Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
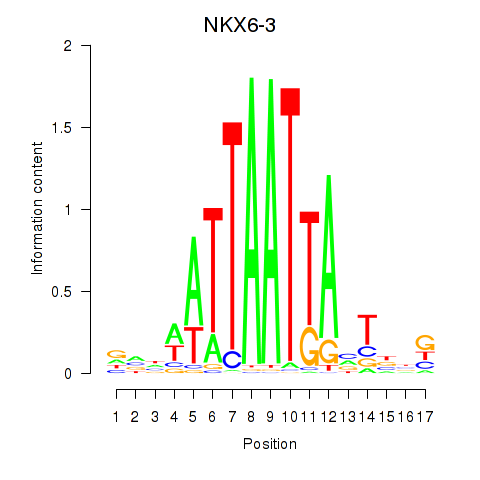
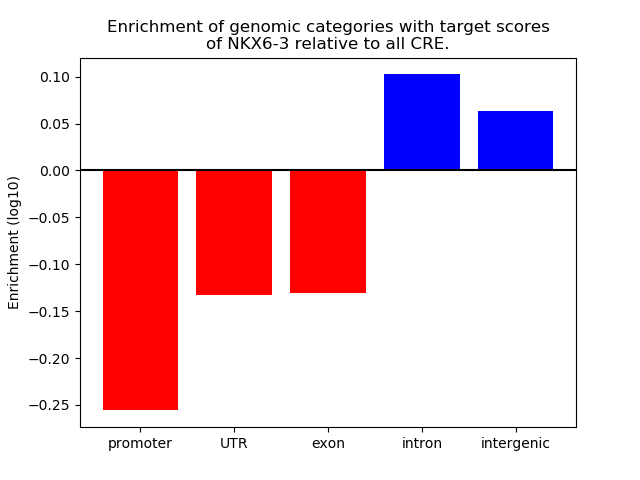
Results for NKX6-3
Z-value: 0.45
Transcription factors associated with NKX6-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX6-3
|
ENSG00000165066.11 | NK6 homeobox 3 |
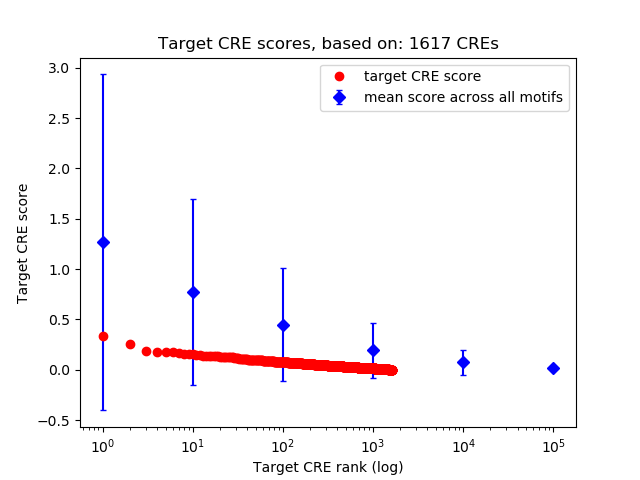
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_41511279_41511901 | NKX6-3 | 2735 | 0.179446 | -0.69 | 4.2e-02 | Click! |
| chr8_41503949_41504289 | NKX6-3 | 759 | 0.542759 | -0.68 | 4.4e-02 | Click! |
| chr8_41510934_41511242 | NKX6-3 | 2233 | 0.206680 | -0.56 | 1.2e-01 | Click! |
Activity of the NKX6-3 motif across conditions
Conditions sorted by the z-value of the NKX6-3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
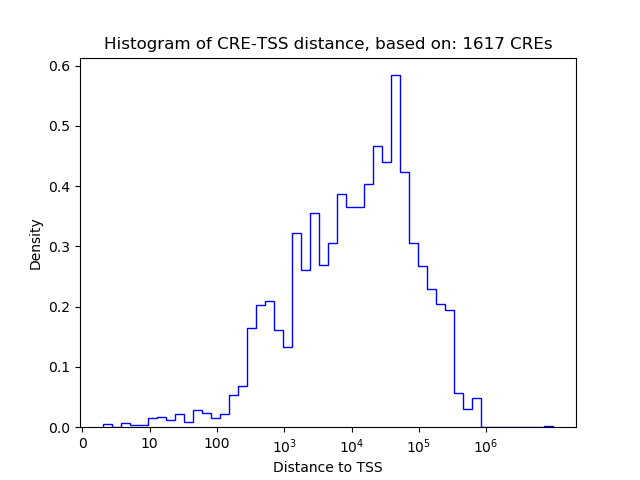
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_114829080_114829277 | 0.33 |
PLS3 |
plastin 3 |
1313 |
0.48 |
| chr7_129933276_129933885 | 0.26 |
CPA4 |
carboxypeptidase A4 |
461 |
0.76 |
| chrX_13906420_13906604 | 0.18 |
GPM6B |
glycoprotein M6B |
664 |
0.81 |
| chr19_58488964_58489115 | 0.18 |
C19orf18 |
chromosome 19 open reading frame 18 |
3137 |
0.15 |
| chr5_147285141_147285292 | 0.18 |
C5orf46 |
chromosome 5 open reading frame 46 |
849 |
0.57 |
| chr18_60385543_60385694 | 0.17 |
PHLPP1 |
PH domain and leucine rich repeat protein phosphatase 1 |
2935 |
0.28 |
| chr10_33798417_33798757 | 0.17 |
NRP1 |
neuropilin 1 |
173397 |
0.03 |
| chr7_98970111_98970529 | 0.16 |
ARPC1B |
actin related protein 2/3 complex, subunit 1B, 41kDa |
1552 |
0.28 |
| chr5_120114861_120115414 | 0.16 |
ENSG00000222609 |
. |
68602 |
0.14 |
| chr5_1883178_1883404 | 0.15 |
IRX4 |
iroquois homeobox 4 |
411 |
0.66 |
| chr11_22724799_22724998 | 0.15 |
ENSG00000222427 |
. |
15001 |
0.2 |
| chr6_21227526_21227677 | 0.15 |
ENSG00000252046 |
. |
233802 |
0.02 |
| chr5_97850230_97850381 | 0.14 |
ENSG00000223053 |
. |
124630 |
0.06 |
| chr3_128207705_128209147 | 0.14 |
RP11-475N22.4 |
|
326 |
0.79 |
| chrX_83589461_83589612 | 0.14 |
ENSG00000221494 |
. |
108700 |
0.07 |
| chr19_6087775_6088269 | 0.14 |
CTC-232P5.3 |
|
20058 |
0.13 |
| chr4_90850866_90851017 | 0.14 |
MMRN1 |
multimerin 1 |
27807 |
0.23 |
| chr2_110872434_110873339 | 0.14 |
MALL |
mal, T-cell differentiation protein-like |
713 |
0.63 |
| chr13_24146066_24146217 | 0.13 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
1338 |
0.59 |
| chr3_69796924_69797075 | 0.13 |
MITF |
microphthalmia-associated transcription factor |
8377 |
0.25 |
| chr5_97850046_97850197 | 0.13 |
ENSG00000223053 |
. |
124814 |
0.06 |
| chr14_101867418_101867569 | 0.13 |
ENSG00000258498 |
. |
159266 |
0.02 |
| chr16_64450080_64450292 | 0.13 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
643395 |
0.0 |
| chr1_82670427_82670613 | 0.13 |
LPHN2 |
latrophilin 2 |
224947 |
0.02 |
| chr2_158039761_158039912 | 0.13 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
74274 |
0.11 |
| chr20_45754385_45754678 | 0.13 |
ENSG00000264901 |
. |
41078 |
0.18 |
| chr6_148831295_148831530 | 0.12 |
ENSG00000223322 |
. |
13964 |
0.3 |
| chr3_54897831_54897982 | 0.12 |
CACNA2D3-AS1 |
CACNA2D3 antisense RNA 1 |
37376 |
0.21 |
| chr3_10315639_10315790 | 0.12 |
TATDN2 |
TatD DNase domain containing 2 |
3110 |
0.16 |
| chr13_79182716_79183828 | 0.12 |
POU4F1 |
POU class 4 homeobox 1 |
5599 |
0.19 |
| chr14_73778052_73778238 | 0.11 |
NUMB |
numb homolog (Drosophila) |
12062 |
0.17 |
| chr7_4046197_4046504 | 0.11 |
SDK1 |
sidekick cell adhesion molecule 1 |
55798 |
0.18 |
| chr9_67293693_67293844 | 0.11 |
RP11-236F9.2 |
|
8470 |
0.29 |
| chr19_19823801_19824098 | 0.11 |
ZNF14 |
zinc finger protein 14 |
19957 |
0.13 |
| chr8_93114852_93115389 | 0.11 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
334 |
0.94 |
| chr7_94025506_94025990 | 0.11 |
COL1A2 |
collagen, type I, alpha 2 |
1875 |
0.46 |
| chr10_60401833_60402026 | 0.10 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
129029 |
0.06 |
| chr14_52123850_52124001 | 0.10 |
FRMD6 |
FERM domain containing 6 |
5227 |
0.22 |
| chr6_85823864_85824548 | 0.10 |
NT5E |
5'-nucleotidase, ecto (CD73) |
335603 |
0.01 |
| chr12_15304261_15304412 | 0.10 |
RERG |
RAS-like, estrogen-regulated, growth inhibitor |
13239 |
0.23 |
| chr1_198238845_198238996 | 0.10 |
NEK7 |
NIMA-related kinase 7 |
48991 |
0.19 |
| chr7_17720120_17720505 | 0.10 |
SNX13 |
sorting nexin 13 |
259779 |
0.02 |
| chr5_140989194_140989345 | 0.10 |
AC008781.7 |
|
8712 |
0.11 |
| chr2_101731109_101731260 | 0.10 |
ENSG00000265860 |
. |
21153 |
0.19 |
| chr9_20041214_20041376 | 0.10 |
ENSG00000266224 |
. |
211544 |
0.02 |
| chr5_92922789_92922940 | 0.10 |
ENSG00000237187 |
. |
1510 |
0.41 |
| chr13_45242490_45242715 | 0.10 |
ENSG00000238932 |
. |
39793 |
0.2 |
| chr10_101689203_101689354 | 0.10 |
DNMBP |
dynamin binding protein |
1475 |
0.38 |
| chr9_132796775_132796926 | 0.10 |
FNBP1 |
formin binding protein 1 |
8591 |
0.2 |
| chr2_230138192_230138530 | 0.10 |
PID1 |
phosphotyrosine interaction domain containing 1 |
2360 |
0.4 |
| chr6_130180797_130180948 | 0.10 |
TMEM244 |
transmembrane protein 244 |
1544 |
0.54 |
| chr2_38828054_38829790 | 0.10 |
AC011247.3 |
|
182 |
0.89 |
| chr12_54473950_54474101 | 0.10 |
RP11-834C11.7 |
|
47 |
0.95 |
| chr3_133020367_133020518 | 0.10 |
RP11-402L6.1 |
|
44410 |
0.17 |
| chr8_41200524_41200675 | 0.10 |
SFRP1 |
secreted frizzled-related protein 1 |
33583 |
0.15 |
| chr5_71643973_71644124 | 0.09 |
PTCD2 |
pentatricopeptide repeat domain 2 |
27823 |
0.16 |
| chr18_52804048_52804199 | 0.09 |
ENSG00000252437 |
. |
9753 |
0.29 |
| chr16_4424352_4424847 | 0.09 |
VASN |
vasorin |
2750 |
0.17 |
| chrX_9269682_9269833 | 0.09 |
TBL1X |
transducin (beta)-like 1X-linked |
161578 |
0.04 |
| chr3_89163237_89163857 | 0.09 |
EPHA3 |
EPH receptor A3 |
6721 |
0.35 |
| chr12_78336292_78336638 | 0.09 |
NAV3 |
neuron navigator 3 |
23591 |
0.28 |
| chr15_56204998_56205389 | 0.09 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
2659 |
0.34 |
| chr5_10551768_10551919 | 0.09 |
ANKRD33B |
ankyrin repeat domain 33B |
12737 |
0.19 |
| chr5_76134459_76134610 | 0.09 |
ENSG00000264391 |
. |
1514 |
0.35 |
| chr7_92224542_92224693 | 0.09 |
FAM133B |
family with sequence similarity 133, member B |
4909 |
0.24 |
| chrX_95690881_95691032 | 0.09 |
ENSG00000223260 |
. |
25175 |
0.28 |
| chr6_45536778_45536929 | 0.09 |
ENSG00000252738 |
. |
76988 |
0.11 |
| chr7_92254665_92254816 | 0.09 |
FAM133B |
family with sequence similarity 133, member B |
35032 |
0.17 |
| chr2_19172713_19172864 | 0.09 |
ENSG00000266738 |
. |
375402 |
0.01 |
| chr6_42284093_42284281 | 0.09 |
ENSG00000221252 |
. |
95947 |
0.06 |
| chr15_73345145_73345907 | 0.09 |
NEO1 |
neogenin 1 |
575 |
0.85 |
| chr18_46620789_46620940 | 0.09 |
DYM |
dymeclin |
2999 |
0.26 |
| chr13_97906946_97907208 | 0.09 |
MBNL2 |
muscleblind-like splicing regulator 2 |
21381 |
0.26 |
| chr18_20407102_20407268 | 0.08 |
RBBP8 |
retinoblastoma binding protein 8 |
28961 |
0.16 |
| chr10_15368154_15368305 | 0.08 |
FAM171A1 |
family with sequence similarity 171, member A1 |
44829 |
0.17 |
| chr17_69531749_69531900 | 0.08 |
ENSG00000222563 |
. |
224515 |
0.02 |
| chr4_53507903_53508112 | 0.08 |
USP46 |
ubiquitin specific peptidase 46 |
14750 |
0.2 |
| chr18_42313149_42313495 | 0.08 |
SETBP1 |
SET binding protein 1 |
36193 |
0.24 |
| chr9_93557702_93558119 | 0.08 |
SYK |
spleen tyrosine kinase |
6159 |
0.34 |
| chr12_11850521_11850672 | 0.08 |
ETV6 |
ets variant 6 |
47808 |
0.17 |
| chr7_13985407_13985644 | 0.08 |
ETV1 |
ets variant 1 |
40541 |
0.2 |
| chr1_51171312_51171463 | 0.08 |
ENSG00000252825 |
. |
44581 |
0.17 |
| chr15_96893541_96893692 | 0.08 |
AC087477.1 |
Uncharacterized protein |
10871 |
0.16 |
| chr1_65005729_65006114 | 0.08 |
ENSG00000264470 |
. |
39609 |
0.19 |
| chr14_77543003_77543154 | 0.08 |
CIPC |
CLOCK-interacting pacemaker |
21362 |
0.15 |
| chr9_12882212_12882363 | 0.08 |
ENSG00000222658 |
. |
3033 |
0.35 |
| chr4_140808579_140808865 | 0.08 |
MAML3 |
mastermind-like 3 (Drosophila) |
2484 |
0.4 |
| chr18_28779723_28779874 | 0.08 |
DSC1 |
desmocollin 1 |
36979 |
0.15 |
| chr13_80658291_80658484 | 0.08 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
255407 |
0.02 |
| chr5_133861629_133862874 | 0.08 |
JADE2 |
jade family PHD finger 2 |
76 |
0.97 |
| chr21_36286009_36286160 | 0.08 |
RUNX1 |
runt-related transcription factor 1 |
23997 |
0.27 |
| chr13_77897361_77897512 | 0.08 |
MYCBP2 |
MYC binding protein 2, E3 ubiquitin protein ligase |
3378 |
0.37 |
| chr12_64580629_64580780 | 0.08 |
ENSG00000212298 |
. |
17869 |
0.15 |
| chr9_89952337_89952906 | 0.08 |
ENSG00000212421 |
. |
77256 |
0.11 |
| chr5_152602934_152603085 | 0.08 |
GRIA1 |
glutamate receptor, ionotropic, AMPA 1 |
267097 |
0.02 |
| chr15_48436377_48436744 | 0.08 |
MYEF2 |
myelin expression factor 2 |
4963 |
0.17 |
| chr6_27060216_27060434 | 0.08 |
ENSG00000222800 |
. |
17434 |
0.13 |
| chrX_107685038_107685189 | 0.08 |
COL4A5 |
collagen, type IV, alpha 5 |
2001 |
0.34 |
| chr14_93720610_93720761 | 0.08 |
UBR7 |
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
35658 |
0.11 |
| chr7_115967399_115967904 | 0.08 |
AC073130.3 |
|
299 |
0.91 |
| chr6_159514146_159514297 | 0.08 |
TAGAP |
T-cell activation RhoGTPase activating protein |
48037 |
0.13 |
| chr4_177930341_177930492 | 0.08 |
ENSG00000222859 |
. |
172180 |
0.03 |
| chr3_37363190_37363371 | 0.07 |
ENSG00000222208 |
. |
19860 |
0.18 |
| chr14_88851403_88852544 | 0.07 |
SPATA7 |
spermatogenesis associated 7 |
15 |
0.98 |
| chrX_133523066_133523217 | 0.07 |
ENSG00000251962 |
. |
14271 |
0.17 |
| chr4_141866142_141866293 | 0.07 |
RNF150 |
ring finger protein 150 |
51973 |
0.17 |
| chr8_116675050_116675578 | 0.07 |
TRPS1 |
trichorhinophalangeal syndrome I |
1409 |
0.6 |
| chr1_215183894_215184045 | 0.07 |
KCNK2 |
potassium channel, subfamily K, member 2 |
4771 |
0.37 |
| chr10_69831641_69832005 | 0.07 |
HERC4 |
HECT and RLD domain containing E3 ubiquitin protein ligase 4 |
2071 |
0.33 |
| chr2_38295840_38295991 | 0.07 |
RMDN2-AS1 |
RMDN2 antisense RNA 1 |
1731 |
0.34 |
| chr15_93362136_93362546 | 0.07 |
FAM174B |
family with sequence similarity 174, member B |
9313 |
0.17 |
| chr15_50427680_50427831 | 0.07 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
16336 |
0.2 |
| chr6_83094369_83094520 | 0.07 |
TPBG |
trophoblast glycoprotein |
20483 |
0.28 |
| chr13_36058740_36058891 | 0.07 |
NBEA |
neurobeachin |
7929 |
0.2 |
| chr10_63812743_63812894 | 0.07 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
3848 |
0.32 |
| chr12_26164349_26164500 | 0.07 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
245 |
0.95 |
| chr6_6005898_6006049 | 0.07 |
NRN1 |
neuritin 1 |
1227 |
0.56 |
| chr11_111849006_111849202 | 0.07 |
DIXDC1 |
DIX domain containing 1 |
1071 |
0.39 |
| chr7_104854367_104854518 | 0.07 |
SRPK2 |
SRSF protein kinase 2 |
55020 |
0.13 |
| chr10_111217087_111217238 | 0.07 |
ENSG00000200217 |
. |
217765 |
0.02 |
| chr2_25978953_25979253 | 0.07 |
ASXL2 |
additional sex combs like 2 (Drosophila) |
24075 |
0.18 |
| chr3_87036535_87037613 | 0.07 |
VGLL3 |
vestigial like 3 (Drosophila) |
2778 |
0.42 |
| chr4_170081571_170081722 | 0.07 |
RP11-327O17.2 |
|
41299 |
0.18 |
| chr11_128600565_128600716 | 0.07 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
34045 |
0.15 |
| chr7_48210671_48210822 | 0.07 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
311 |
0.94 |
| chr7_85112676_85112827 | 0.07 |
SEMA3D |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D |
296580 |
0.01 |
| chr6_125684129_125684280 | 0.07 |
RP11-735G4.1 |
|
11266 |
0.27 |
| chr9_100743461_100743673 | 0.07 |
ANP32B |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member B |
2076 |
0.25 |
| chr7_70201396_70202343 | 0.07 |
AUTS2 |
autism susceptibility candidate 2 |
7744 |
0.34 |
| chr10_33670460_33670611 | 0.07 |
NRP1 |
neuropilin 1 |
45345 |
0.19 |
| chr12_67041467_67041618 | 0.07 |
GRIP1 |
glutamate receptor interacting protein 1 |
413 |
0.92 |
| chr10_34189936_34190141 | 0.07 |
ENSG00000199200 |
. |
300964 |
0.01 |
| chr10_74600201_74600352 | 0.07 |
OIT3 |
oncoprotein induced transcript 3 |
53063 |
0.12 |
| chr6_9139698_9139849 | 0.07 |
SLC35B3 |
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
703979 |
0.0 |
| chr14_20895899_20896050 | 0.07 |
KLHL33 |
kelch-like family member 33 |
7827 |
0.08 |
| chr8_90742457_90742611 | 0.07 |
RIPK2 |
receptor-interacting serine-threonine kinase 2 |
27441 |
0.25 |
| chr7_87720149_87720440 | 0.07 |
ADAM22 |
ADAM metallopeptidase domain 22 |
72052 |
0.11 |
| chr3_30553614_30553765 | 0.07 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
94305 |
0.09 |
| chr4_15685369_15685921 | 0.07 |
FAM200B |
family with sequence similarity 200, member B |
2216 |
0.26 |
| chr17_73348111_73348291 | 0.07 |
RP11-16C1.3 |
|
7715 |
0.11 |
| chr10_24754741_24755189 | 0.07 |
KIAA1217 |
KIAA1217 |
495 |
0.84 |
| chr6_143026333_143026484 | 0.07 |
RP1-67K17.3 |
|
43172 |
0.2 |
| chr15_82414957_82415108 | 0.07 |
RP11-597K23.2 |
Uncharacterized protein |
34000 |
0.16 |
| chr10_103442163_103442336 | 0.07 |
FBXW4 |
F-box and WD repeat domain containing 4 |
12767 |
0.18 |
| chr4_139143471_139143622 | 0.07 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
19957 |
0.26 |
| chr5_140420029_140420180 | 0.07 |
PCDHB1 |
protocadherin beta 1 |
10875 |
0.12 |
| chr4_14728626_14728777 | 0.07 |
ENSG00000202449 |
. |
36367 |
0.24 |
| chr2_191517520_191517671 | 0.07 |
NAB1 |
NGFI-A binding protein 1 (EGR1 binding protein 1) |
856 |
0.66 |
| chr20_17538596_17539086 | 0.07 |
BFSP1 |
beaded filament structural protein 1, filensin |
642 |
0.72 |
| chr4_81118792_81119547 | 0.07 |
PRDM8 |
PR domain containing 8 |
511 |
0.82 |
| chr20_657176_657327 | 0.07 |
SCRT2 |
scratch family zinc finger 2 |
428 |
0.66 |
| chr2_189361179_189361330 | 0.07 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
73511 |
0.13 |
| chr1_79297788_79297939 | 0.06 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
85707 |
0.1 |
| chr4_134070427_134071051 | 0.06 |
PCDH10 |
protocadherin 10 |
269 |
0.96 |
| chr4_112244997_112245242 | 0.06 |
ENSG00000200963 |
. |
7449 |
0.34 |
| chr4_135119262_135119413 | 0.06 |
PABPC4L |
poly(A) binding protein, cytoplasmic 4-like |
3566 |
0.39 |
| chr15_77575085_77575236 | 0.06 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
27266 |
0.24 |
| chr3_44045083_44045345 | 0.06 |
ENSG00000252980 |
. |
67365 |
0.13 |
| chr12_109122720_109122871 | 0.06 |
CORO1C |
coronin, actin binding protein, 1C |
1532 |
0.34 |
| chr11_36253008_36253159 | 0.06 |
COMMD9 |
COMM domain containing 9 |
57891 |
0.13 |
| chr1_8138513_8138990 | 0.06 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
52383 |
0.13 |
| chr7_95107815_95108198 | 0.06 |
ASB4 |
ankyrin repeat and SOCS box containing 4 |
7207 |
0.2 |
| chr5_106805601_106805833 | 0.06 |
EFNA5 |
ephrin-A5 |
200611 |
0.03 |
| chr12_12551242_12551393 | 0.06 |
LOH12CR1 |
loss of heterozygosity, 12, chromosomal region 1 |
40965 |
0.13 |
| chr20_50384318_50384901 | 0.06 |
ATP9A |
ATPase, class II, type 9A |
258 |
0.94 |
| chr3_49424014_49424165 | 0.06 |
RHOA-IT1 |
RHOA intronic transcript 1 (non-protein coding) |
19650 |
0.09 |
| chr14_56711281_56711464 | 0.06 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
125545 |
0.06 |
| chr4_119950142_119950384 | 0.06 |
SYNPO2 |
synaptopodin 2 |
5637 |
0.3 |
| chr5_103680813_103681290 | 0.06 |
ENSG00000239808 |
. |
84524 |
0.11 |
| chr5_30944702_30944853 | 0.06 |
CDH6 |
cadherin 6, type 2, K-cadherin (fetal kidney) |
249080 |
0.02 |
| chr10_104102183_104102477 | 0.06 |
NFKB2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
51537 |
0.08 |
| chr8_48282315_48282609 | 0.06 |
SPIDR |
scaffolding protein involved in DNA repair |
70501 |
0.12 |
| chr21_28640922_28641073 | 0.06 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
302165 |
0.01 |
| chr4_177824622_177824937 | 0.06 |
VEGFC |
vascular endothelial growth factor C |
110898 |
0.07 |
| chr7_27141205_27141470 | 0.06 |
HOXA2 |
homeobox A2 |
1093 |
0.25 |
| chr12_86222890_86223044 | 0.06 |
RASSF9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
7381 |
0.28 |
| chr6_56595678_56595829 | 0.06 |
DST |
dystonin |
54924 |
0.16 |
| chr1_8537218_8537608 | 0.06 |
ENSG00000221083 |
. |
34565 |
0.16 |
| chrX_12908437_12908588 | 0.06 |
TLR8 |
toll-like receptor 8 |
16227 |
0.17 |
| chr11_103677106_103677422 | 0.06 |
ENSG00000264200 |
. |
43370 |
0.2 |
| chr12_32635785_32635936 | 0.06 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
3046 |
0.28 |
| chr6_111922654_111922805 | 0.06 |
C6orf3 |
|
1651 |
0.34 |
| chr15_51367983_51368134 | 0.06 |
RP11-394B5.2 |
|
6275 |
0.24 |
| chr8_29388025_29388176 | 0.06 |
RP4-676L2.1 |
|
177413 |
0.03 |
| chr2_141580056_141580207 | 0.06 |
ENSG00000201892 |
. |
169981 |
0.04 |
| chr6_19691587_19692480 | 0.06 |
ENSG00000200957 |
. |
49273 |
0.18 |
| chr12_72670200_72670351 | 0.06 |
ENSG00000236333 |
. |
1588 |
0.43 |
| chr21_39807445_39807798 | 0.06 |
ERG |
v-ets avian erythroblastosis virus E26 oncogene homolog |
62724 |
0.14 |
| chr1_164665790_164665949 | 0.06 |
RP11-477H21.2 |
|
16547 |
0.21 |
| chr12_109118598_109118801 | 0.06 |
CORO1C |
coronin, actin binding protein, 1C |
5628 |
0.17 |
| chr2_46933523_46933944 | 0.06 |
SOCS5 |
suppressor of cytokine signaling 5 |
7407 |
0.22 |
| chr4_119130339_119130490 | 0.06 |
ENSG00000269893 |
. |
69931 |
0.13 |
| chr1_100317573_100317724 | 0.06 |
AGL |
amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
578 |
0.74 |
| chr12_10902988_10903139 | 0.06 |
YBX3 |
Y box binding protein 3 |
27152 |
0.13 |
| chr11_126517104_126517255 | 0.06 |
RP11-115C10.1 |
|
5568 |
0.3 |
| chr3_18024994_18025145 | 0.06 |
TBC1D5 |
TBC1 domain family, member 5 |
240925 |
0.02 |
| chr17_20388317_20388497 | 0.06 |
LGALS9B |
lectin, galactoside-binding, soluble, 9B |
17559 |
0.13 |
| chr1_8166685_8166836 | 0.06 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
80392 |
0.08 |
| chr21_38367175_38367326 | 0.06 |
HLCS |
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
4714 |
0.17 |
| chr11_60686819_60686970 | 0.06 |
TMEM109 |
transmembrane protein 109 |
256 |
0.81 |
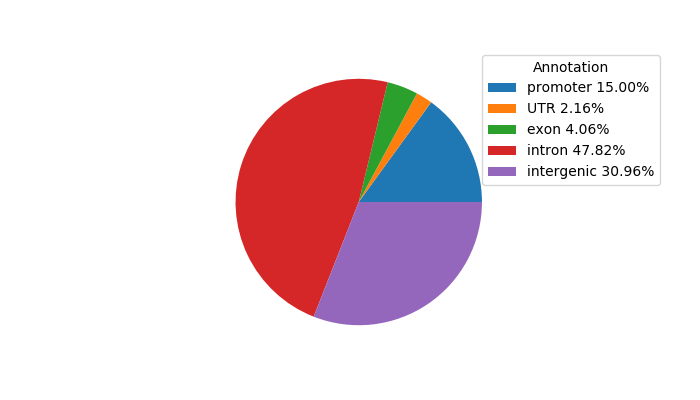
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |