Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
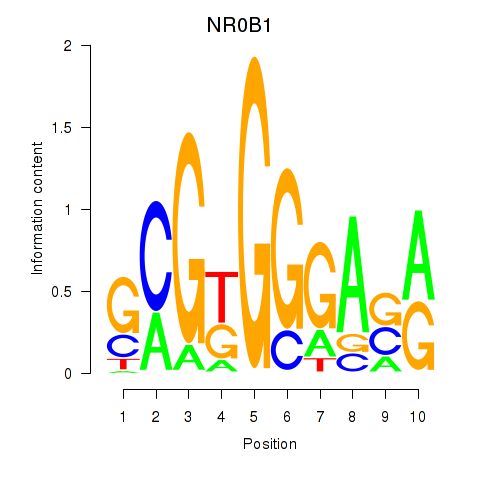
Results for NR0B1
Z-value: 1.12
Transcription factors associated with NR0B1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR0B1
|
ENSG00000169297.6 | nuclear receptor subfamily 0 group B member 1 |
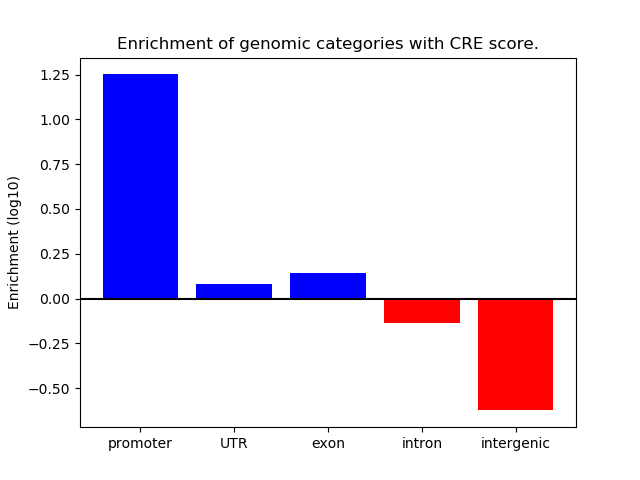
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chrX_30326264_30326415 | NR0B1 | 266 | 0.915917 | 0.54 | 1.3e-01 | Click! |
| chrX_30327007_30327420 | NR0B1 | 282 | 0.908658 | 0.41 | 2.8e-01 | Click! |
| chrX_30326616_30326849 | NR0B1 | 127 | 0.962905 | 0.34 | 3.8e-01 | Click! |
| chrX_30302316_30302467 | NR0B1 | 24214 | 0.143050 | 0.27 | 4.8e-01 | Click! |
Activity of the NR0B1 motif across conditions
Conditions sorted by the z-value of the NR0B1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
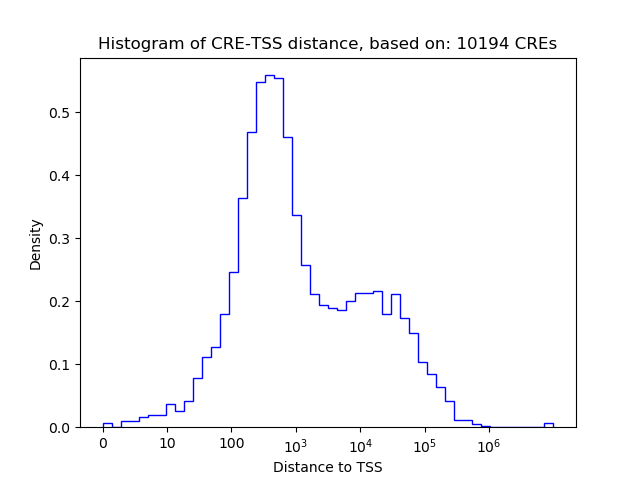
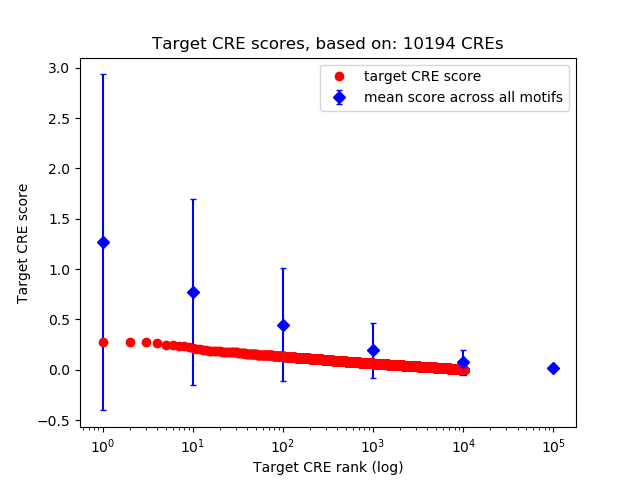
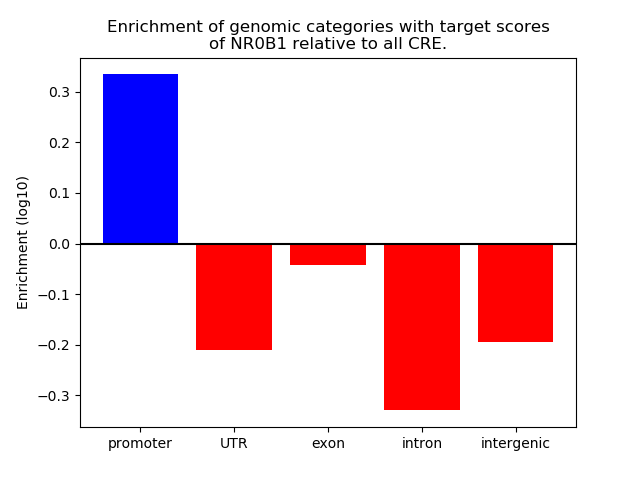
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_133562382_133562833 | 0.28 |
EYA4 |
eyes absent homolog 4 (Drosophila) |
51 |
0.99 |
| chr7_5465601_5465997 | 0.27 |
TNRC18 |
trinucleotide repeat containing 18 |
754 |
0.57 |
| chr13_38444003_38444421 | 0.27 |
TRPC4 |
transient receptor potential cation channel, subfamily C, member 4 |
273 |
0.94 |
| chr4_187645380_187645697 | 0.27 |
FAT1 |
FAT atypical cadherin 1 |
529 |
0.87 |
| chr1_82268816_82269203 | 0.25 |
LPHN2 |
latrophilin 2 |
2927 |
0.41 |
| chr10_79396329_79396588 | 0.24 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
747 |
0.74 |
| chr7_102153061_102153212 | 0.23 |
RASA4B |
RAS p21 protein activator 4B |
5021 |
0.1 |
| chr12_51319016_51319581 | 0.23 |
METTL7A |
methyltransferase like 7A |
764 |
0.59 |
| chr19_41726166_41726324 | 0.23 |
AXL |
AXL receptor tyrosine kinase |
1105 |
0.33 |
| chr12_64238267_64238814 | 0.21 |
SRGAP1 |
SLIT-ROBO Rho GTPase activating protein 1 |
1 |
0.97 |
| chr12_54745959_54746186 | 0.21 |
RP11-753H16.3 |
|
1373 |
0.19 |
| chr9_4491201_4491352 | 0.20 |
SLC1A1 |
solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 |
832 |
0.71 |
| chr2_238395551_238395789 | 0.20 |
MLPH |
melanophilin |
141 |
0.96 |
| chr11_65324098_65324348 | 0.20 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
1013 |
0.31 |
| chr10_128076524_128076793 | 0.19 |
ADAM12 |
ADAM metallopeptidase domain 12 |
366 |
0.92 |
| chr1_19971146_19971510 | 0.19 |
NBL1 |
neuroblastoma 1, DAN family BMP antagonist |
520 |
0.75 |
| chr2_217547252_217547403 | 0.19 |
AC007563.5 |
|
11860 |
0.16 |
| chr10_52833546_52833721 | 0.19 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
301 |
0.92 |
| chr16_49892788_49893452 | 0.19 |
ZNF423 |
zinc finger protein 423 |
1290 |
0.54 |
| chr9_138986890_138987195 | 0.19 |
NACC2 |
NACC family member 2, BEN and BTB (POZ) domain containing |
89 |
0.97 |
| chr18_21851258_21851803 | 0.18 |
OSBPL1A |
oxysterol binding protein-like 1A |
666 |
0.6 |
| chr2_200320797_200320948 | 0.18 |
SATB2 |
SATB homeobox 2 |
61 |
0.97 |
| chr4_157892760_157892940 | 0.18 |
PDGFC |
platelet derived growth factor C |
304 |
0.92 |
| chr14_51338563_51338822 | 0.18 |
ABHD12B |
abhydrolase domain containing 12B |
186 |
0.93 |
| chr20_23401803_23402072 | 0.18 |
NAPB |
N-ethylmaleimide-sensitive factor attachment protein, beta |
161 |
0.92 |
| chr9_131486459_131486739 | 0.18 |
RP11-545E17.3 |
|
125 |
0.59 |
| chr2_200327065_200327302 | 0.18 |
SATB2 |
SATB homeobox 2 |
2648 |
0.32 |
| chr7_94025506_94025990 | 0.17 |
COL1A2 |
collagen, type I, alpha 2 |
1875 |
0.46 |
| chr4_1303323_1303652 | 0.17 |
MAEA |
macrophage erythroblast attacher |
88 |
0.96 |
| chr12_66217596_66217877 | 0.17 |
HMGA2 |
high mobility group AT-hook 2 |
175 |
0.96 |
| chr9_89560215_89560647 | 0.17 |
GAS1 |
growth arrest-specific 1 |
1673 |
0.54 |
| chr12_80794329_80794553 | 0.17 |
PTPRQ |
protein tyrosine phosphatase, receptor type, Q |
5333 |
0.26 |
| chr21_9540266_9540489 | 0.17 |
ENSG00000238411 |
. |
142814 |
0.05 |
| chr17_39968419_39968821 | 0.17 |
LEPREL4 |
leprecan-like 4 |
169 |
0.65 |
| chr7_27244730_27244900 | 0.16 |
HOTTIP |
HOXA distal transcript antisense RNA |
4759 |
0.08 |
| chr8_29970817_29970976 | 0.16 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
11440 |
0.15 |
| chr4_145566489_145566777 | 0.16 |
HHIP-AS1 |
HHIP antisense RNA 1 |
431 |
0.56 |
| chr12_115103708_115103959 | 0.16 |
ENSG00000199220 |
. |
7966 |
0.23 |
| chr2_97018957_97019230 | 0.16 |
NCAPH |
non-SMC condensin I complex, subunit H |
11125 |
0.15 |
| chr3_73674636_73674822 | 0.16 |
PDZRN3 |
PDZ domain containing ring finger 3 |
638 |
0.7 |
| chr11_69460305_69460629 | 0.16 |
CCND1 |
cyclin D1 |
4493 |
0.24 |
| chr12_6861300_6861451 | 0.16 |
MLF2 |
myeloid leukemia factor 2 |
469 |
0.6 |
| chr1_240256340_240256520 | 0.16 |
FMN2 |
formin 2 |
1250 |
0.57 |
| chr19_36247080_36247289 | 0.16 |
HSPB6 |
heat shock protein, alpha-crystallin-related, B6 |
726 |
0.32 |
| chr2_164593136_164593407 | 0.16 |
FIGN |
fidgetin |
749 |
0.81 |
| chr5_28926507_28926682 | 0.16 |
ENSG00000252601 |
. |
144009 |
0.05 |
| chr1_32228873_32229294 | 0.15 |
BAI2 |
brain-specific angiogenesis inhibitor 2 |
440 |
0.77 |
| chr13_108520858_108521069 | 0.15 |
FAM155A |
family with sequence similarity 155, member A |
1880 |
0.44 |
| chr8_49831422_49831573 | 0.15 |
SNAI2 |
snail family zinc finger 2 |
2491 |
0.43 |
| chr7_19155713_19156221 | 0.15 |
TWIST1 |
twist family bHLH transcription factor 1 |
1328 |
0.36 |
| chr11_35547343_35547568 | 0.15 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
124 |
0.98 |
| chr11_117070981_117071292 | 0.15 |
TAGLN |
transgelin |
165 |
0.93 |
| chr12_120242014_120242193 | 0.15 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
916 |
0.66 |
| chr10_128076891_128077106 | 0.15 |
ADAM12 |
ADAM metallopeptidase domain 12 |
26 |
0.99 |
| chr16_4422135_4423224 | 0.15 |
VASN |
vasorin |
830 |
0.49 |
| chr8_49293229_49293438 | 0.15 |
ENSG00000252710 |
. |
72743 |
0.13 |
| chr17_45608577_45608728 | 0.15 |
NPEPPS |
aminopeptidase puromycin sensitive |
41 |
0.97 |
| chr2_177001025_177001411 | 0.15 |
HOXD3 |
homeobox D3 |
467 |
0.41 |
| chr6_1821988_1822154 | 0.15 |
FOXC1 |
forkhead box C1 |
211390 |
0.02 |
| chr4_186695661_186695838 | 0.15 |
SORBS2 |
sorbin and SH3 domain containing 2 |
681 |
0.77 |
| chr20_53092813_53093232 | 0.15 |
DOK5 |
docking protein 5 |
765 |
0.8 |
| chr2_71175523_71175730 | 0.15 |
AC007040.7 |
|
56 |
0.94 |
| chr7_69064480_69064650 | 0.15 |
AUTS2 |
autism susceptibility candidate 2 |
32 |
0.99 |
| chr9_79306692_79307074 | 0.15 |
PRUNE2 |
prune homolog 2 (Drosophila) |
226 |
0.95 |
| chr13_35324652_35324864 | 0.15 |
NBEA |
neurobeachin |
191666 |
0.03 |
| chr11_63933058_63933366 | 0.15 |
MACROD1 |
MACRO domain containing 1 |
321 |
0.79 |
| chr15_55881115_55881360 | 0.15 |
PYGO1 |
pygopus family PHD finger 1 |
92 |
0.98 |
| chr8_89338531_89338740 | 0.15 |
RP11-586K2.1 |
|
430 |
0.76 |
| chr6_152699487_152699714 | 0.15 |
SYNE1-AS1 |
SYNE1 antisense RNA 1 |
2081 |
0.29 |
| chr22_38071896_38072177 | 0.14 |
LGALS1 |
lectin, galactoside-binding, soluble, 1 |
421 |
0.7 |
| chr7_855867_856218 | 0.14 |
SUN1 |
Sad1 and UNC84 domain containing 1 |
210 |
0.94 |
| chr5_33794654_33794915 | 0.14 |
ENSG00000201623 |
. |
93483 |
0.08 |
| chr8_109799930_109800109 | 0.14 |
TMEM74 |
transmembrane protein 74 |
175 |
0.97 |
| chr10_123914477_123914646 | 0.14 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
8380 |
0.28 |
| chr1_86042054_86042529 | 0.14 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
1642 |
0.36 |
| chr2_66659997_66660626 | 0.14 |
MEIS1-AS3 |
MEIS1 antisense RNA 3 |
291 |
0.87 |
| chr10_96117964_96118115 | 0.14 |
NOC3L |
nucleolar complex associated 3 homolog (S. cerevisiae) |
3672 |
0.25 |
| chr1_1292304_1292633 | 0.14 |
MXRA8 |
matrix-remodelling associated 8 |
1447 |
0.18 |
| chr4_111397030_111397405 | 0.14 |
ENPEP |
glutamyl aminopeptidase (aminopeptidase A) |
12 |
0.98 |
| chr6_72596608_72597181 | 0.14 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
167 |
0.98 |
| chr17_76920735_76921015 | 0.14 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
594 |
0.68 |
| chr11_121971856_121972438 | 0.14 |
RP11-166D19.1 |
|
247 |
0.88 |
| chr5_163723747_163723929 | 0.14 |
CTC-340A15.2 |
|
86 |
0.99 |
| chr10_81203565_81203736 | 0.14 |
ZCCHC24 |
zinc finger, CCHC domain containing 24 |
325 |
0.9 |
| chr11_118480914_118481065 | 0.14 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
2631 |
0.17 |
| chr17_30184995_30185438 | 0.14 |
COPRS |
coordinator of PRMT5, differentiation stimulator |
734 |
0.68 |
| chr17_37353212_37353473 | 0.14 |
CACNB1 |
calcium channel, voltage-dependent, beta 1 subunit |
613 |
0.61 |
| chr13_96293607_96293758 | 0.14 |
DZIP1 |
DAZ interacting zinc finger protein 1 |
1835 |
0.39 |
| chr14_89017580_89017758 | 0.14 |
PTPN21 |
protein tyrosine phosphatase, non-receptor type 21 |
135 |
0.68 |
| chr9_132427262_132427515 | 0.14 |
PRRX2 |
paired related homeobox 2 |
532 |
0.7 |
| chr16_3162631_3163041 | 0.14 |
ZNF205 |
zinc finger protein 205 |
40 |
0.8 |
| chr12_20521473_20521679 | 0.14 |
PDE3A |
phosphodiesterase 3A, cGMP-inhibited |
603 |
0.72 |
| chr4_108746206_108746434 | 0.14 |
SGMS2 |
sphingomyelin synthase 2 |
44 |
0.98 |
| chr8_103668410_103668650 | 0.14 |
KLF10 |
Kruppel-like factor 10 |
400 |
0.88 |
| chr4_37910177_37910528 | 0.13 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
17632 |
0.23 |
| chr20_50045192_50045343 | 0.13 |
ENSG00000266761 |
. |
24247 |
0.22 |
| chr19_47507472_47507696 | 0.13 |
NPAS1 |
neuronal PAS domain protein 1 |
15493 |
0.16 |
| chr2_217558888_217559156 | 0.13 |
AC007563.5 |
|
165 |
0.81 |
| chr8_23584501_23584839 | 0.13 |
RP11-175E9.1 |
|
20016 |
0.15 |
| chr18_11908463_11908810 | 0.13 |
MPPE1 |
metallophosphoesterase 1 |
5 |
0.55 |
| chr10_93472276_93472444 | 0.13 |
PPP1R3C |
protein phosphatase 1, regulatory subunit 3C |
79549 |
0.09 |
| chr20_57458594_57458745 | 0.13 |
ENSG00000225806 |
. |
5178 |
0.15 |
| chr19_16178450_16178920 | 0.13 |
TPM4 |
tropomyosin 4 |
175 |
0.95 |
| chr5_143265670_143265821 | 0.13 |
HMHB1 |
histocompatibility (minor) HB-1 |
74019 |
0.12 |
| chr13_35516657_35516808 | 0.13 |
NBEA |
neurobeachin |
65 |
0.99 |
| chr18_46456625_46456828 | 0.13 |
SMAD7 |
SMAD family member 7 |
18149 |
0.23 |
| chr2_204398111_204398262 | 0.13 |
RAPH1 |
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
30 |
0.99 |
| chr2_46303415_46303611 | 0.13 |
AC017006.2 |
|
2454 |
0.39 |
| chr13_24477531_24477691 | 0.13 |
C1QTNF9B |
C1q and tumor necrosis factor related protein 9B |
817 |
0.59 |
| chr14_90989296_90989447 | 0.13 |
ENSG00000252748 |
. |
42114 |
0.16 |
| chr17_39969274_39969651 | 0.13 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
279 |
0.68 |
| chr12_106532127_106532580 | 0.13 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
1458 |
0.48 |
| chr11_69456449_69456721 | 0.13 |
CCND1 |
cyclin D1 |
611 |
0.77 |
| chr11_27721206_27721474 | 0.13 |
BDNF |
brain-derived neurotrophic factor |
126 |
0.97 |
| chr9_16828717_16828972 | 0.13 |
BNC2 |
basonuclin 2 |
3442 |
0.37 |
| chr19_40854561_40855376 | 0.13 |
PLD3 |
phospholipase D family, member 3 |
357 |
0.56 |
| chr17_36413093_36413244 | 0.13 |
RP11-1407O15.2 |
TBC1 domain family member 3 |
9 |
0.97 |
| chr8_50822371_50822736 | 0.13 |
SNTG1 |
syntrophin, gamma 1 |
204 |
0.97 |
| chr5_121647230_121647613 | 0.13 |
SNCAIP |
synuclein, alpha interacting protein |
14 |
0.52 |
| chr2_133427812_133428020 | 0.13 |
LYPD1 |
LY6/PLAUR domain containing 1 |
138 |
0.97 |
| chr10_75758418_75758588 | 0.13 |
VCL |
vinculin |
629 |
0.7 |
| chrX_153095524_153095861 | 0.13 |
PDZD4 |
PDZ domain containing 4 |
121 |
0.93 |
| chr12_96183088_96183298 | 0.13 |
NTN4 |
netrin 4 |
533 |
0.73 |
| chr4_153702075_153702226 | 0.13 |
ARFIP1 |
ADP-ribosylation factor interacting protein 1 |
1013 |
0.44 |
| chr16_56709683_56709834 | 0.13 |
MT1H |
metallothionein 1H |
6006 |
0.08 |
| chr3_13008489_13008743 | 0.13 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
552 |
0.83 |
| chr12_49393262_49393666 | 0.13 |
DDN |
dendrin |
372 |
0.63 |
| chr7_145813457_145813835 | 0.13 |
CNTNAP2 |
contactin associated protein-like 2 |
193 |
0.96 |
| chr19_13734648_13734872 | 0.13 |
CACNA1A |
calcium channel, voltage-dependent, P/Q type, alpha 1A subunit |
44 |
0.98 |
| chr17_47091446_47091597 | 0.13 |
RP11-501C14.6 |
|
434 |
0.71 |
| chr20_34742202_34742447 | 0.12 |
EPB41L1 |
erythrocyte membrane protein band 4.1-like 1 |
340 |
0.81 |
| chr2_223155041_223155406 | 0.12 |
CCDC140 |
coiled-coil domain containing 140 |
7643 |
0.2 |
| chr11_86665468_86665750 | 0.12 |
FZD4 |
frizzled family receptor 4 |
824 |
0.7 |
| chr15_21360217_21360545 | 0.12 |
ENSG00000243059 |
. |
51914 |
0.14 |
| chr6_19691587_19692480 | 0.12 |
ENSG00000200957 |
. |
49273 |
0.18 |
| chr12_27617019_27617170 | 0.12 |
SMCO2 |
single-pass membrane protein with coiled-coil domains 2 |
2649 |
0.29 |
| chr20_35172707_35172858 | 0.12 |
MYL9 |
myosin, light chain 9, regulatory |
2882 |
0.2 |
| chr17_40932315_40932466 | 0.12 |
WNK4 |
WNK lysine deficient protein kinase 4 |
306 |
0.77 |
| chr2_74230374_74230525 | 0.12 |
TET3 |
tet methylcytosine dioxygenase 3 |
609 |
0.71 |
| chr2_218283085_218283236 | 0.12 |
ENSG00000251982 |
. |
166881 |
0.04 |
| chr7_100304309_100304512 | 0.12 |
POP7 |
processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) |
451 |
0.69 |
| chr17_1080749_1080900 | 0.12 |
ABR |
active BCR-related |
2254 |
0.29 |
| chr11_61355070_61355300 | 0.12 |
SYT7 |
synaptotagmin VII |
6565 |
0.2 |
| chr19_42456013_42456185 | 0.12 |
RABAC1 |
Rab acceptor 1 (prenylated) |
6967 |
0.14 |
| chr11_89867468_89867846 | 0.12 |
NAALAD2 |
N-acetylated alpha-linked acidic dipeptidase 2 |
40 |
0.98 |
| chr2_74230833_74231311 | 0.12 |
TET3 |
tet methylcytosine dioxygenase 3 |
1232 |
0.43 |
| chrX_100794184_100794359 | 0.12 |
ARMCX1 |
armadillo repeat containing, X-linked 1 |
11243 |
0.15 |
| chr2_206062809_206062987 | 0.12 |
NRP2 |
neuropilin 2 |
484326 |
0.01 |
| chr6_131456080_131456669 | 0.12 |
AKAP7 |
A kinase (PRKA) anchor protein 7 |
432 |
0.9 |
| chr4_187648163_187648374 | 0.12 |
FAT1 |
FAT atypical cadherin 1 |
392 |
0.92 |
| chr8_23711375_23711668 | 0.12 |
STC1 |
stanniocalcin 1 |
303 |
0.92 |
| chr19_14048794_14049313 | 0.12 |
PODNL1 |
podocan-like 1 |
144 |
0.92 |
| chr2_190043861_190044108 | 0.12 |
COL5A2 |
collagen, type V, alpha 2 |
621 |
0.79 |
| chr3_134125279_134125630 | 0.12 |
ENSG00000221313 |
. |
3528 |
0.24 |
| chr14_105218972_105219273 | 0.12 |
SIVA1 |
SIVA1, apoptosis-inducing factor |
315 |
0.83 |
| chr4_151502633_151502921 | 0.12 |
MAB21L2 |
mab-21-like 2 (C. elegans) |
300 |
0.91 |
| chr17_38600759_38601009 | 0.12 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
1171 |
0.39 |
| chr16_51184635_51185066 | 0.12 |
SALL1 |
spalt-like transcription factor 1 |
302 |
0.89 |
| chr2_203014256_203014407 | 0.12 |
AC079354.5 |
|
29783 |
0.12 |
| chr5_64558662_64558851 | 0.12 |
ENSG00000207439 |
. |
139560 |
0.05 |
| chr13_51565289_51565511 | 0.12 |
ENSG00000222920 |
. |
36588 |
0.18 |
| chr17_74235938_74236314 | 0.12 |
RNF157 |
ring finger protein 157 |
256 |
0.89 |
| chr15_96886136_96886425 | 0.12 |
ENSG00000222651 |
. |
9790 |
0.16 |
| chr11_117070436_117070728 | 0.12 |
TAGLN |
transgelin |
389 |
0.8 |
| chr17_37558067_37558428 | 0.12 |
CTB-131K11.1 |
|
201 |
0.65 |
| chr16_87028312_87028580 | 0.12 |
RP11-899L11.3 |
|
221075 |
0.02 |
| chr10_88728773_88729195 | 0.12 |
ENSG00000272734 |
. |
49 |
0.52 |
| chr14_52536506_52536657 | 0.12 |
NID2 |
nidogen 2 (osteonidogen) |
869 |
0.66 |
| chr15_61051494_61051741 | 0.12 |
RP11-554D20.1 |
|
4488 |
0.26 |
| chr3_23244042_23244389 | 0.12 |
UBE2E2-AS1 |
UBE2E2 antisense RNA 1 (head to head) |
146 |
0.7 |
| chr1_23894723_23894953 | 0.12 |
ID3 |
inhibitor of DNA binding 3, dominant negative helix-loop-helix protein |
8553 |
0.18 |
| chrX_70287985_70288182 | 0.12 |
SNX12 |
sorting nexin 12 |
170 |
0.92 |
| chr1_33815013_33815190 | 0.12 |
PHC2 |
polyhomeotic homolog 2 (Drosophila) |
385 |
0.67 |
| chr20_62959593_62959798 | 0.12 |
PCMTD2 |
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 |
62830 |
0.12 |
| chr13_29157926_29158077 | 0.12 |
POMP |
proteasome maturation protein |
75240 |
0.1 |
| chr16_56658952_56660008 | 0.12 |
MT1E |
metallothionein 1E |
93 |
0.91 |
| chr8_6264084_6264351 | 0.11 |
MCPH1 |
microcephalin 1 |
53 |
0.63 |
| chr9_12775957_12776199 | 0.11 |
LURAP1L |
leucine rich adaptor protein 1-like |
1058 |
0.54 |
| chr22_43010694_43010914 | 0.11 |
POLDIP3 |
polymerase (DNA-directed), delta interacting protein 3 |
59 |
0.86 |
| chr1_155830135_155830448 | 0.11 |
SYT11 |
synaptotagmin XI |
944 |
0.35 |
| chr2_20248255_20248470 | 0.11 |
LAPTM4A |
lysosomal protein transmembrane 4 alpha |
3427 |
0.2 |
| chr21_47401799_47402727 | 0.11 |
COL6A1 |
collagen, type VI, alpha 1 |
612 |
0.75 |
| chr6_157610005_157610158 | 0.11 |
ENSG00000252609 |
. |
102350 |
0.07 |
| chr2_91751048_91751437 | 0.11 |
IGKV1OR2-118 |
immunoglobulin kappa variable 1/OR2-118 (pseudogene) |
72146 |
0.11 |
| chr6_105585236_105585620 | 0.11 |
BVES-AS1 |
BVES antisense RNA 1 |
134 |
0.78 |
| chr11_75022648_75022930 | 0.11 |
ARRB1 |
arrestin, beta 1 |
5036 |
0.16 |
| chr19_3095677_3095900 | 0.11 |
GNA11 |
guanine nucleotide binding protein (G protein), alpha 11 (Gq class) |
1380 |
0.29 |
| chr17_39683598_39683785 | 0.11 |
KRT19 |
keratin 19 |
860 |
0.38 |
| chr6_113886088_113886398 | 0.11 |
ENSG00000266650 |
. |
37874 |
0.19 |
| chr11_133939469_133939706 | 0.11 |
JAM3 |
junctional adhesion molecule 3 |
621 |
0.79 |
| chr11_12398813_12399038 | 0.11 |
PARVA |
parvin, alpha |
185 |
0.96 |
| chr4_160188472_160188729 | 0.11 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
287 |
0.93 |
| chr3_124839242_124839433 | 0.11 |
SLC12A8 |
solute carrier family 12, member 8 |
361 |
0.8 |
| chr5_140888543_140888704 | 0.11 |
DIAPH1 |
diaphanous-related formin 1 |
17091 |
0.09 |
| chr4_82136118_82136337 | 0.11 |
PRKG2 |
protein kinase, cGMP-dependent, type II |
9 |
0.99 |
| chr10_77158473_77158721 | 0.11 |
ENSG00000237149 |
. |
2680 |
0.25 |
| chr5_175793030_175793335 | 0.11 |
ARL10 |
ADP-ribosylation factor-like 10 |
711 |
0.45 |
| chr4_380286_380451 | 0.11 |
ENSG00000211482 |
. |
23230 |
0.15 |
| chr14_61083017_61083244 | 0.11 |
ENSG00000253014 |
. |
9345 |
0.21 |
| chr16_86544208_86544564 | 0.11 |
FOXF1 |
forkhead box F1 |
253 |
0.93 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.1 | 0.4 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.1 | 0.3 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.1 | 0.3 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.1 | 0.2 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.1 | 0.2 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.2 | GO:0032353 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.1 | 0.1 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.1 | 0.2 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.2 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.3 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.2 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.2 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.1 | 0.1 | GO:0002689 | negative regulation of leukocyte chemotaxis(GO:0002689) |
| 0.0 | 0.1 | GO:0021825 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) substrate-dependent cerebral cortex tangential migration(GO:0021825) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.0 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.3 | GO:0033033 | negative regulation of myeloid cell apoptotic process(GO:0033033) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.0 | GO:0060632 | regulation of microtubule-based movement(GO:0060632) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0033087 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:1900121 | regulation of receptor binding(GO:1900120) negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.4 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.2 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0044332 | Wnt signaling pathway involved in dorsal/ventral axis specification(GO:0044332) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.4 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.2 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.1 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0003417 | endochondral bone growth(GO:0003416) growth plate cartilage development(GO:0003417) bone growth(GO:0098868) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.0 | 0.1 | GO:0090081 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.0 | GO:0044415 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.3 | GO:0051965 | positive regulation of synapse assembly(GO:0051965) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.2 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.0 | GO:0060197 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.1 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.0 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.1 | GO:0031953 | negative regulation of protein autophosphorylation(GO:0031953) |
| 0.0 | 0.1 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.0 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.2 | GO:0071320 | cellular response to cAMP(GO:0071320) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0070474 | uterine smooth muscle contraction(GO:0070471) regulation of uterine smooth muscle contraction(GO:0070472) positive regulation of uterine smooth muscle contraction(GO:0070474) |
| 0.0 | 0.2 | GO:0042074 | cell migration involved in gastrulation(GO:0042074) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0046599 | regulation of centriole replication(GO:0046599) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.3 | GO:0010171 | body morphogenesis(GO:0010171) |
| 0.0 | 0.1 | GO:0046476 | glycosylceramide biosynthetic process(GO:0046476) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.2 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0045715 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) |
| 0.0 | 0.0 | GO:1903960 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.0 | GO:0042816 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.2 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.1 | GO:0014051 | gamma-aminobutyric acid secretion(GO:0014051) |
| 0.0 | 0.2 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0034695 | response to prostaglandin E(GO:0034695) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:0006451 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0031272 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:0009148 | pyrimidine nucleoside triphosphate biosynthetic process(GO:0009148) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0032400 | melanosome localization(GO:0032400) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0008053 | mitochondrial fusion(GO:0008053) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0033173 | calcineurin-NFAT signaling cascade(GO:0033173) |
| 0.0 | 0.1 | GO:0045725 | positive regulation of glycogen biosynthetic process(GO:0045725) positive regulation of glycogen metabolic process(GO:0070875) |
| 0.0 | 0.1 | GO:0000470 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) maturation of LSU-rRNA(GO:0000470) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0048293 | isotype switching to IgE isotypes(GO:0048289) regulation of isotype switching to IgE isotypes(GO:0048293) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.1 | GO:0051571 | positive regulation of histone H3-K4 methylation(GO:0051571) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0044091 | membrane biogenesis(GO:0044091) |
| 0.0 | 0.1 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.0 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.2 | GO:0060761 | negative regulation of response to cytokine stimulus(GO:0060761) |
| 0.0 | 0.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.0 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.0 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.2 | GO:1901863 | positive regulation of striated muscle tissue development(GO:0045844) positive regulation of muscle organ development(GO:0048636) positive regulation of muscle tissue development(GO:1901863) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0007090 | obsolete regulation of S phase of mitotic cell cycle(GO:0007090) |
| 0.0 | 0.2 | GO:0010812 | negative regulation of cell-substrate adhesion(GO:0010812) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0006910 | phagocytosis, recognition(GO:0006910) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0006047 | UDP-N-acetylglucosamine metabolic process(GO:0006047) |
| 0.0 | 0.2 | GO:0032011 | ARF protein signal transduction(GO:0032011) |
| 0.0 | 0.1 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 0.0 | GO:0046022 | regulation of transcription from RNA polymerase II promoter, mitotic(GO:0046021) positive regulation of transcription from RNA polymerase II promoter during mitosis(GO:0046022) |
| 0.0 | 0.3 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.0 | GO:0072193 | ureter development(GO:0072189) ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.1 | GO:0007199 | G-protein coupled receptor signaling pathway coupled to cGMP nucleotide second messenger(GO:0007199) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0033240 | positive regulation of cellular amine metabolic process(GO:0033240) |
| 0.0 | 0.0 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.2 | GO:0043631 | RNA polyadenylation(GO:0043631) |
| 0.0 | 0.0 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.1 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0021889 | olfactory bulb interneuron differentiation(GO:0021889) |
| 0.0 | 0.2 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0050732 | negative regulation of peptidyl-tyrosine phosphorylation(GO:0050732) |
| 0.0 | 0.2 | GO:0006890 | retrograde vesicle-mediated transport, Golgi to ER(GO:0006890) |
| 0.0 | 0.0 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.0 | GO:0014824 | tonic smooth muscle contraction(GO:0014820) artery smooth muscle contraction(GO:0014824) |
| 0.0 | 0.0 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.0 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.0 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.0 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0061117 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.0 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.3 | GO:0051925 | obsolete regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051925) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.0 | GO:0045329 | amino-acid betaine biosynthetic process(GO:0006578) carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.0 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.1 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.0 | GO:0006998 | nuclear envelope organization(GO:0006998) |
| 0.0 | 0.0 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.0 | GO:0017000 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0060976 | coronary vasculature development(GO:0060976) |
| 0.0 | 0.0 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.0 | GO:0070857 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0016556 | mRNA modification(GO:0016556) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0072216 | positive regulation of metanephros development(GO:0072216) |
| 0.0 | 0.0 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.0 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.0 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.2 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.1 | GO:0002483 | antigen processing and presentation of endogenous peptide antigen(GO:0002483) antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.0 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0046146 | tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.0 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.1 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0071062 | alphav-beta3 integrin-vitronectin complex(GO:0071062) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.7 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.0 | 0.1 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) ribonuclease P complex(GO:0030677) multimeric ribonuclease P complex(GO:0030681) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.2 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.0 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.1 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.0 | GO:0044215 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
| 0.0 | 0.1 | GO:0030122 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0042555 | MCM complex(GO:0042555) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.0 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.0 | GO:0031211 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.6 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.0 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.2 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.1 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.1 | 0.2 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.2 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.1 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.1 | 0.3 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.1 | 0.2 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.1 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.2 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.2 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0001077 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0003910 | DNA ligase activity(GO:0003909) DNA ligase (ATP) activity(GO:0003910) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0000295 | adenine nucleotide transmembrane transporter activity(GO:0000295) purine nucleotide transmembrane transporter activity(GO:0015216) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.3 | GO:0030275 | LRR domain binding(GO:0030275) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.0 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.2 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.1 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0004505 | phenylalanine 4-monooxygenase activity(GO:0004505) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.6 | GO:0004177 | aminopeptidase activity(GO:0004177) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.0 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0055106 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.0 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.4 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0000977 | transcription regulatory region sequence-specific DNA binding(GO:0000976) RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 0.0 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.0 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.0 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0016417 | S-acyltransferase activity(GO:0016417) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.5 | GO:0017048 | Rho GTPase binding(GO:0017048) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.0 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.1 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.0 | 0.1 | GO:0046965 | retinoid X receptor binding(GO:0046965) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.1 | GO:0004550 | nucleoside diphosphate kinase activity(GO:0004550) |
| 0.0 | 0.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.1 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.0 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.0 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0005041 | low-density lipoprotein receptor activity(GO:0005041) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.4 | GO:0008013 | beta-catenin binding(GO:0008013) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.2 | GO:0004435 | phosphatidylinositol phospholipase C activity(GO:0004435) |
| 0.0 | 0.0 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.1 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0016742 | hydroxymethyl-, formyl- and related transferase activity(GO:0016742) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.0 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.5 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.4 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.6 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.3 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | ST GAQ PATHWAY | G alpha q Pathway |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.0 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.1 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.0 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.5 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.4 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | Genes involved in Cell death signalling via NRAGE, NRIF and NADE |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.0 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.0 | REACTOME NUCLEAR EVENTS KINASE AND TRANSCRIPTION FACTOR ACTIVATION | Genes involved in Nuclear Events (kinase and transcription factor activation) |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.2 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.1 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.0 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.1 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.0 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.1 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |