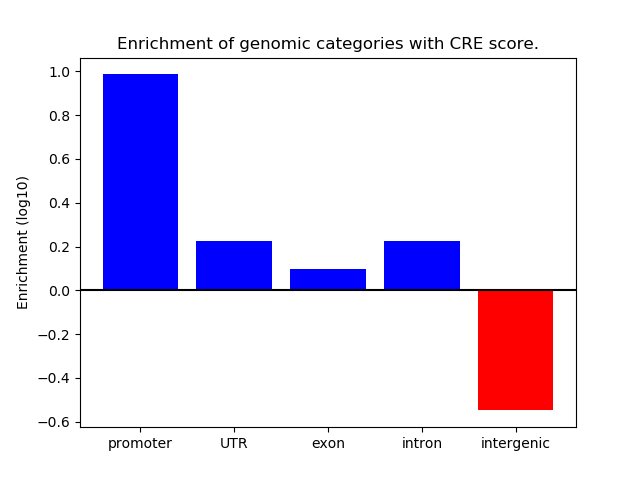
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_23510263_23510414 | NKX3-1 | 30064 | 0.135650 | -0.58 | 9.9e-02 | Click! |
| chr8_23540502_23540896 | NKX3-1 | 259 | 0.909043 | 0.48 | 1.9e-01 | Click! |
| chr8_23539616_23540460 | NKX3-1 | 364 | 0.854562 | -0.15 | 7.0e-01 | Click! |
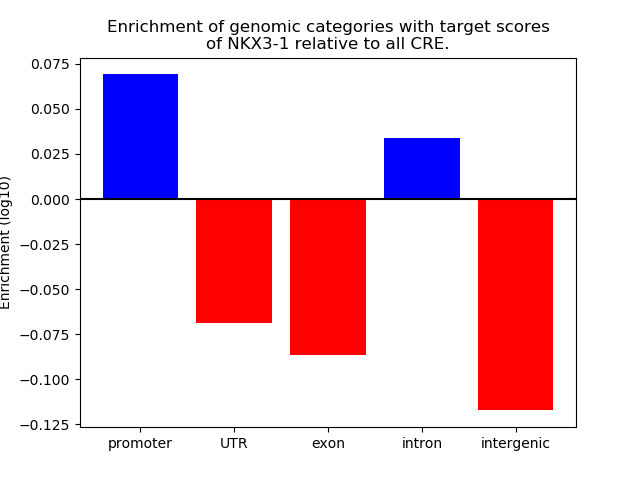
Activity of the NKX3-1 motif across conditions
Conditions sorted by the z-value of the NKX3-1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
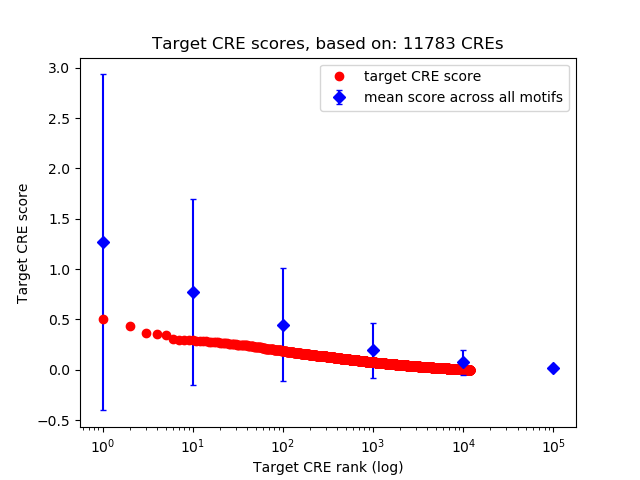
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_24831599_24831750 | 0.50 |
RCAN3 |
RCAN family member 3 |
2287 |
0.27 |
| chr17_33864237_33864745 | 0.44 |
SLFN12L |
schlafen family member 12-like |
389 |
0.76 |
| chr1_160615359_160615600 | 0.36 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
1332 |
0.39 |
| chr7_150148744_150149359 | 0.35 |
GIMAP8 |
GTPase, IMAP family member 8 |
1333 |
0.41 |
| chr14_22999291_22999566 | 0.35 |
TRAJ15 |
T cell receptor alpha joining 15 |
848 |
0.46 |
| chr13_97875674_97876020 | 0.30 |
MBNL2 |
muscleblind-like splicing regulator 2 |
1238 |
0.62 |
| chr7_150413706_150414064 | 0.30 |
GIMAP1 |
GTPase, IMAP family member 1 |
240 |
0.91 |
| chr1_226921433_226921633 | 0.30 |
ITPKB |
inositol-trisphosphate 3-kinase B |
3626 |
0.29 |
| chr15_38854880_38855238 | 0.30 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
1777 |
0.36 |
| chr14_22987346_22987542 | 0.29 |
TRAJ15 |
T cell receptor alpha joining 15 |
11136 |
0.1 |
| chr12_40010191_40010342 | 0.29 |
ABCD2 |
ATP-binding cassette, sub-family D (ALD), member 2 |
3287 |
0.3 |
| chr4_109086470_109087224 | 0.29 |
LEF1 |
lymphoid enhancer-binding factor 1 |
610 |
0.72 |
| chr20_57721382_57721625 | 0.29 |
ZNF831 |
zinc finger protein 831 |
44572 |
0.15 |
| chr13_24541098_24541249 | 0.28 |
SPATA13 |
spermatogenesis associated 13 |
12771 |
0.19 |
| chr13_50657780_50657986 | 0.28 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
1576 |
0.37 |
| chr14_102291594_102292005 | 0.28 |
CTD-2017C7.1 |
|
14069 |
0.14 |
| chr5_58333022_58333173 | 0.28 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
2242 |
0.33 |
| chr14_53593743_53594005 | 0.27 |
DDHD1 |
DDHD domain containing 1 |
22930 |
0.17 |
| chr6_160940752_160940978 | 0.27 |
LPAL2 |
lipoprotein, Lp(a)-like 2, pseudogene |
8826 |
0.3 |
| chr5_54357840_54358274 | 0.27 |
ENSG00000240535 |
. |
19385 |
0.12 |
| chr14_22982783_22983297 | 0.27 |
TRAJ15 |
T cell receptor alpha joining 15 |
15540 |
0.1 |
| chr2_86081133_86081567 | 0.27 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
13427 |
0.17 |
| chr3_18477317_18477468 | 0.26 |
SATB1 |
SATB homeobox 1 |
647 |
0.74 |
| chr1_156785806_156786457 | 0.26 |
SH2D2A |
SH2 domain containing 2A |
495 |
0.51 |
| chr2_214004488_214004639 | 0.26 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
8790 |
0.3 |
| chr8_128808074_128808411 | 0.26 |
ENSG00000249859 |
. |
34 |
0.99 |
| chr1_84612029_84612357 | 0.26 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
2239 |
0.42 |
| chrY_7143368_7143656 | 0.26 |
PRKY |
protein kinase, Y-linked, pseudogene |
1158 |
0.57 |
| chr6_156717396_156717547 | 0.26 |
ENSG00000212295 |
. |
17587 |
0.3 |
| chr4_153459156_153459307 | 0.26 |
ENSG00000268471 |
. |
1651 |
0.34 |
| chr12_6736731_6736882 | 0.25 |
LPAR5 |
lysophosphatidic acid receptor 5 |
4009 |
0.1 |
| chr3_111261912_111262220 | 0.25 |
CD96 |
CD96 molecule |
1069 |
0.62 |
| chr1_100889106_100889257 | 0.25 |
ENSG00000216067 |
. |
44850 |
0.13 |
| chr2_204801338_204802214 | 0.25 |
ICOS |
inducible T-cell co-stimulator |
273 |
0.95 |
| chr3_188115136_188115434 | 0.25 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
157633 |
0.04 |
| chr10_30993847_30994006 | 0.25 |
SVILP1 |
supervillin pseudogene 1 |
9076 |
0.27 |
| chr11_128388558_128388952 | 0.25 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
3341 |
0.29 |
| chr14_69248337_69248488 | 0.24 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
9548 |
0.22 |
| chr5_67577677_67577828 | 0.24 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
1617 |
0.52 |
| chr9_130736775_130737070 | 0.24 |
FAM102A |
family with sequence similarity 102, member A |
5870 |
0.12 |
| chr13_95874473_95874624 | 0.24 |
ENSG00000238463 |
. |
11950 |
0.25 |
| chr13_99216323_99216474 | 0.24 |
STK24 |
serine/threonine kinase 24 |
12719 |
0.21 |
| chr14_22977741_22978031 | 0.24 |
TRAJ15 |
T cell receptor alpha joining 15 |
20694 |
0.09 |
| chr1_43422633_43422983 | 0.24 |
SLC2A1 |
solute carrier family 2 (facilitated glucose transporter), member 1 |
1692 |
0.36 |
| chr1_160616059_160616370 | 0.24 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
597 |
0.7 |
| chr6_109801750_109801916 | 0.24 |
ZBTB24 |
zinc finger and BTB domain containing 24 |
2607 |
0.17 |
| chr12_14924776_14925101 | 0.23 |
HIST4H4 |
histone cluster 4, H4 |
873 |
0.43 |
| chr19_17517338_17517489 | 0.23 |
MVB12A |
multivesicular body subunit 12A |
459 |
0.54 |
| chr7_50267269_50267784 | 0.23 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
76798 |
0.1 |
| chr6_106548995_106549243 | 0.23 |
RP1-134E15.3 |
|
1104 |
0.52 |
| chr7_45068352_45068655 | 0.23 |
CCM2 |
cerebral cavernous malformation 2 |
1232 |
0.4 |
| chr7_38389357_38389573 | 0.23 |
AMPH |
amphiphysin |
113248 |
0.07 |
| chr1_84610970_84611403 | 0.23 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
1232 |
0.6 |
| chr11_128388975_128389288 | 0.23 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
2965 |
0.31 |
| chr1_110046634_110046785 | 0.22 |
AMIGO1 |
adhesion molecule with Ig-like domain 1 |
5595 |
0.11 |
| chr6_159234769_159235011 | 0.22 |
EZR-AS1 |
EZR antisense RNA 1 |
4153 |
0.2 |
| chr2_214010651_214010802 | 0.22 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
2627 |
0.41 |
| chr2_68962051_68962936 | 0.22 |
ARHGAP25 |
Rho GTPase activating protein 25 |
479 |
0.86 |
| chr14_61799079_61799314 | 0.22 |
PRKCH |
protein kinase C, eta |
5565 |
0.21 |
| chr2_198756913_198757256 | 0.22 |
PLCL1 |
phospholipase C-like 1 |
82102 |
0.1 |
| chrX_78343760_78343985 | 0.22 |
GPR174 |
G protein-coupled receptor 174 |
82597 |
0.11 |
| chr1_112167264_112167415 | 0.21 |
RAP1A |
RAP1A, member of RAS oncogene family |
4570 |
0.2 |
| chr5_156621256_156621554 | 0.21 |
ITK |
IL2-inducible T-cell kinase |
13568 |
0.12 |
| chr21_33033050_33033234 | 0.21 |
SOD1 |
superoxide dismutase 1, soluble |
1163 |
0.44 |
| chr1_179801278_179801429 | 0.21 |
RP11-12M5.3 |
|
14027 |
0.16 |
| chr7_8011713_8012079 | 0.21 |
AC006042.7 |
|
2362 |
0.25 |
| chr22_24796948_24797099 | 0.21 |
ADORA2A |
adenosine A2a receptor |
22542 |
0.18 |
| chr10_18790313_18790509 | 0.21 |
RP11-383B4.4 |
|
31854 |
0.18 |
| chr14_22948364_22948515 | 0.21 |
ENSG00000251002 |
. |
1839 |
0.18 |
| chr1_89737700_89738180 | 0.21 |
GBP5 |
guanylate binding protein 5 |
547 |
0.77 |
| chr3_34525180_34525331 | 0.21 |
PDCD6IP |
programmed cell death 6 interacting protein |
685140 |
0.0 |
| chr9_123885039_123885250 | 0.21 |
CNTRL |
centriolin |
1065 |
0.57 |
| chr17_40424104_40424391 | 0.21 |
AC003104.1 |
|
454 |
0.71 |
| chrX_40033281_40033432 | 0.21 |
BCOR |
BCL6 corepressor |
3217 |
0.38 |
| chr17_38019970_38020260 | 0.21 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
264 |
0.89 |
| chr6_106550239_106550540 | 0.20 |
RP1-134E15.3 |
|
2374 |
0.32 |
| chr6_143248743_143249008 | 0.20 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
17463 |
0.25 |
| chr11_76794049_76794313 | 0.20 |
CAPN5 |
calpain 5 |
16141 |
0.16 |
| chr19_16471836_16472011 | 0.20 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
841 |
0.55 |
| chr2_143934651_143934928 | 0.20 |
RP11-190J23.1 |
|
5048 |
0.3 |
| chr15_81585953_81586104 | 0.20 |
IL16 |
interleukin 16 |
3226 |
0.26 |
| chr2_38826300_38826455 | 0.20 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
791 |
0.61 |
| chr4_154360305_154360456 | 0.20 |
KIAA0922 |
KIAA0922 |
27118 |
0.19 |
| chr2_127783302_127783743 | 0.20 |
ENSG00000238788 |
. |
8525 |
0.27 |
| chr1_101784802_101784953 | 0.20 |
RP4-575N6.5 |
|
76163 |
0.09 |
| chr13_42033808_42033959 | 0.20 |
RGCC |
regulator of cell cycle |
2188 |
0.25 |
| chr2_143888975_143889165 | 0.20 |
ARHGAP15 |
Rho GTPase activating protein 15 |
2187 |
0.4 |
| chr18_33550373_33550669 | 0.20 |
C18orf21 |
chromosome 18 open reading frame 21 |
1525 |
0.4 |
| chr6_35267244_35267414 | 0.20 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
1700 |
0.35 |
| chr21_43345808_43345959 | 0.19 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
916 |
0.55 |
| chr15_45007683_45007937 | 0.19 |
B2M |
beta-2-microglobulin |
4095 |
0.17 |
| chr20_57426029_57426307 | 0.19 |
GNAS-AS1 |
GNAS antisense RNA 1 |
210 |
0.89 |
| chr2_235399576_235400049 | 0.19 |
ARL4C |
ADP-ribosylation factor-like 4C |
5432 |
0.35 |
| chr12_45268809_45268960 | 0.19 |
NELL2 |
NEL-like 2 (chicken) |
367 |
0.9 |
| chr13_30950896_30951047 | 0.19 |
KATNAL1 |
katanin p60 subunit A-like 1 |
69350 |
0.11 |
| chr11_73692681_73692970 | 0.19 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
842 |
0.53 |
| chr12_121570606_121571339 | 0.19 |
P2RX7 |
purinergic receptor P2X, ligand-gated ion channel, 7 |
198 |
0.94 |
| chr12_9767730_9767881 | 0.19 |
KLRB1 |
killer cell lectin-like receptor subfamily B, member 1 |
7323 |
0.14 |
| chr18_48492673_48492824 | 0.19 |
ELAC1 |
elaC ribonuclease Z 1 |
1613 |
0.3 |
| chr12_9760266_9760417 | 0.19 |
KLRB1 |
killer cell lectin-like receptor subfamily B, member 1 |
141 |
0.94 |
| chr19_23945488_23945685 | 0.19 |
RPSAP58 |
ribosomal protein SA pseudogene 58 |
221 |
0.93 |
| chr4_38511777_38511989 | 0.19 |
RP11-617D20.1 |
|
114313 |
0.06 |
| chrY_7542080_7542231 | 0.19 |
RFTN1P1 |
raftlin, lipid raft linker 1 pseudogene 1 |
102593 |
0.08 |
| chr18_21572812_21573447 | 0.19 |
TTC39C |
tetratricopeptide repeat domain 39C |
392 |
0.85 |
| chr8_23312978_23313222 | 0.19 |
ENTPD4 |
ectonucleoside triphosphate diphosphohydrolase 4 |
2060 |
0.32 |
| chr4_143321559_143321710 | 0.19 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
30778 |
0.27 |
| chr6_149816889_149817128 | 0.19 |
ZC3H12D |
zinc finger CCCH-type containing 12D |
10811 |
0.15 |
| chrX_71003919_71004210 | 0.18 |
ENSG00000221684 |
. |
24759 |
0.22 |
| chr10_130834757_130834924 | 0.18 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
430608 |
0.01 |
| chrX_70917042_70917365 | 0.18 |
ENSG00000221684 |
. |
62102 |
0.11 |
| chr19_40169930_40170081 | 0.18 |
LGALS17A |
|
9 |
0.96 |
| chr6_154570961_154571273 | 0.18 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
2301 |
0.45 |
| chr15_56537627_56537839 | 0.18 |
TEX9 |
testis expressed 9 |
1526 |
0.37 |
| chr13_99216166_99216317 | 0.18 |
STK24 |
serine/threonine kinase 24 |
12876 |
0.21 |
| chr2_214010385_214010590 | 0.18 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
2866 |
0.4 |
| chr16_81748402_81748687 | 0.18 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
24158 |
0.24 |
| chr2_175708437_175708594 | 0.18 |
CHN1 |
chimerin 1 |
2618 |
0.35 |
| chr9_273119_273416 | 0.18 |
DOCK8 |
dedicator of cytokinesis 8 |
197 |
0.59 |
| chr17_2699734_2700214 | 0.18 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
198 |
0.94 |
| chr12_104888025_104888586 | 0.18 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
37526 |
0.2 |
| chr6_112083181_112083332 | 0.18 |
FYN |
FYN oncogene related to SRC, FGR, YES |
2136 |
0.42 |
| chr10_6626003_6626633 | 0.18 |
PRKCQ |
protein kinase C, theta |
4055 |
0.36 |
| chrY_23417410_23417561 | 0.17 |
CYorf17 |
chromosome Y open reading frame 17 |
130761 |
0.05 |
| chr6_117869427_117870101 | 0.17 |
GOPC |
golgi-associated PDZ and coiled-coil motif containing |
53763 |
0.12 |
| chr13_75900551_75900966 | 0.17 |
TBC1D4 |
TBC1 domain family, member 4 |
14909 |
0.24 |
| chrX_13680096_13680380 | 0.17 |
TCEANC |
transcription elongation factor A (SII) N-terminal and central domain containing |
6824 |
0.2 |
| chr4_154419256_154419548 | 0.17 |
KIAA0922 |
KIAA0922 |
31901 |
0.2 |
| chr7_150147681_150147832 | 0.17 |
GIMAP8 |
GTPase, IMAP family member 8 |
38 |
0.97 |
| chr14_60650630_60650881 | 0.17 |
DHRS7 |
dehydrogenase/reductase (SDR family) member 7 |
14181 |
0.21 |
| chr2_23755902_23756169 | 0.17 |
AC011239.1 |
Uncharacterized protein |
8821 |
0.3 |
| chr14_65880630_65880947 | 0.17 |
FUT8 |
fucosyltransferase 8 (alpha (1,6) fucosyltransferase) |
1048 |
0.63 |
| chr7_150417044_150417195 | 0.17 |
GIMAP1 |
GTPase, IMAP family member 1 |
3474 |
0.2 |
| chr6_24926936_24927087 | 0.17 |
FAM65B |
family with sequence similarity 65, member B |
9177 |
0.23 |
| chr14_99733165_99733625 | 0.17 |
AL109767.1 |
|
4110 |
0.24 |
| chr4_70625874_70626025 | 0.17 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
367 |
0.91 |
| chr11_60744031_60744182 | 0.17 |
CD6 |
CD6 molecule |
4768 |
0.13 |
| chr8_8727017_8727210 | 0.17 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
24042 |
0.18 |
| chr17_28049935_28050167 | 0.17 |
RP11-82O19.1 |
|
38070 |
0.1 |
| chr3_13057259_13057549 | 0.17 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
28868 |
0.23 |
| chr20_45983472_45983818 | 0.17 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
756 |
0.54 |
| chr6_143164322_143164558 | 0.17 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
6256 |
0.3 |
| chr2_197030684_197030835 | 0.17 |
STK17B |
serine/threonine kinase 17b |
4965 |
0.22 |
| chr14_106326931_106327329 | 0.17 |
ENSG00000265714 |
. |
1106 |
0.09 |
| chr4_26885727_26885945 | 0.17 |
STIM2 |
stromal interaction molecule 2 |
22755 |
0.22 |
| chr6_170920732_170921165 | 0.17 |
PDCD2 |
programmed cell death 2 |
27200 |
0.19 |
| chr4_154388914_154389185 | 0.17 |
KIAA0922 |
KIAA0922 |
1548 |
0.49 |
| chr11_64636867_64637025 | 0.17 |
EHD1 |
EH-domain containing 1 |
6192 |
0.1 |
| chr6_118880020_118880242 | 0.17 |
PLN |
phospholamban |
10670 |
0.3 |
| chr2_162915924_162916075 | 0.17 |
AC008063.2 |
|
13767 |
0.2 |
| chr8_71564383_71564535 | 0.17 |
ENSG00000243532 |
. |
2659 |
0.21 |
| chr6_106547367_106547905 | 0.17 |
RP1-134E15.3 |
|
379 |
0.74 |
| chr2_191274324_191274475 | 0.17 |
MFSD6 |
major facilitator superfamily domain containing 6 |
1318 |
0.47 |
| chr10_6110992_6111224 | 0.16 |
IL2RA |
interleukin 2 receptor, alpha |
6820 |
0.16 |
| chr3_110345992_110346143 | 0.16 |
ENSG00000221206 |
. |
75255 |
0.12 |
| chr15_100345675_100345826 | 0.16 |
CTD-2054N24.2 |
Uncharacterized protein |
1478 |
0.31 |
| chr9_125799790_125799941 | 0.16 |
GPR21 |
G protein-coupled receptor 21 |
3059 |
0.22 |
| chr10_61667108_61667259 | 0.16 |
CCDC6 |
coiled-coil domain containing 6 |
769 |
0.74 |
| chr2_202127394_202127951 | 0.16 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
993 |
0.56 |
| chr4_40205321_40205527 | 0.16 |
RHOH |
ras homolog family member H |
3460 |
0.27 |
| chr4_154388544_154388898 | 0.16 |
KIAA0922 |
KIAA0922 |
1220 |
0.57 |
| chrX_118819066_118819466 | 0.16 |
SEPT6 |
septin 6 |
7526 |
0.18 |
| chr16_17463239_17463649 | 0.16 |
XYLT1 |
xylosyltransferase I |
101294 |
0.09 |
| chr7_26193443_26193677 | 0.16 |
NFE2L3 |
nuclear factor, erythroid 2-like 3 |
1700 |
0.36 |
| chr4_122164507_122165136 | 0.16 |
TNIP3 |
TNFAIP3 interacting protein 3 |
16200 |
0.22 |
| chr10_32665664_32666047 | 0.16 |
EPC1 |
enhancer of polycomb homolog 1 (Drosophila) |
1871 |
0.27 |
| chr12_104854827_104855220 | 0.16 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
4244 |
0.32 |
| chr11_70211468_70211619 | 0.16 |
AP000487.6 |
|
2069 |
0.21 |
| chr2_192540821_192541028 | 0.16 |
NABP1 |
nucleic acid binding protein 1 |
1938 |
0.47 |
| chr1_160611300_160611550 | 0.16 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
5386 |
0.18 |
| chr15_65187513_65187769 | 0.16 |
ENSG00000264929 |
. |
4797 |
0.17 |
| chr3_56788528_56788714 | 0.16 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
20974 |
0.24 |
| chr4_4765123_4765274 | 0.16 |
MSX1 |
msh homeobox 1 |
96195 |
0.08 |
| chr6_161351612_161351773 | 0.16 |
RP3-428L16.1 |
|
420 |
0.89 |
| chr6_149415675_149415826 | 0.16 |
RP11-162J8.3 |
|
62041 |
0.12 |
| chr2_62702174_62702386 | 0.16 |
ENSG00000241625 |
. |
16033 |
0.23 |
| chr10_75011688_75012124 | 0.16 |
MRPS16 |
mitochondrial ribosomal protein S16 |
484 |
0.47 |
| chr10_70360809_70361174 | 0.16 |
TET1 |
tet methylcytosine dioxygenase 1 |
40578 |
0.13 |
| chr7_50355345_50355813 | 0.16 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
7261 |
0.3 |
| chr22_44577376_44577610 | 0.16 |
PARVG |
parvin, gamma |
215 |
0.96 |
| chrX_123482778_123482929 | 0.15 |
SH2D1A |
SH2 domain containing 1A |
2414 |
0.44 |
| chr1_226922484_226922931 | 0.15 |
ITPKB |
inositol-trisphosphate 3-kinase B |
2452 |
0.34 |
| chr14_92339571_92339902 | 0.15 |
FBLN5 |
fibulin 5 |
4650 |
0.23 |
| chr4_109080254_109080405 | 0.15 |
LEF1 |
lymphoid enhancer-binding factor 1 |
7128 |
0.24 |
| chr4_96645430_96645581 | 0.15 |
PDHA2 |
pyruvate dehydrogenase (lipoamide) alpha 2 |
115734 |
0.06 |
| chr6_170403243_170403627 | 0.15 |
RP11-302L19.1 |
|
74306 |
0.11 |
| chr18_66440168_66440319 | 0.15 |
RP11-106E15.1 |
|
18423 |
0.2 |
| chr5_55315108_55315259 | 0.15 |
ENSG00000238326 |
. |
18734 |
0.15 |
| chr13_42849044_42849223 | 0.15 |
AKAP11 |
A kinase (PRKA) anchor protein 11 |
2844 |
0.38 |
| chr14_91488738_91489174 | 0.15 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
37522 |
0.15 |
| chr19_55766409_55766580 | 0.15 |
PPP6R1 |
protein phosphatase 6, regulatory subunit 1 |
643 |
0.49 |
| chr19_42432968_42433149 | 0.15 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
5812 |
0.14 |
| chr5_1800151_1800360 | 0.15 |
MRPL36 |
mitochondrial ribosomal protein L36 |
246 |
0.88 |
| chr3_186745964_186746115 | 0.15 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
2768 |
0.33 |
| chr4_78738053_78738204 | 0.15 |
CNOT6L |
CCR4-NOT transcription complex, subunit 6-like |
2089 |
0.42 |
| chr8_10981801_10982129 | 0.15 |
AF131215.3 |
|
1609 |
0.27 |
| chr11_128380502_128380688 | 0.15 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
5306 |
0.26 |
| chr13_75900993_75901212 | 0.15 |
TBC1D4 |
TBC1 domain family, member 4 |
14565 |
0.24 |
| chr2_143898305_143898456 | 0.15 |
ARHGAP15 |
Rho GTPase activating protein 15 |
11497 |
0.25 |
| chr20_33999186_33999337 | 0.15 |
UQCC1 |
ubiquinol-cytochrome c reductase complex assembly factor 1 |
505 |
0.71 |
| chr15_22547843_22548176 | 0.15 |
ENSG00000221641 |
. |
34729 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 | 0.2 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.1 | 0.2 | GO:0090382 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.1 | 0.4 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.2 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.2 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.2 | GO:0071025 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.2 | GO:0051569 | regulation of histone H3-K4 methylation(GO:0051569) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.2 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.3 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
| 0.0 | 0.1 | GO:0043368 | positive T cell selection(GO:0043368) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0002475 | antigen processing and presentation via MHC class Ib(GO:0002475) |
| 0.0 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.1 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0002420 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0043628 | ncRNA 3'-end processing(GO:0043628) |
| 0.0 | 0.1 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0051136 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.0 | GO:0002839 | regulation of response to tumor cell(GO:0002834) positive regulation of response to tumor cell(GO:0002836) regulation of immune response to tumor cell(GO:0002837) positive regulation of immune response to tumor cell(GO:0002839) |
| 0.0 | 0.3 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.7 | GO:0006997 | nucleus organization(GO:0006997) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.0 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.1 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.2 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0002329 | immature B cell differentiation(GO:0002327) pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.1 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.2 | GO:0000303 | response to superoxide(GO:0000303) |
| 0.0 | 0.2 | GO:0019059 | obsolete initiation of viral infection(GO:0019059) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.2 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0006102 | isocitrate metabolic process(GO:0006102) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.0 | 0.1 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.0 | GO:0032351 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.0 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.1 | GO:0051642 | centrosome localization(GO:0051642) |
| 0.0 | 0.1 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.1 | GO:0046415 | urate metabolic process(GO:0046415) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:0031935 | regulation of chromatin silencing(GO:0031935) negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0006301 | postreplication repair(GO:0006301) |
| 0.0 | 0.0 | GO:0033345 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0009120 | deoxyribonucleoside metabolic process(GO:0009120) |
| 0.0 | 0.1 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.0 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.1 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous antigen(GO:0019883) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0034390 | smooth muscle cell apoptotic process(GO:0034390) regulation of smooth muscle cell apoptotic process(GO:0034391) negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.9 | GO:0050852 | T cell receptor signaling pathway(GO:0050852) |
| 0.0 | 0.1 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.3 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0030889 | negative regulation of B cell proliferation(GO:0030889) |
| 0.0 | 0.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.0 | 0.1 | GO:0016255 | attachment of GPI anchor to protein(GO:0016255) |
| 0.0 | 0.0 | GO:0071034 | CUT catabolic process(GO:0071034) CUT metabolic process(GO:0071043) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.1 | GO:0098534 | centriole replication(GO:0007099) centriole assembly(GO:0098534) |
| 0.0 | 0.0 | GO:0031507 | heterochromatin assembly(GO:0031507) heterochromatin organization(GO:0070828) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0010833 | telomere maintenance via telomere lengthening(GO:0010833) |
| 0.0 | 0.0 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.1 | GO:0006264 | mitochondrial DNA replication(GO:0006264) |
| 0.0 | 0.0 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.0 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.0 | GO:0070265 | necrotic cell death(GO:0070265) |
| 0.0 | 0.0 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0000002 | mitochondrial genome maintenance(GO:0000002) |
| 0.0 | 0.1 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.0 | GO:0021819 | cerebral cortex radial glia guided migration(GO:0021801) layer formation in cerebral cortex(GO:0021819) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.1 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.0 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.1 | GO:0045954 | positive regulation of natural killer cell mediated immunity(GO:0002717) positive regulation of natural killer cell mediated cytotoxicity(GO:0045954) |
| 0.0 | 0.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0000724 | double-strand break repair via homologous recombination(GO:0000724) recombinational repair(GO:0000725) |
| 0.0 | 0.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.2 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 | 0.0 | GO:0072600 | protein targeting to Golgi(GO:0000042) establishment of protein localization to Golgi(GO:0072600) |
| 0.0 | 0.1 | GO:0070076 | histone lysine demethylation(GO:0070076) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0006787 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0006065 | UDP-glucuronate biosynthetic process(GO:0006065) |
| 0.0 | 0.2 | GO:0043484 | regulation of RNA splicing(GO:0043484) |
| 0.0 | 0.0 | GO:0032469 | endoplasmic reticulum calcium ion homeostasis(GO:0032469) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0072487 | MSL complex(GO:0072487) |
| 0.1 | 0.3 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.1 | 0.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.3 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.2 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.1 | GO:0005683 | U7 snRNP(GO:0005683) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.3 | GO:0005762 | organellar large ribosomal subunit(GO:0000315) mitochondrial large ribosomal subunit(GO:0005762) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.2 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.2 | GO:0030130 | clathrin coat of trans-Golgi network vesicle(GO:0030130) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.0 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0070419 | nonhomologous end joining complex(GO:0070419) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.0 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.1 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.1 | 0.6 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.2 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.0 | GO:0010521 | telomerase inhibitor activity(GO:0010521) |
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.2 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.1 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 1.0 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0004448 | isocitrate dehydrogenase activity(GO:0004448) |
| 0.0 | 0.2 | GO:0030276 | clathrin binding(GO:0030276) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.0 | GO:0016406 | carnitine O-acyltransferase activity(GO:0016406) |
| 0.0 | 0.3 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.5 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0090482 | vitamin transmembrane transporter activity(GO:0090482) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.0 | 0.1 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.0 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.1 | GO:0016421 | CoA carboxylase activity(GO:0016421) |
| 0.0 | 0.1 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.0 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.0 | GO:0035173 | histone kinase activity(GO:0035173) |
| 0.0 | 0.3 | GO:0001614 | purinergic nucleotide receptor activity(GO:0001614) nucleotide receptor activity(GO:0016502) |
| 0.0 | 0.1 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.1 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.1 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.2 | GO:0005246 | calcium channel regulator activity(GO:0005246) |
| 0.0 | 0.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.0 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.0 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.1 | GO:0015211 | purine nucleoside transmembrane transporter activity(GO:0015211) |
| 0.0 | 0.0 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.0 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.0 | GO:0042156 | obsolete zinc-mediated transcriptional activator activity(GO:0042156) |
| 0.0 | 0.1 | GO:0004437 | obsolete inositol or phosphatidylinositol phosphatase activity(GO:0004437) |
| 0.0 | 0.1 | GO:1901677 | organophosphate ester transmembrane transporter activity(GO:0015605) phosphate transmembrane transporter activity(GO:1901677) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.2 | GO:0016409 | palmitoyltransferase activity(GO:0016409) |
| 0.0 | 0.0 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.1 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.0 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.5 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.0 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.5 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.6 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.0 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.0 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | PID BETA CATENIN DEG PATHWAY | Degradation of beta catenin |
| 0.0 | 0.0 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.5 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME ACTIVATION OF CHAPERONE GENES BY ATF6 ALPHA | Genes involved in Activation of Chaperone Genes by ATF6-alpha |
| 0.1 | 0.1 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.1 | 1.0 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME G BETA GAMMA SIGNALLING THROUGH PI3KGAMMA | Genes involved in G beta:gamma signalling through PI3Kgamma |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.0 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.1 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.1 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.0 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.1 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 1.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.6 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |
| 0.0 | 0.0 | REACTOME PEPTIDE CHAIN ELONGATION | Genes involved in Peptide chain elongation |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.1 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.2 | REACTOME PERK REGULATED GENE EXPRESSION | Genes involved in PERK regulated gene expression |
| 0.0 | 0.1 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.0 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |