Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
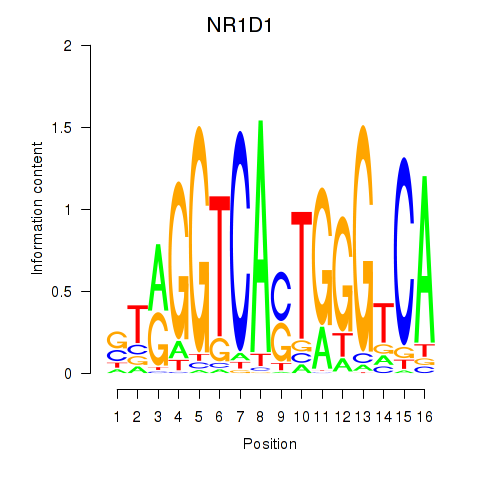
Results for NR1D1
Z-value: 0.83
Transcription factors associated with NR1D1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1D1
|
ENSG00000126368.5 | nuclear receptor subfamily 1 group D member 1 |
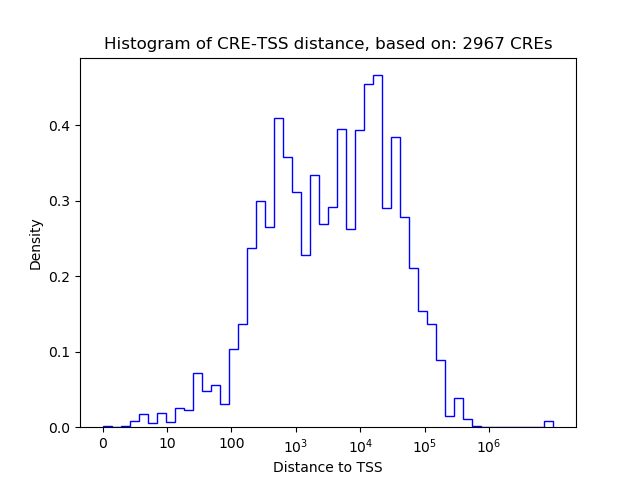
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr17_38255735_38256962 | NR1D1 | 630 | 0.599245 | -0.70 | 3.6e-02 | Click! |
| chr17_38267322_38267473 | NR1D1 | 10419 | 0.112073 | -0.65 | 5.6e-02 | Click! |
| chr17_38264116_38264342 | NR1D1 | 7251 | 0.119112 | -0.60 | 8.8e-02 | Click! |
| chr17_38252430_38252581 | NR1D1 | 4473 | 0.134797 | 0.52 | 1.5e-01 | Click! |
| chr17_38254187_38254627 | NR1D1 | 2571 | 0.176473 | -0.49 | 1.8e-01 | Click! |
Activity of the NR1D1 motif across conditions
Conditions sorted by the z-value of the NR1D1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
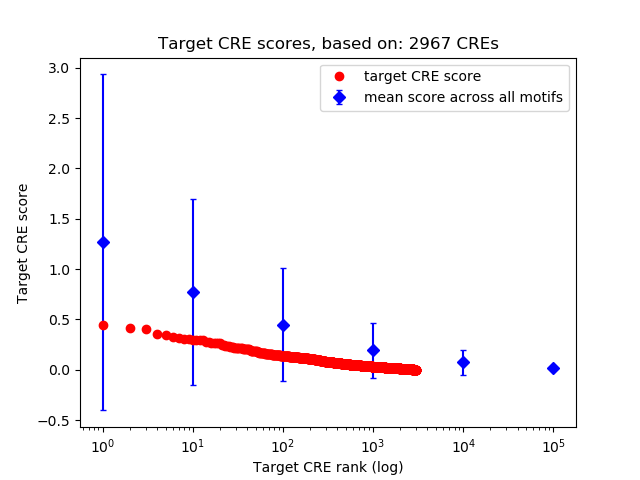
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_49627195_49627425 | 0.45 |
TUBA1C |
tubulin, alpha 1c |
5601 |
0.13 |
| chr1_249097926_249098077 | 0.42 |
SH3BP5L |
SH3-binding domain protein 5-like |
13271 |
0.15 |
| chr2_207972488_207972682 | 0.40 |
ENSG00000253008 |
. |
2212 |
0.31 |
| chr2_201559806_201559957 | 0.35 |
AOX1 |
aldehyde oxidase 1 |
32251 |
0.16 |
| chr7_508860_509011 | 0.34 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
49210 |
0.14 |
| chr19_20067439_20067590 | 0.33 |
AC007204.1 |
Uncharacterized protein |
18875 |
0.14 |
| chr15_50404394_50404634 | 0.31 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
2187 |
0.37 |
| chr11_48057635_48057786 | 0.31 |
AC103828.1 |
|
20303 |
0.19 |
| chr1_249097665_249097816 | 0.30 |
SH3BP5L |
SH3-binding domain protein 5-like |
13532 |
0.15 |
| chr12_123754396_123755524 | 0.30 |
CDK2AP1 |
cyclin-dependent kinase 2 associated protein 1 |
728 |
0.52 |
| chr10_75626090_75626284 | 0.30 |
CAMK2G |
calcium/calmodulin-dependent protein kinase II gamma |
5575 |
0.13 |
| chr1_38412223_38412952 | 0.29 |
INPP5B |
inositol polyphosphate-5-phosphatase, 75kDa |
136 |
0.94 |
| chr1_249097046_249097197 | 0.29 |
SH3BP5L |
SH3-binding domain protein 5-like |
14151 |
0.15 |
| chr7_26141005_26141262 | 0.28 |
ENSG00000266430 |
. |
41174 |
0.15 |
| chr9_71590240_71590555 | 0.28 |
PRKACG |
protein kinase, cAMP-dependent, catalytic, gamma |
38642 |
0.15 |
| chr3_32433354_32434295 | 0.26 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
293 |
0.93 |
| chr17_32582682_32583141 | 0.26 |
AC005549.3 |
Uncharacterized protein |
605 |
0.41 |
| chr7_45039935_45040086 | 0.26 |
CCM2 |
cerebral cavernous malformation 2 |
223 |
0.91 |
| chr2_232419167_232419527 | 0.26 |
NMUR1 |
neuromedin U receptor 1 |
24141 |
0.13 |
| chr2_232186481_232186632 | 0.26 |
ENSG00000263641 |
. |
40863 |
0.13 |
| chr12_54758511_54758662 | 0.25 |
GPR84 |
G protein-coupled receptor 84 |
315 |
0.77 |
| chr12_111843881_111845308 | 0.25 |
SH2B3 |
SH2B adaptor protein 3 |
842 |
0.62 |
| chr7_130071040_130071220 | 0.24 |
CEP41 |
centrosomal protein 41kDa |
4310 |
0.17 |
| chr17_7959293_7959452 | 0.24 |
ALOX15B |
arachidonate 15-lipoxygenase, type B |
16898 |
0.09 |
| chr1_9124419_9124586 | 0.24 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
5102 |
0.18 |
| chr12_71003154_71004126 | 0.23 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
16 |
0.99 |
| chr11_82608865_82609016 | 0.22 |
C11orf82 |
chromosome 11 open reading frame 82 |
2077 |
0.29 |
| chr9_35489468_35490886 | 0.22 |
RUSC2 |
RUN and SH3 domain containing 2 |
53 |
0.97 |
| chr19_41932700_41933275 | 0.22 |
B3GNT8 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 8 |
1648 |
0.18 |
| chr4_175344904_175345055 | 0.22 |
ENSG00000265846 |
. |
33 |
0.99 |
| chr5_176783043_176783194 | 0.21 |
RGS14 |
regulator of G-protein signaling 14 |
1720 |
0.2 |
| chr5_98362367_98362646 | 0.21 |
ENSG00000200351 |
. |
90055 |
0.09 |
| chr22_38029329_38029754 | 0.21 |
SH3BP1 |
SH3-domain binding protein 1 |
5941 |
0.11 |
| chr1_212731391_212732317 | 0.21 |
ATF3 |
activating transcription factor 3 |
6822 |
0.21 |
| chr6_25206992_25207143 | 0.21 |
ENSG00000264238 |
. |
3586 |
0.22 |
| chr9_7976512_7976663 | 0.21 |
TMEM261 |
transmembrane protein 261 |
176520 |
0.04 |
| chr4_8263855_8264006 | 0.21 |
HTRA3 |
HtrA serine peptidase 3 |
7562 |
0.22 |
| chr7_81267749_81267900 | 0.20 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
124488 |
0.06 |
| chr17_20491916_20492779 | 0.20 |
CDRT15L2 |
CMT1A duplicated region transcript 15-like 2 |
9310 |
0.21 |
| chr4_159738305_159738456 | 0.20 |
FNIP2 |
folliculin interacting protein 2 |
10980 |
0.21 |
| chr19_4473895_4474732 | 0.20 |
HDGFRP2 |
Hepatoma-derived growth factor-related protein 2 |
576 |
0.53 |
| chr9_139559538_139560114 | 0.20 |
EGFL7 |
EGF-like-domain, multiple 7 |
418 |
0.68 |
| chr2_54808515_54808696 | 0.20 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
23074 |
0.17 |
| chr17_71287404_71287786 | 0.19 |
CDC42EP4 |
CDC42 effector protein (Rho GTPase binding) 4 |
19952 |
0.15 |
| chr20_62238249_62238400 | 0.19 |
GMEB2 |
glucocorticoid modulatory element binding protein 2 |
2069 |
0.19 |
| chrX_119259306_119259683 | 0.19 |
RP4-755D9.1 |
|
5032 |
0.19 |
| chr3_4520313_4520464 | 0.19 |
SUMF1 |
sulfatase modifying factor 1 |
11423 |
0.2 |
| chr1_21851738_21851889 | 0.19 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
15948 |
0.2 |
| chr9_139837883_139838667 | 0.18 |
FBXW5 |
F-box and WD repeat domain containing 5 |
711 |
0.36 |
| chr6_134438521_134438724 | 0.18 |
ENSG00000266875 |
. |
16089 |
0.21 |
| chr19_43027430_43027581 | 0.18 |
CEACAM1 |
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
4469 |
0.2 |
| chr1_16554480_16554631 | 0.18 |
ANO7P1 |
anoctamin 7 pseudogene 1 |
33 |
0.95 |
| chr6_165722551_165723097 | 0.18 |
C6orf118 |
chromosome 6 open reading frame 118 |
272 |
0.95 |
| chr10_102108013_102108269 | 0.17 |
SCD |
stearoyl-CoA desaturase (delta-9-desaturase) |
1260 |
0.33 |
| chr1_27951514_27951705 | 0.17 |
FGR |
feline Gardner-Rasheed sarcoma viral oncogene homolog |
1036 |
0.44 |
| chrX_117347179_117347330 | 0.17 |
KLHL13 |
kelch-like family member 13 |
95951 |
0.08 |
| chr7_134469863_134470014 | 0.17 |
CALD1 |
caldesmon 1 |
5509 |
0.31 |
| chr13_45753588_45753739 | 0.17 |
KCTD4 |
potassium channel tetramerization domain containing 4 |
15189 |
0.17 |
| chr11_114178829_114179071 | 0.17 |
NNMT |
nicotinamide N-methyltransferase |
10077 |
0.21 |
| chr1_206689423_206689574 | 0.16 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
8619 |
0.15 |
| chr14_23235943_23236779 | 0.16 |
OXA1L |
oxidase (cytochrome c) assembly 1-like |
433 |
0.52 |
| chr15_75336418_75336696 | 0.16 |
PPCDC |
phosphopantothenoylcysteine decarboxylase |
941 |
0.52 |
| chr17_38279440_38280347 | 0.16 |
MSL1 |
male-specific lethal 1 homolog (Drosophila) |
937 |
0.43 |
| chr22_43358143_43358294 | 0.16 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
2313 |
0.35 |
| chr9_91604330_91604481 | 0.16 |
C9orf47 |
chromosome 9 open reading frame 47 |
1373 |
0.4 |
| chr22_17739469_17739620 | 0.16 |
CECR1 |
cat eye syndrome chromosome region, candidate 1 |
36665 |
0.16 |
| chr10_11784750_11785238 | 0.16 |
ECHDC3 |
enoyl CoA hydratase domain containing 3 |
549 |
0.83 |
| chr4_151204575_151204726 | 0.15 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
19213 |
0.26 |
| chr2_109253274_109253425 | 0.15 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
15627 |
0.21 |
| chr7_143082052_143082462 | 0.15 |
ZYX |
zyxin |
2164 |
0.18 |
| chr10_108923955_108924106 | 0.15 |
SORCS1 |
sortilin-related VPS10 domain containing receptor 1 |
254 |
0.96 |
| chrX_9310842_9311361 | 0.15 |
TBL1X |
transducin (beta)-like 1X-linked |
120234 |
0.06 |
| chr1_36042987_36043882 | 0.15 |
RP4-728D4.2 |
|
104 |
0.95 |
| chr9_92054863_92055014 | 0.15 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
3540 |
0.31 |
| chr10_102759781_102760726 | 0.15 |
LZTS2 |
leucine zipper, putative tumor suppressor 2 |
643 |
0.52 |
| chr10_82264222_82264373 | 0.15 |
RP11-137H2.4 |
|
31401 |
0.16 |
| chr17_75120485_75120909 | 0.15 |
SEC14L1 |
SEC14-like 1 (S. cerevisiae) |
3258 |
0.26 |
| chr5_146256062_146256213 | 0.15 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
2068 |
0.41 |
| chr11_36412455_36412606 | 0.15 |
RP11-514F3.5 |
|
4459 |
0.22 |
| chr19_49465738_49466010 | 0.15 |
FTL |
ferritin, light polypeptide |
2684 |
0.11 |
| chr1_160635203_160635434 | 0.15 |
RP11-404F10.2 |
|
5250 |
0.17 |
| chr2_152215166_152215348 | 0.15 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
1151 |
0.45 |
| chr14_33403030_33403481 | 0.15 |
NPAS3 |
neuronal PAS domain protein 3 |
884 |
0.76 |
| chr13_49435905_49436056 | 0.15 |
ENSG00000265585 |
. |
68368 |
0.11 |
| chr11_124627874_124628025 | 0.15 |
RP11-677M14.3 |
|
1076 |
0.32 |
| chr1_8557419_8557570 | 0.15 |
ENSG00000221083 |
. |
14484 |
0.21 |
| chr2_5367157_5367308 | 0.15 |
ENSG00000207192 |
. |
374365 |
0.01 |
| chr15_74222160_74222311 | 0.15 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
1646 |
0.29 |
| chr11_124752266_124752462 | 0.14 |
RP11-664I21.5 |
|
1223 |
0.3 |
| chr3_150995573_150995821 | 0.14 |
P2RY14 |
purinergic receptor P2Y, G-protein coupled, 14 |
460 |
0.8 |
| chr3_128943995_128944272 | 0.14 |
COPG1 |
coatomer protein complex, subunit gamma 1 |
24316 |
0.13 |
| chr17_32583150_32583439 | 0.14 |
AC005549.3 |
Uncharacterized protein |
988 |
0.32 |
| chr21_40378227_40378378 | 0.14 |
ENSG00000272015 |
. |
111593 |
0.06 |
| chr20_62478644_62478873 | 0.14 |
AL158091.1 |
Protein LOC100509861 |
3485 |
0.11 |
| chr8_128003605_128003873 | 0.14 |
ENSG00000212451 |
. |
319972 |
0.01 |
| chr2_129038014_129038165 | 0.14 |
HS6ST1 |
heparan sulfate 6-O-sulfotransferase 1 |
38062 |
0.18 |
| chr1_41706915_41707336 | 0.14 |
SCMH1 |
sex comb on midleg homolog 1 (Drosophila) |
657 |
0.8 |
| chr12_46568114_46568265 | 0.14 |
SLC38A1 |
solute carrier family 38, member 1 |
93295 |
0.09 |
| chr15_64231749_64232239 | 0.14 |
RP11-111E14.1 |
|
11521 |
0.22 |
| chr7_36195851_36196117 | 0.14 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
3107 |
0.3 |
| chr6_157388469_157388620 | 0.14 |
RP1-137K2.2 |
|
52296 |
0.17 |
| chr7_116228317_116228468 | 0.14 |
AC006159.4 |
|
17104 |
0.18 |
| chr21_40181513_40181664 | 0.14 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
28 |
0.99 |
| chr3_47034351_47034502 | 0.14 |
NBEAL2 |
neurobeachin-like 2 |
2385 |
0.28 |
| chr5_67515256_67515407 | 0.14 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
3727 |
0.29 |
| chr12_117500658_117500867 | 0.14 |
TESC |
tescalcin |
16126 |
0.25 |
| chr2_7148515_7148666 | 0.14 |
RNF144A |
ring finger protein 144A |
11519 |
0.24 |
| chr12_115137987_115138212 | 0.14 |
TBX3 |
T-box 3 |
16130 |
0.21 |
| chr19_42144659_42144810 | 0.14 |
CEACAM4 |
carcinoembryonic antigen-related cell adhesion molecule 4 |
11292 |
0.14 |
| chr17_79396214_79396830 | 0.14 |
RP11-1055B8.7 |
BAH and coiled-coil domain-containing protein 1 |
8874 |
0.11 |
| chr9_140215453_140215604 | 0.14 |
NRARP |
NOTCH-regulated ankyrin repeat protein |
18825 |
0.09 |
| chr11_72452757_72453026 | 0.14 |
ARAP1 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 |
10557 |
0.12 |
| chr10_15138786_15140121 | 0.14 |
RPP38 |
ribonuclease P/MRP 38kDa subunit |
34 |
0.62 |
| chr22_35726479_35726752 | 0.13 |
ENSG00000266320 |
. |
5018 |
0.18 |
| chr17_79375573_79375724 | 0.13 |
ENSG00000266392 |
. |
1070 |
0.35 |
| chr3_119041182_119042341 | 0.13 |
ARHGAP31-AS1 |
ARHGAP31 antisense RNA 1 |
154 |
0.94 |
| chr7_32699949_32700100 | 0.13 |
ENSG00000207573 |
. |
72569 |
0.11 |
| chr18_64350086_64350429 | 0.13 |
ENSG00000221536 |
. |
29519 |
0.24 |
| chr19_18525487_18525638 | 0.13 |
SSBP4 |
single stranded DNA binding protein 4 |
4112 |
0.1 |
| chr21_47472818_47473242 | 0.13 |
AP001471.1 |
|
44414 |
0.11 |
| chr2_175546375_175546968 | 0.13 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
928 |
0.66 |
| chr10_30163714_30163865 | 0.13 |
SVIL |
supervillin |
139056 |
0.05 |
| chr14_89632118_89632412 | 0.13 |
FOXN3 |
forkhead box N3 |
14823 |
0.28 |
| chr14_103584850_103585001 | 0.13 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
4873 |
0.18 |
| chr7_12151395_12151884 | 0.13 |
TMEM106B |
transmembrane protein 106B |
99228 |
0.09 |
| chr12_6493326_6493955 | 0.13 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
221 |
0.89 |
| chr10_134388453_134389007 | 0.13 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
32700 |
0.18 |
| chr2_40786004_40786155 | 0.13 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
46504 |
0.21 |
| chr20_33864845_33865490 | 0.13 |
MMP24-AS1 |
MMP24 antisense RNA 1 |
663 |
0.38 |
| chr2_16083664_16083952 | 0.13 |
MYCNOS |
MYCN opposite strand |
1437 |
0.36 |
| chr7_26577801_26577952 | 0.13 |
KIAA0087 |
KIAA0087 |
531 |
0.86 |
| chr19_47732183_47732562 | 0.13 |
BBC3 |
BCL2 binding component 3 |
2079 |
0.22 |
| chr5_141215789_141215991 | 0.13 |
PCDH1 |
protocadherin 1 |
33264 |
0.16 |
| chr12_26348638_26349346 | 0.13 |
SSPN |
sarcospan |
386 |
0.88 |
| chr1_27882496_27882647 | 0.13 |
RP1-159A19.4 |
|
30255 |
0.13 |
| chr8_58716546_58716697 | 0.13 |
FAM110B |
family with sequence similarity 110, member B |
190492 |
0.03 |
| chr3_37534801_37534952 | 0.13 |
ITGA9 |
integrin, alpha 9 |
41266 |
0.16 |
| chr10_94452035_94452186 | 0.13 |
HHEX |
hematopoietically expressed homeobox |
498 |
0.8 |
| chr18_74189003_74189166 | 0.13 |
ZNF516 |
zinc finger protein 516 |
13651 |
0.17 |
| chr6_166900831_166900982 | 0.13 |
ENSG00000222958 |
. |
22015 |
0.16 |
| chr1_11786795_11786946 | 0.13 |
AGTRAP |
angiotensin II receptor-associated protein |
9271 |
0.12 |
| chr18_3794614_3794765 | 0.12 |
ENSG00000238790 |
. |
16628 |
0.17 |
| chr11_2404413_2404564 | 0.12 |
CD81 |
CD81 molecule |
798 |
0.47 |
| chr17_79421896_79422047 | 0.12 |
ENSG00000266189 |
. |
3757 |
0.13 |
| chr20_62739628_62739779 | 0.12 |
NPBWR2 |
neuropeptides B/W receptor 2 |
1179 |
0.33 |
| chr4_99535433_99535584 | 0.12 |
TSPAN5 |
tetraspanin 5 |
43278 |
0.16 |
| chr10_45874400_45874551 | 0.12 |
ALOX5 |
arachidonate 5-lipoxygenase |
4800 |
0.27 |
| chr5_154093938_154094089 | 0.12 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
1551 |
0.33 |
| chr8_52798262_52798413 | 0.12 |
RP11-110G21.2 |
|
10597 |
0.18 |
| chr8_87052179_87052395 | 0.12 |
PSKH2 |
protein serine kinase H2 |
29639 |
0.19 |
| chr3_133287095_133287246 | 0.12 |
CDV3 |
CDV3 homolog (mouse) |
5404 |
0.22 |
| chr6_46458824_46459774 | 0.12 |
RCAN2 |
regulator of calcineurin 2 |
200 |
0.78 |
| chr9_69261149_69261300 | 0.12 |
CBWD6 |
COBW domain containing 6 |
1285 |
0.46 |
| chr10_15503569_15503720 | 0.12 |
FAM171A1 |
family with sequence similarity 171, member A1 |
90583 |
0.09 |
| chr19_8567892_8568043 | 0.12 |
PRAM1 |
PML-RARA regulated adaptor molecule 1 |
29 |
0.96 |
| chr2_38265076_38265999 | 0.12 |
RMDN2-AS1 |
RMDN2 antisense RNA 1 |
2053 |
0.33 |
| chr20_43987050_43987201 | 0.12 |
SYS1 |
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
3452 |
0.15 |
| chr2_27296685_27296836 | 0.12 |
OST4 |
oligosaccharyltransferase 4 homolog (S. cerevisiae) |
2119 |
0.12 |
| chr21_43882491_43882642 | 0.12 |
SLC37A1 |
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
33562 |
0.1 |
| chr7_38296522_38296673 | 0.12 |
STARD3NL |
STARD3 N-terminal like |
78600 |
0.11 |
| chrX_153962751_153963096 | 0.12 |
GAB3 |
GRB2-associated binding protein 3 |
16409 |
0.12 |
| chr12_4397764_4397915 | 0.12 |
CCND2-AS1 |
CCND2 antisense RNA 1 |
12489 |
0.16 |
| chr11_123106721_123106884 | 0.12 |
CLMP |
CXADR-like membrane protein |
40813 |
0.14 |
| chr17_1090163_1090534 | 0.12 |
ABR |
active BCR-related |
268 |
0.91 |
| chr1_156124803_156124988 | 0.12 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
733 |
0.51 |
| chr1_98515040_98515403 | 0.12 |
ENSG00000225206 |
. |
3494 |
0.38 |
| chr12_71551766_71551917 | 0.12 |
TSPAN8 |
tetraspanin 8 |
38 |
0.98 |
| chrX_150565701_150566113 | 0.12 |
VMA21 |
VMA21 vacuolar H+-ATPase homolog (S. cerevisiae) |
231 |
0.96 |
| chr1_28209156_28209307 | 0.12 |
THEMIS2 |
thymocyte selection associated family member 2 |
2989 |
0.16 |
| chr10_104411475_104411626 | 0.12 |
TRIM8 |
tripartite motif containing 8 |
6906 |
0.18 |
| chr16_27248668_27248966 | 0.12 |
NSMCE1 |
non-SMC element 1 homolog (S. cerevisiae) |
4418 |
0.19 |
| chr15_89680666_89681045 | 0.12 |
ENSG00000239151 |
. |
16845 |
0.18 |
| chr2_232226788_232226939 | 0.12 |
ENSG00000263641 |
. |
556 |
0.75 |
| chr22_43369638_43369789 | 0.12 |
PACSIN2 |
protein kinase C and casein kinase substrate in neurons 2 |
13808 |
0.21 |
| chr4_1006679_1006830 | 0.12 |
FGFRL1 |
fibroblast growth factor receptor-like 1 |
515 |
0.71 |
| chr13_38445031_38445182 | 0.12 |
TRPC4 |
transient receptor potential cation channel, subfamily C, member 4 |
544 |
0.83 |
| chr21_39543980_39544194 | 0.12 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
14959 |
0.18 |
| chr8_27219392_27219619 | 0.12 |
PTK2B |
protein tyrosine kinase 2 beta |
18663 |
0.21 |
| chr16_88980980_88981131 | 0.12 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
12132 |
0.11 |
| chr2_105458782_105459564 | 0.11 |
LINC01158 |
long intergenic non-protein coding RNA 1158 |
8738 |
0.14 |
| chr7_134854914_134855228 | 0.11 |
C7orf49 |
chromosome 7 open reading frame 49 |
342 |
0.81 |
| chr1_88368784_88369082 | 0.11 |
ENSG00000199318 |
. |
449877 |
0.01 |
| chr6_109288373_109288524 | 0.11 |
RP11-787I22.3 |
|
31237 |
0.15 |
| chr2_131094823_131095064 | 0.11 |
IMP4 |
IMP4, U3 small nucleolar ribonucleoprotein, homolog (yeast) |
4855 |
0.17 |
| chr22_40659716_40659867 | 0.11 |
TNRC6B |
trinucleotide repeat containing 6B |
1216 |
0.56 |
| chr7_93812562_93812751 | 0.11 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
178962 |
0.03 |
| chr3_66535668_66535819 | 0.11 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
15613 |
0.29 |
| chr11_128564573_128565732 | 0.11 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
766 |
0.51 |
| chr5_148747021_148747172 | 0.11 |
RP11-394O4.3 |
|
4641 |
0.13 |
| chr4_185268616_185268767 | 0.11 |
ENSG00000244512 |
. |
68602 |
0.09 |
| chr11_93868067_93868416 | 0.11 |
PANX1 |
pannexin 1 |
6146 |
0.28 |
| chr15_91285368_91285519 | 0.11 |
ENSG00000200677 |
. |
13099 |
0.14 |
| chr7_101525780_101526170 | 0.11 |
CTA-339C12.1 |
|
57966 |
0.12 |
| chr2_242605024_242605218 | 0.11 |
ATG4B |
autophagy related 4B, cysteine peptidase |
890 |
0.44 |
| chr16_89042423_89042673 | 0.11 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
853 |
0.55 |
| chr8_134706257_134706408 | 0.11 |
ENSG00000212273 |
. |
49364 |
0.19 |
| chr17_40729237_40729659 | 0.11 |
PSMC3IP |
PSMC3 interacting protein |
266 |
0.8 |
| chr11_64512499_64513555 | 0.11 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
99 |
0.95 |
| chr15_101995655_101995861 | 0.11 |
PCSK6 |
proprotein convertase subtilisin/kexin type 6 |
11890 |
0.25 |
| chr19_22817145_22817689 | 0.11 |
ZNF492 |
zinc finger protein 492 |
291 |
0.89 |
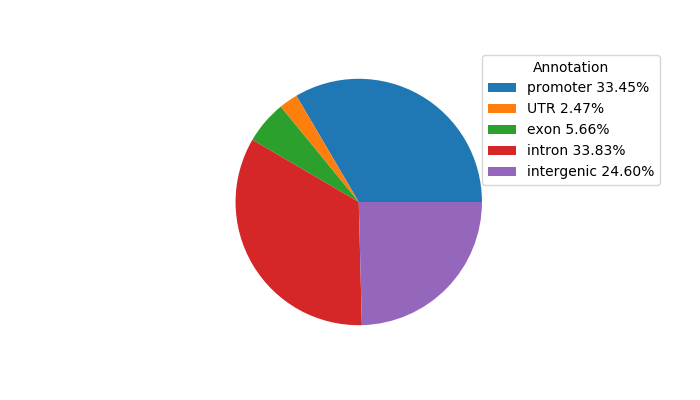
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.2 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.2 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.1 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0035581 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.1 | GO:0043116 | negative regulation of vascular permeability(GO:0043116) |
| 0.0 | 0.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0010544 | negative regulation of platelet activation(GO:0010544) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.0 | 0.0 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0046839 | phospholipid dephosphorylation(GO:0046839) |
| 0.0 | 0.0 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0043984 | histone H4-K16 acetylation(GO:0043984) |
| 0.0 | 0.1 | GO:0051299 | centrosome separation(GO:0051299) |
| 0.0 | 0.0 | GO:0072010 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0042629 | mast cell granule(GO:0042629) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.2 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0016215 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.2 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.0 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.0 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |