Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
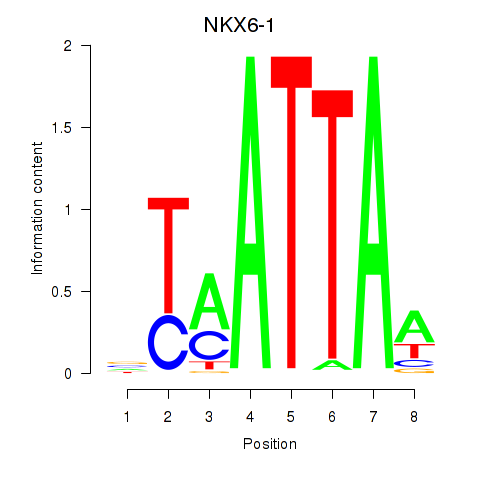
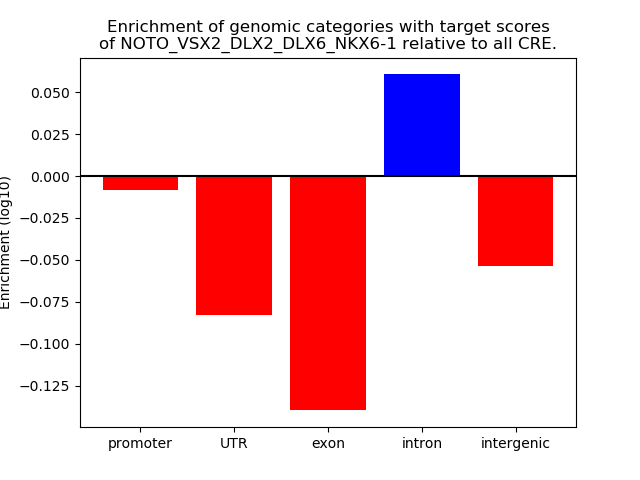
Results for NOTO_VSX2_DLX2_DLX6_NKX6-1
Z-value: 0.31
Transcription factors associated with NOTO_VSX2_DLX2_DLX6_NKX6-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
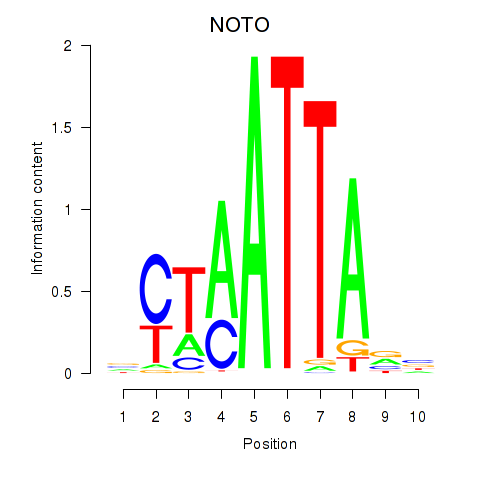
NOTO
|
ENSG00000214513.3 | notochord homeobox |
|
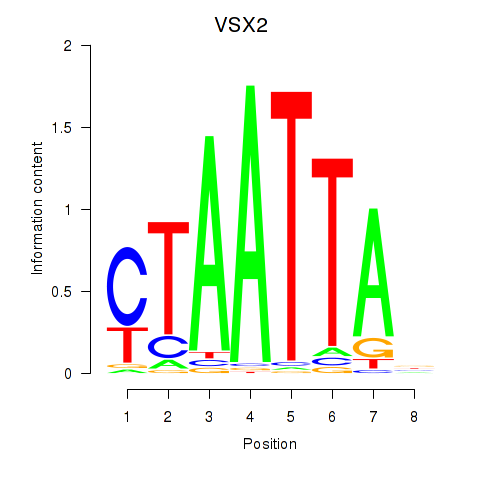
VSX2
|
ENSG00000119614.2 | visual system homeobox 2 |
|
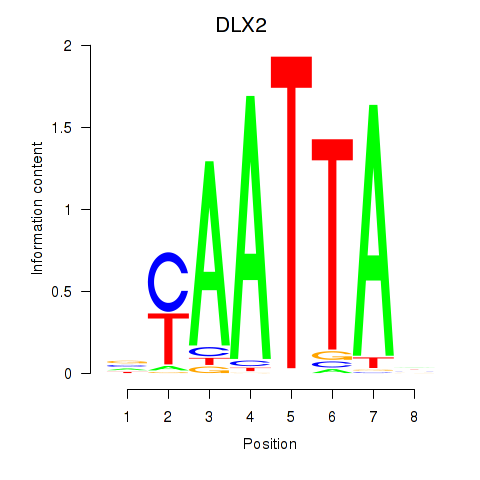
DLX2
|
ENSG00000115844.6 | distal-less homeobox 2 |
|
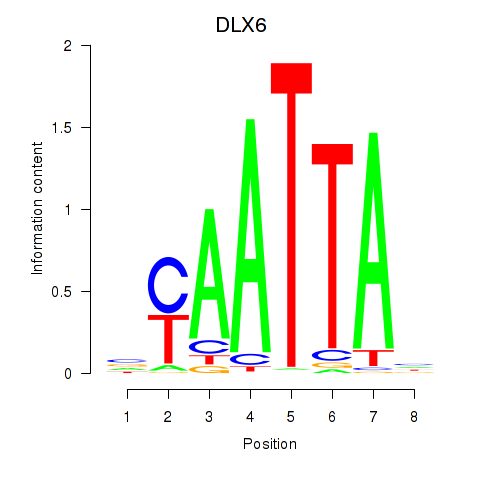
DLX6
|
ENSG00000006377.9 | distal-less homeobox 6 |
|
NKX6-1
|
ENSG00000163623.5 | NK6 homeobox 1 |
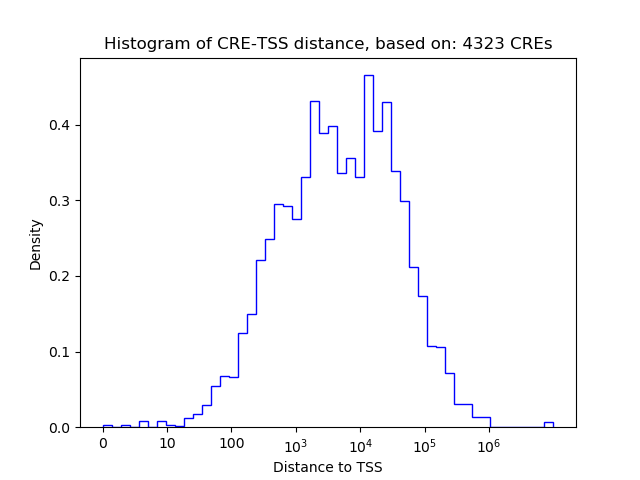
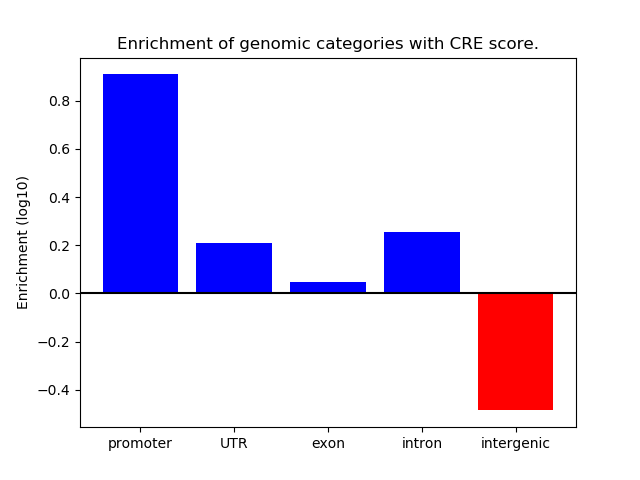
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_172970805_172971712 | DLX2 | 3630 | 0.254786 | -0.76 | 1.8e-02 | Click! |
| chr2_172963527_172963843 | DLX2 | 3943 | 0.244525 | -0.64 | 6.4e-02 | Click! |
| chr2_172970602_172970787 | DLX2 | 3066 | 0.273655 | -0.59 | 9.2e-02 | Click! |
| chr2_172961462_172961931 | DLX2 | 5932 | 0.218659 | -0.53 | 1.4e-01 | Click! |
| chr2_172960519_172960695 | DLX2 | 7021 | 0.211664 | -0.52 | 1.5e-01 | Click! |
| chr7_96635533_96635717 | DLX6 | 335 | 0.793174 | -0.51 | 1.6e-01 | Click! |
| chr7_96634475_96634654 | DLX6 | 296 | 0.789736 | -0.47 | 2.0e-01 | Click! |
| chr7_96635226_96635397 | DLX6 | 21 | 0.957943 | -0.41 | 2.7e-01 | Click! |
| chr7_96634662_96635012 | DLX6 | 23 | 0.954019 | -0.38 | 3.2e-01 | Click! |
| chr7_96635926_96636202 | DLX6 | 673 | 0.484864 | -0.38 | 3.2e-01 | Click! |
| chr4_85414354_85414505 | NKX6-1 | 3674 | 0.350707 | -0.61 | 7.8e-02 | Click! |
| chr4_85419922_85420185 | NKX6-1 | 450 | 0.893788 | -0.61 | 8.3e-02 | Click! |
| chr4_85420275_85420452 | NKX6-1 | 760 | 0.772953 | -0.59 | 9.2e-02 | Click! |
| chr4_85419328_85419920 | NKX6-1 | 21 | 0.987510 | -0.58 | 9.9e-02 | Click! |
| chr4_85418590_85418741 | NKX6-1 | 562 | 0.851816 | -0.47 | 2.0e-01 | Click! |
| chr2_73429434_73429594 | NOTO | 128 | 0.949734 | -0.49 | 1.8e-01 | Click! |
| chr2_73402539_73402866 | NOTO | 26684 | 0.129677 | -0.38 | 3.1e-01 | Click! |
| chr2_73404796_73404997 | NOTO | 24490 | 0.132908 | -0.31 | 4.1e-01 | Click! |
| chr2_73429933_73430225 | NOTO | 693 | 0.616065 | 0.30 | 4.3e-01 | Click! |
| chr2_73403004_73403567 | NOTO | 26101 | 0.130556 | -0.16 | 6.8e-01 | Click! |
| chr14_74718219_74718381 | VSX2 | 12125 | 0.152141 | -0.49 | 1.8e-01 | Click! |
| chr14_74725090_74725251 | VSX2 | 18995 | 0.134493 | -0.41 | 2.7e-01 | Click! |
| chr14_74707042_74707610 | VSX2 | 1151 | 0.458714 | 0.26 | 5.0e-01 | Click! |
| chr14_74684578_74684763 | VSX2 | 21505 | 0.151615 | -0.26 | 5.1e-01 | Click! |
| chr14_74684411_74684562 | VSX2 | 21689 | 0.151271 | 0.22 | 5.6e-01 | Click! |
Activity of the NOTO_VSX2_DLX2_DLX6_NKX6-1 motif across conditions
Conditions sorted by the z-value of the NOTO_VSX2_DLX2_DLX6_NKX6-1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
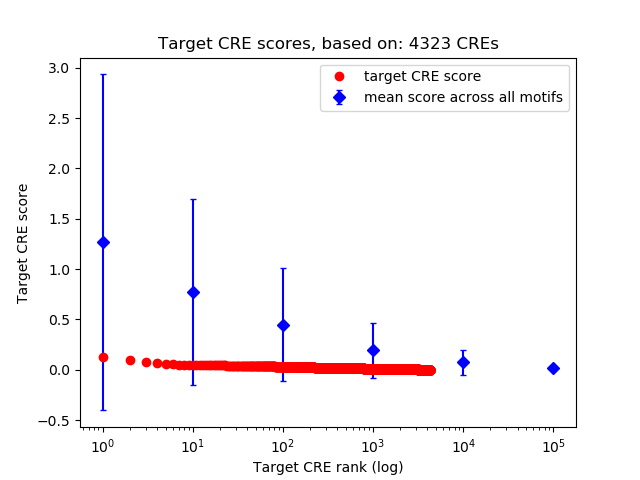
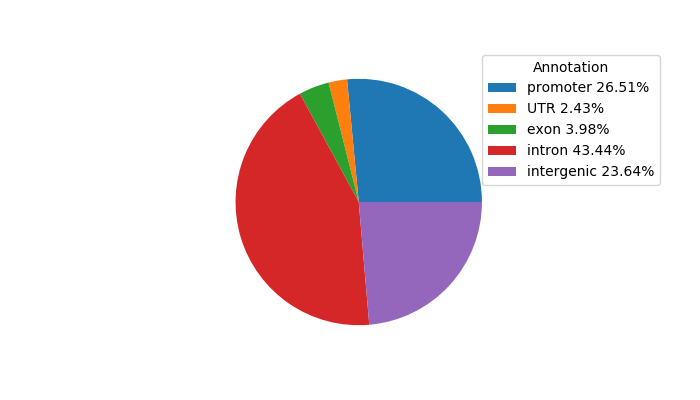
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_78400540_78401325 | 0.13 |
GPR174 |
G protein-coupled receptor 174 |
25537 |
0.27 |
| chr6_154568354_154568864 | 0.10 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
53 |
0.99 |
| chr7_50345819_50346403 | 0.08 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
1733 |
0.5 |
| chr5_126094279_126094440 | 0.06 |
ENSG00000252185 |
. |
3250 |
0.29 |
| chr6_159464766_159465080 | 0.06 |
TAGAP |
T-cell activation RhoGTPase activating protein |
1127 |
0.51 |
| chr2_61111507_61111691 | 0.05 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
2808 |
0.27 |
| chr2_143887292_143887566 | 0.05 |
ARHGAP15 |
Rho GTPase activating protein 15 |
546 |
0.84 |
| chr6_25041553_25042305 | 0.05 |
RP3-425P12.5 |
|
138 |
0.72 |
| chr14_88471433_88472132 | 0.05 |
GPR65 |
G protein-coupled receptor 65 |
314 |
0.88 |
| chr7_106507255_106507406 | 0.05 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
1406 |
0.54 |
| chr3_151961363_151961565 | 0.05 |
MBNL1 |
muscleblind-like splicing regulator 1 |
24365 |
0.2 |
| chr7_50348449_50348781 | 0.05 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
297 |
0.94 |
| chr12_9912992_9913534 | 0.05 |
CD69 |
CD69 molecule |
234 |
0.92 |
| chr5_118606885_118607036 | 0.05 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
2511 |
0.28 |
| chr13_99957828_99957979 | 0.05 |
GPR183 |
G protein-coupled receptor 183 |
1756 |
0.38 |
| chr4_36245078_36245884 | 0.05 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
80 |
0.74 |
| chr5_118606012_118606179 | 0.05 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
1646 |
0.37 |
| chr15_75080868_75081404 | 0.05 |
ENSG00000264386 |
. |
38 |
0.76 |
| chr14_22974481_22974956 | 0.04 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
18547 |
0.09 |
| chr1_198627689_198627922 | 0.04 |
RP11-553K8.5 |
|
8385 |
0.25 |
| chr7_37011253_37011404 | 0.04 |
ELMO1 |
engulfment and cell motility 1 |
13337 |
0.2 |
| chr2_193992778_193992929 | 0.04 |
TMEFF2 |
transmembrane protein with EGF-like and two follistatin-like domains 2 |
932418 |
0.0 |
| chr1_180387724_180387875 | 0.04 |
ENSG00000265435 |
. |
19726 |
0.22 |
| chr1_198615140_198615291 | 0.04 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
6923 |
0.25 |
| chrX_78620994_78621299 | 0.04 |
ITM2A |
integral membrane protein 2A |
1710 |
0.55 |
| chr10_33425652_33425891 | 0.04 |
ENSG00000263576 |
. |
38207 |
0.16 |
| chr21_32567568_32567719 | 0.04 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
65104 |
0.13 |
| chr1_234544831_234544982 | 0.04 |
COA6 |
cytochrome c oxidase assembly factor 6 homolog (S. cerevisiae) |
35459 |
0.12 |
| chr6_26198173_26198564 | 0.04 |
HIST1H3D |
histone cluster 1, H3d |
890 |
0.22 |
| chr2_68961577_68961872 | 0.04 |
ARHGAP25 |
Rho GTPase activating protein 25 |
189 |
0.96 |
| chr3_43329409_43329595 | 0.04 |
SNRK |
SNF related kinase |
1424 |
0.43 |
| chr1_29253817_29254129 | 0.04 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
12882 |
0.18 |
| chrX_11780001_11780185 | 0.04 |
MSL3 |
male-specific lethal 3 homolog (Drosophila) |
2346 |
0.44 |
| chr6_159464058_159464225 | 0.04 |
TAGAP |
T-cell activation RhoGTPase activating protein |
1909 |
0.34 |
| chr2_215664874_215665025 | 0.04 |
BARD1 |
BRCA1 associated RING domain 1 |
9448 |
0.21 |
| chr10_46075820_46076565 | 0.04 |
MARCH8 |
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
13747 |
0.25 |
| chr17_75456326_75456913 | 0.04 |
SEPT9 |
septin 9 |
4171 |
0.18 |
| chr2_158299843_158300108 | 0.04 |
CYTIP |
cytohesin 1 interacting protein |
679 |
0.68 |
| chr12_15112961_15113206 | 0.04 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
1117 |
0.46 |
| chr17_29637369_29637894 | 0.04 |
EVI2B |
ecotropic viral integration site 2B |
3471 |
0.16 |
| chr12_54891959_54892179 | 0.04 |
NCKAP1L |
NCK-associated protein 1-like |
499 |
0.73 |
| chr1_28975271_28975442 | 0.04 |
ENSG00000270103 |
. |
244 |
0.88 |
| chr3_16883621_16883873 | 0.04 |
PLCL2 |
phospholipase C-like 2 |
42705 |
0.19 |
| chr10_129861850_129862130 | 0.04 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
16156 |
0.26 |
| chrY_2804248_2804778 | 0.04 |
ZFY |
zinc finger protein, Y-linked |
967 |
0.67 |
| chr9_125662893_125663901 | 0.04 |
RC3H2 |
ring finger and CCCH-type domains 2 |
3008 |
0.16 |
| chr5_126147731_126147991 | 0.04 |
LMNB1 |
lamin B1 |
34980 |
0.19 |
| chr1_197730742_197731002 | 0.04 |
RP11-448G4.4 |
|
4423 |
0.25 |
| chr7_43688967_43689164 | 0.04 |
COA1 |
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
830 |
0.67 |
| chr1_112013416_112013602 | 0.04 |
C1orf162 |
chromosome 1 open reading frame 162 |
2905 |
0.14 |
| chr19_42390602_42391041 | 0.04 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
2306 |
0.19 |
| chr7_33078848_33079779 | 0.04 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
1208 |
0.42 |
| chr5_130882612_130883040 | 0.04 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
14100 |
0.29 |
| chr1_100852790_100852941 | 0.04 |
ENSG00000216067 |
. |
8534 |
0.21 |
| chr3_151963607_151963763 | 0.04 |
MBNL1 |
muscleblind-like splicing regulator 1 |
22144 |
0.21 |
| chr16_31271922_31272073 | 0.04 |
ITGAM |
integrin, alpha M (complement component 3 receptor 3 subunit) |
682 |
0.53 |
| chr13_46745397_46745548 | 0.04 |
ENSG00000240767 |
. |
1589 |
0.32 |
| chr7_115668295_115668446 | 0.04 |
TFEC |
transcription factor EC |
2425 |
0.43 |
| chr4_144312492_144312643 | 0.04 |
GAB1 |
GRB2-associated binding protein 1 |
96 |
0.98 |
| chr12_47600587_47601314 | 0.04 |
PCED1B |
PC-esterase domain containing 1B |
9102 |
0.22 |
| chr4_154411300_154411567 | 0.04 |
KIAA0922 |
KIAA0922 |
23932 |
0.22 |
| chr2_174129320_174129481 | 0.04 |
MLK7-AS1 |
MLK7 antisense RNA 1 |
6948 |
0.3 |
| chr17_10601145_10601947 | 0.04 |
ADPRM |
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
635 |
0.41 |
| chr1_24862040_24862255 | 0.04 |
ENSG00000266551 |
. |
5943 |
0.18 |
| chr11_128589269_128589524 | 0.04 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
23478 |
0.17 |
| chr17_28085648_28085799 | 0.04 |
RP11-82O19.1 |
|
2398 |
0.25 |
| chr3_98253310_98253461 | 0.03 |
GPR15 |
G protein-coupled receptor 15 |
2642 |
0.21 |
| chr10_26771389_26771611 | 0.03 |
ENSG00000199733 |
. |
27018 |
0.21 |
| chr16_50776896_50777610 | 0.03 |
RP11-327F22.1 |
|
527 |
0.39 |
| chr15_55547653_55548107 | 0.03 |
RAB27A |
RAB27A, member RAS oncogene family |
6647 |
0.22 |
| chr21_15917916_15918619 | 0.03 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
395 |
0.89 |
| chr14_22977314_22977663 | 0.03 |
TRAJ15 |
T cell receptor alpha joining 15 |
21092 |
0.09 |
| chr12_95071864_95072096 | 0.03 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
27642 |
0.23 |
| chr1_150584633_150584820 | 0.03 |
ENSG00000253047 |
. |
11591 |
0.1 |
| chr16_81030456_81031156 | 0.03 |
CMC2 |
C-x(9)-C motif containing 2 |
1458 |
0.31 |
| chr10_112632655_112633305 | 0.03 |
PDCD4-AS1 |
PDCD4 antisense RNA 1 |
989 |
0.38 |
| chr11_128392558_128393039 | 0.03 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
593 |
0.77 |
| chr13_41372571_41372761 | 0.03 |
SLC25A15 |
solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 |
5372 |
0.18 |
| chr12_131796680_131796831 | 0.03 |
ENSG00000212251 |
. |
12459 |
0.23 |
| chrX_129243329_129244377 | 0.03 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
483 |
0.82 |
| chr19_6481304_6482171 | 0.03 |
DENND1C |
DENN/MADD domain containing 1C |
27 |
0.95 |
| chr1_200987607_200987758 | 0.03 |
KIF21B |
kinesin family member 21B |
4854 |
0.21 |
| chr18_56326442_56326593 | 0.03 |
MALT1 |
mucosa associated lymphoid tissue lymphoma translocation gene 1 |
12101 |
0.14 |
| chr1_200865204_200865355 | 0.03 |
C1orf106 |
chromosome 1 open reading frame 106 |
1330 |
0.42 |
| chr22_31615175_31615326 | 0.03 |
ENSG00000199695 |
. |
3589 |
0.13 |
| chr12_8666250_8666401 | 0.03 |
CLEC4D |
C-type lectin domain family 4, member D |
189 |
0.93 |
| chr3_108543496_108543820 | 0.03 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
2039 |
0.42 |
| chr14_70111468_70111619 | 0.03 |
KIAA0247 |
KIAA0247 |
33230 |
0.18 |
| chr1_206732541_206732911 | 0.03 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
2233 |
0.27 |
| chr14_22747289_22747440 | 0.03 |
ENSG00000238634 |
. |
136477 |
0.04 |
| chr9_102849500_102849651 | 0.03 |
ERP44 |
endoplasmic reticulum protein 44 |
11747 |
0.18 |
| chr13_46755198_46755455 | 0.03 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
1133 |
0.45 |
| chr3_32996346_32996497 | 0.03 |
CCR4 |
chemokine (C-C motif) receptor 4 |
3355 |
0.33 |
| chr3_69140694_69141185 | 0.03 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
6767 |
0.15 |
| chr15_52844361_52844512 | 0.03 |
ARPP19 |
cAMP-regulated phosphoprotein, 19kDa |
5505 |
0.2 |
| chr21_32555155_32555306 | 0.03 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
52691 |
0.16 |
| chr1_206753637_206754165 | 0.03 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
23408 |
0.13 |
| chrX_9801459_9801610 | 0.03 |
ENSG00000206844 |
. |
41207 |
0.15 |
| chrX_13106517_13106711 | 0.03 |
FAM9C |
family with sequence similarity 9, member C |
43813 |
0.18 |
| chr9_117721707_117721858 | 0.03 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
29085 |
0.23 |
| chr1_39594945_39595096 | 0.03 |
ENSG00000206654 |
. |
8305 |
0.17 |
| chr12_65779210_65779361 | 0.03 |
MSRB3 |
methionine sulfoxide reductase B3 |
58630 |
0.15 |
| chr2_99345755_99346000 | 0.03 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
834 |
0.68 |
| chr5_81074467_81075538 | 0.03 |
SSBP2 |
single-stranded DNA binding protein 2 |
27930 |
0.25 |
| chr1_231748956_231749347 | 0.03 |
LINC00582 |
long intergenic non-protein coding RNA 582 |
1315 |
0.46 |
| chr18_57572017_57572168 | 0.03 |
PMAIP1 |
phorbol-12-myristate-13-acetate-induced protein 1 |
4844 |
0.29 |
| chr21_32559881_32560032 | 0.03 |
TIAM1 |
T-cell lymphoma invasion and metastasis 1 |
57417 |
0.15 |
| chr17_21157480_21157631 | 0.03 |
C17orf103 |
chromosome 17 open reading frame 103 |
833 |
0.56 |
| chr10_70850849_70851349 | 0.03 |
SRGN |
serglycin |
3225 |
0.25 |
| chrX_118818753_118818904 | 0.03 |
SEPT6 |
septin 6 |
7964 |
0.18 |
| chr20_1357596_1357769 | 0.03 |
RP11-314N13.3 |
|
15416 |
0.13 |
| chr17_74564111_74564262 | 0.03 |
RP11-666A8.9 |
|
1688 |
0.17 |
| chr14_22955963_22956254 | 0.03 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
63 |
0.95 |
| chr1_236284065_236284436 | 0.03 |
GPR137B |
G protein-coupled receptor 137B |
21582 |
0.19 |
| chr4_36243817_36244102 | 0.03 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
1602 |
0.35 |
| chr7_94035982_94036133 | 0.03 |
COL1A2 |
collagen, type I, alpha 2 |
12184 |
0.27 |
| chr7_37488846_37488997 | 0.03 |
ELMO1 |
engulfment and cell motility 1 |
69 |
0.98 |
| chr15_44101218_44101369 | 0.03 |
HYPK |
huntingtin interacting protein K |
8496 |
0.08 |
| chrX_35788537_35788688 | 0.03 |
MAGEB16 |
melanoma antigen family B, 16 |
27847 |
0.25 |
| chr13_49065751_49065929 | 0.03 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
41373 |
0.17 |
| chr17_75663137_75663644 | 0.03 |
SEPT9 |
septin 9 |
185395 |
0.03 |
| chr11_104904746_104905919 | 0.03 |
CASP1 |
caspase 1, apoptosis-related cysteine peptidase |
508 |
0.77 |
| chr4_91603417_91603568 | 0.03 |
CCSER1 |
coiled-coil serine-rich protein 1 |
32830 |
0.25 |
| chr10_7526759_7527322 | 0.03 |
ENSG00000207453 |
. |
1731 |
0.45 |
| chr8_27223967_27224118 | 0.03 |
PTK2B |
protein tyrosine kinase 2 beta |
14126 |
0.22 |
| chr3_63954713_63955178 | 0.03 |
ATXN7 |
ataxin 7 |
1525 |
0.34 |
| chr1_111215065_111215351 | 0.03 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
2447 |
0.28 |
| chr12_65013240_65013416 | 0.03 |
ENSG00000207546 |
. |
2961 |
0.17 |
| chr11_20179312_20179463 | 0.03 |
DBX1 |
developing brain homeobox 1 |
2483 |
0.35 |
| chr10_1779633_1779784 | 0.03 |
ADARB2 |
adenosine deaminase, RNA-specific, B2 (non-functional) |
38 |
0.99 |
| chr14_22182172_22182323 | 0.03 |
OR4E2 |
olfactory receptor, family 4, subfamily E, member 2 |
48950 |
0.13 |
| chr10_134261699_134261957 | 0.03 |
C10orf91 |
chromosome 10 open reading frame 91 |
3135 |
0.24 |
| chr1_90290694_90290845 | 0.03 |
LRRC8D |
leucine rich repeat containing 8 family, member D |
3289 |
0.28 |
| chr9_184434_184636 | 0.03 |
CBWD1 |
COBW domain containing 1 |
5463 |
0.2 |
| chr3_124774787_124775424 | 0.03 |
HEG1 |
heart development protein with EGF-like domains 1 |
303 |
0.88 |
| chr2_136876136_136876323 | 0.03 |
CXCR4 |
chemokine (C-X-C motif) receptor 4 |
494 |
0.87 |
| chr17_37933702_37934037 | 0.03 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
609 |
0.66 |
| chr4_40201423_40201705 | 0.03 |
RHOH |
ras homolog family member H |
400 |
0.87 |
| chr18_74837172_74837323 | 0.03 |
MBP |
myelin basic protein |
2421 |
0.42 |
| chr14_90187580_90187856 | 0.03 |
ENSG00000200312 |
. |
8863 |
0.23 |
| chr1_206749362_206749620 | 0.03 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
18998 |
0.14 |
| chr9_92032170_92032321 | 0.03 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
1503 |
0.49 |
| chr17_56736328_56736479 | 0.03 |
ENSG00000202077 |
. |
263 |
0.86 |
| chr2_64369947_64371114 | 0.03 |
AC074289.1 |
|
157 |
0.93 |
| chr2_197032053_197032740 | 0.03 |
STK17B |
serine/threonine kinase 17b |
3328 |
0.25 |
| chr2_175458036_175458370 | 0.03 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
4290 |
0.21 |
| chr9_70851276_70851465 | 0.03 |
CBWD3 |
COBW domain containing 3 |
5027 |
0.19 |
| chr3_195962656_195962807 | 0.03 |
SLC51A |
solute carrier family 51, alpha subunit |
5855 |
0.13 |
| chr10_30745504_30745655 | 0.03 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
17828 |
0.21 |
| chr6_36008402_36008553 | 0.03 |
MAPK14 |
mitogen-activated protein kinase 14 |
11611 |
0.19 |
| chr1_198652510_198652661 | 0.03 |
RP11-553K8.5 |
|
16395 |
0.24 |
| chr5_150593261_150593549 | 0.03 |
GM2A |
GM2 ganglioside activator |
1694 |
0.36 |
| chr8_82022711_82022967 | 0.03 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
1464 |
0.56 |
| chr10_6626003_6626633 | 0.03 |
PRKCQ |
protein kinase C, theta |
4055 |
0.36 |
| chr15_69074948_69075145 | 0.03 |
ENSG00000265195 |
. |
19218 |
0.22 |
| chrY_1551045_1551310 | 0.03 |
NA |
NA |
> 106 |
NA |
| chr5_74615521_74615672 | 0.03 |
HMGCR |
3-hydroxy-3-methylglutaryl-CoA reductase |
16558 |
0.19 |
| chr7_17039488_17039788 | 0.03 |
AGR3 |
anterior gradient 3 |
118027 |
0.06 |
| chr19_48832910_48833240 | 0.03 |
EMP3 |
epithelial membrane protein 3 |
4210 |
0.13 |
| chr9_70495582_70495733 | 0.03 |
CBWD5 |
COBW domain containing 5 |
5411 |
0.29 |
| chr1_66806017_66806168 | 0.03 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
8220 |
0.31 |
| chr8_134086469_134086645 | 0.03 |
SLA |
Src-like-adaptor |
13954 |
0.23 |
| chr9_80524074_80524374 | 0.03 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
86309 |
0.1 |
| chr17_18204377_18204528 | 0.03 |
TOP3A |
topoisomerase (DNA) III alpha |
1531 |
0.27 |
| chr14_75885513_75886422 | 0.03 |
RP11-293M10.6 |
|
8426 |
0.2 |
| chr4_26269987_26270138 | 0.03 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
4167 |
0.36 |
| chr9_69267993_69268163 | 0.03 |
CBWD6 |
COBW domain containing 6 |
5485 |
0.22 |
| chr17_75450125_75450790 | 0.03 |
SEPT9 |
septin 9 |
295 |
0.88 |
| chr6_167526422_167526606 | 0.03 |
CCR6 |
chemokine (C-C motif) receptor 6 |
1219 |
0.48 |
| chr3_128899189_128899802 | 0.03 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
3247 |
0.18 |
| chr3_107697576_107697727 | 0.03 |
CD47 |
CD47 molecule |
79557 |
0.11 |
| chr13_31312055_31312206 | 0.03 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
2485 |
0.4 |
| chr1_21349011_21349508 | 0.03 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
28136 |
0.19 |
| chr3_47182364_47182515 | 0.03 |
SETD2 |
SET domain containing 2 |
22627 |
0.16 |
| chr18_21572812_21573447 | 0.02 |
TTC39C |
tetratricopeptide repeat domain 39C |
392 |
0.85 |
| chr4_90226799_90226950 | 0.02 |
GPRIN3 |
GPRIN family member 3 |
2287 |
0.45 |
| chr3_135909805_135909956 | 0.02 |
MSL2 |
male-specific lethal 2 homolog (Drosophila) |
3516 |
0.32 |
| chr4_38623548_38623699 | 0.02 |
RP11-617D20.1 |
|
2573 |
0.31 |
| chr11_75063727_75063930 | 0.02 |
ARRB1 |
arrestin, beta 1 |
955 |
0.48 |
| chr4_25865391_25865542 | 0.02 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
84 |
0.98 |
| chr22_18260542_18260943 | 0.02 |
BID |
BH3 interacting domain death agonist |
3311 |
0.23 |
| chr15_43531850_43532099 | 0.02 |
ENSG00000202211 |
. |
7904 |
0.13 |
| chr8_8203302_8203453 | 0.02 |
SGK223 |
Tyrosine-protein kinase SgK223 |
35880 |
0.18 |
| chr2_197133486_197133637 | 0.02 |
AC020571.3 |
|
8404 |
0.2 |
| chr12_90537525_90537676 | 0.02 |
ENSG00000252823 |
. |
389764 |
0.01 |
| chr9_79265748_79266757 | 0.02 |
PRUNE2 |
prune homolog 2 (Drosophila) |
1213 |
0.53 |
| chr10_54463336_54463751 | 0.02 |
RP11-556E13.1 |
|
51626 |
0.16 |
| chr6_20174159_20174424 | 0.02 |
RP11-239H6.2 |
|
38027 |
0.18 |
| chr14_91863541_91863979 | 0.02 |
CCDC88C |
coiled-coil domain containing 88C |
19930 |
0.21 |
| chr14_75990505_75990656 | 0.02 |
BATF |
basic leucine zipper transcription factor, ATF-like |
1677 |
0.38 |
| chr6_170597990_170598273 | 0.02 |
DLL1 |
delta-like 1 (Drosophila) |
1430 |
0.33 |
| chr10_73847731_73848399 | 0.02 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
21 |
0.98 |
| chr1_158802859_158803010 | 0.02 |
MNDA |
myeloid cell nuclear differentiation antigen |
1827 |
0.35 |
| chr11_104940824_104940975 | 0.02 |
CARD16 |
caspase recruitment domain family, member 16 |
24796 |
0.15 |
| chr4_106070591_106071096 | 0.02 |
TET2 |
tet methylcytosine dioxygenase 2 |
2229 |
0.34 |
| chr8_66843424_66843611 | 0.02 |
PDE7A |
phosphodiesterase 7A |
89330 |
0.09 |
| chrX_1600809_1601304 | 0.02 |
ASMTL |
acetylserotonin O-methyltransferase-like |
28401 |
0.16 |
| chr17_29644876_29645027 | 0.02 |
CTD-2370N5.3 |
|
707 |
0.55 |
| chr11_82882204_82882391 | 0.02 |
PCF11 |
PCF11 cleavage and polyadenylation factor subunit |
4 |
0.97 |
| chr2_175498105_175498391 | 0.02 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
1059 |
0.58 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |