Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
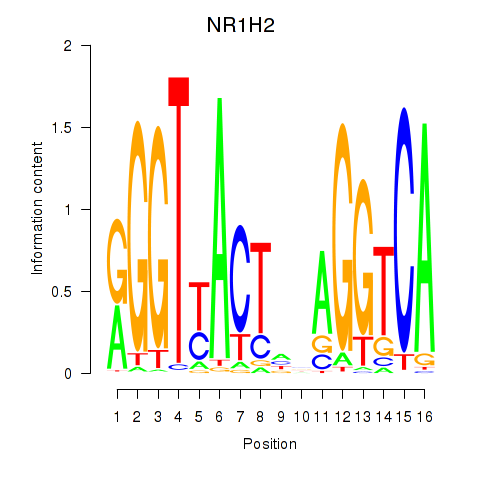
Results for NR1H2
Z-value: 0.82
Transcription factors associated with NR1H2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1H2
|
ENSG00000131408.9 | nuclear receptor subfamily 1 group H member 2 |
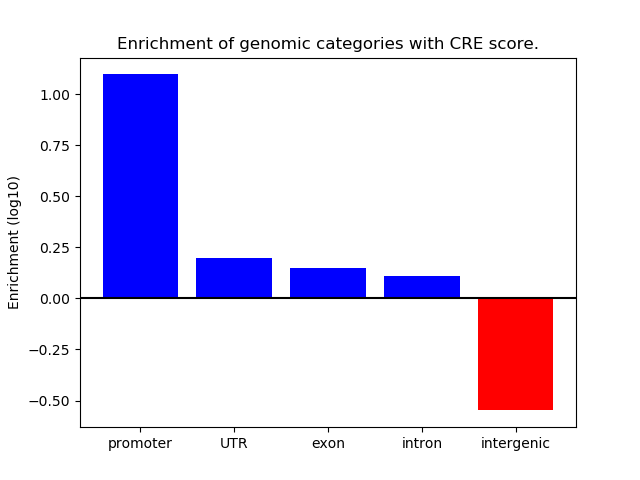
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_50879369_50879585 | NR1H2 | 238 | 0.836200 | 0.69 | 4.1e-02 | Click! |
| chr19_50832551_50833183 | NR1H2 | 43 | 0.732339 | 0.46 | 2.1e-01 | Click! |
| chr19_50879744_50879984 | NR1H2 | 134 | 0.908262 | 0.44 | 2.3e-01 | Click! |
| chr19_50833878_50834029 | NR1H2 | 1004 | 0.310566 | 0.38 | 3.2e-01 | Click! |
| chr19_50834536_50834687 | NR1H2 | 1662 | 0.198914 | 0.37 | 3.2e-01 | Click! |
Activity of the NR1H2 motif across conditions
Conditions sorted by the z-value of the NR1H2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
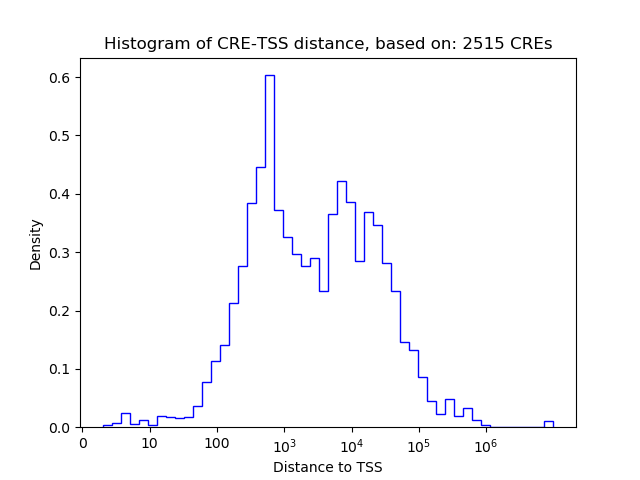
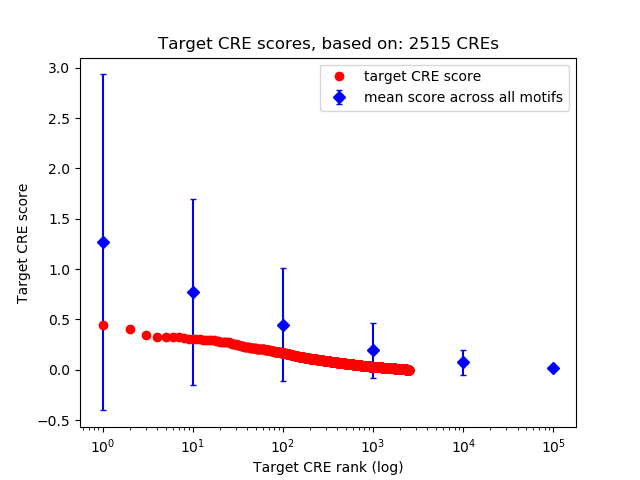
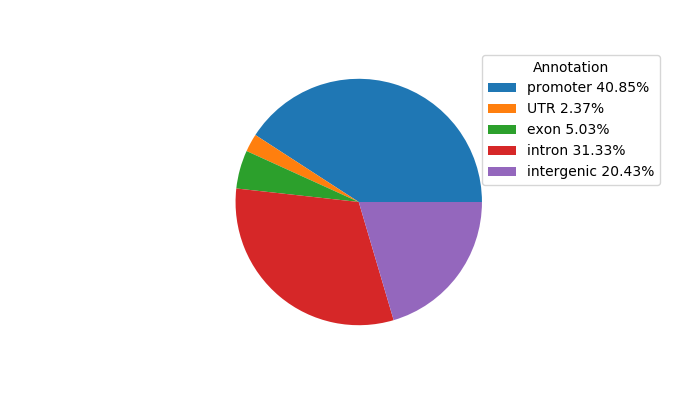
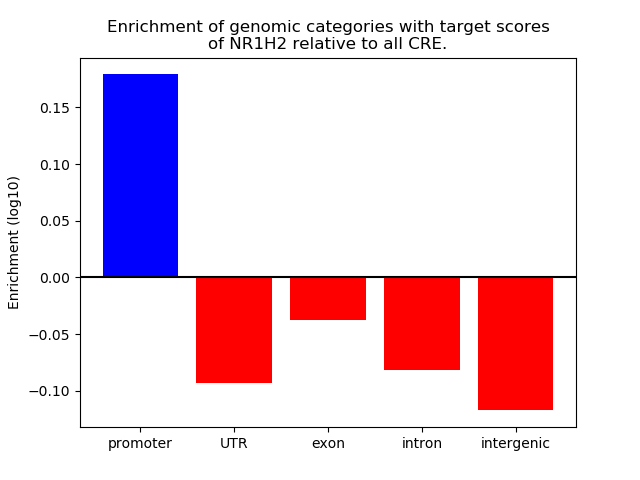
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_7145795_7146333 | 0.44 |
RREB1 |
ras responsive element binding protein 1 |
6586 |
0.22 |
| chr7_43804203_43804455 | 0.41 |
BLVRA |
biliverdin reductase A |
539 |
0.79 |
| chr12_54361938_54362089 | 0.35 |
HOTAIR |
HOX transcript antisense RNA |
685 |
0.4 |
| chr11_47380887_47381038 | 0.33 |
MYBPC3 |
myosin binding protein C, cardiac |
6709 |
0.11 |
| chr19_36398415_36398818 | 0.33 |
TYROBP |
TYRO protein tyrosine kinase binding protein |
533 |
0.58 |
| chr13_110757726_110757877 | 0.33 |
ENSG00000265885 |
. |
7303 |
0.31 |
| chr3_30655384_30655643 | 0.33 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
7420 |
0.31 |
| chr19_49004358_49004509 | 0.32 |
LMTK3 |
lemur tyrosine kinase 3 |
10628 |
0.1 |
| chr10_134369040_134369355 | 0.31 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
17554 |
0.21 |
| chr7_106506867_106507018 | 0.31 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
1018 |
0.65 |
| chr2_27109742_27109997 | 0.31 |
DPYSL5 |
dihydropyrimidinase-like 5 |
38493 |
0.13 |
| chr14_69422245_69422436 | 0.30 |
ACTN1 |
actinin, alpha 1 |
8075 |
0.23 |
| chr19_55766822_55767012 | 0.30 |
PPP6R1 |
protein phosphatase 6, regulatory subunit 1 |
220 |
0.84 |
| chr9_140130864_140131015 | 0.30 |
TUBB4B |
tubulin, beta 4B class IVb |
4726 |
0.06 |
| chr3_12219302_12219453 | 0.30 |
TIMP4 |
TIMP metallopeptidase inhibitor 4 |
18526 |
0.25 |
| chr1_10447601_10448200 | 0.29 |
ENSG00000265945 |
. |
2159 |
0.22 |
| chr19_43882588_43882879 | 0.29 |
CTC-490G23.4 |
|
9611 |
0.16 |
| chr7_50345023_50345395 | 0.29 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
831 |
0.74 |
| chr2_139261843_139261994 | 0.28 |
SPOPL |
speckle-type POZ protein-like |
2547 |
0.45 |
| chr1_1173779_1173930 | 0.28 |
B3GALT6 |
UDP-Gal:betaGal beta 1,3-galactosyltransferase polypeptide 6 |
6225 |
0.08 |
| chr3_134324631_134324870 | 0.28 |
EPHB1 |
EPH receptor B1 |
9457 |
0.22 |
| chr9_129624166_129624406 | 0.28 |
ZBTB34 |
zinc finger and BTB domain containing 34 |
1342 |
0.48 |
| chr16_84809531_84809682 | 0.27 |
USP10 |
ubiquitin specific peptidase 10 |
7733 |
0.21 |
| chr1_2321939_2322090 | 0.27 |
MORN1 |
MORN repeat containing 1 |
579 |
0.52 |
| chr10_77189804_77189955 | 0.27 |
RP11-399K21.10 |
|
1467 |
0.43 |
| chr16_85604560_85604711 | 0.27 |
GSE1 |
Gse1 coiled-coil protein |
40380 |
0.16 |
| chr5_140996135_140996687 | 0.26 |
AC008781.7 |
|
1570 |
0.23 |
| chr14_106326572_106326804 | 0.26 |
ENSG00000265714 |
. |
664 |
0.18 |
| chr7_129418799_129419025 | 0.26 |
ENSG00000207691 |
. |
4058 |
0.17 |
| chr4_182284557_182284708 | 0.26 |
ENSG00000251742 |
. |
471629 |
0.01 |
| chr1_155220015_155220168 | 0.25 |
FAM189B |
family with sequence similarity 189, member B |
4608 |
0.07 |
| chr19_3135427_3135578 | 0.25 |
GNA15 |
guanine nucleotide binding protein (G protein), alpha 15 (Gq class) |
689 |
0.53 |
| chr4_187491209_187491867 | 0.24 |
MTNR1A |
melatonin receptor 1A |
14817 |
0.17 |
| chr1_40426265_40426496 | 0.24 |
MFSD2A |
major facilitator superfamily domain containing 2A |
5558 |
0.17 |
| chr11_16134687_16134838 | 0.24 |
ENSG00000221556 |
. |
11100 |
0.3 |
| chr3_126075576_126075794 | 0.23 |
KLF15 |
Kruppel-like factor 15 |
600 |
0.75 |
| chr16_30710458_30711178 | 0.23 |
SRCAP |
Snf2-related CREBBP activator protein |
356 |
0.48 |
| chr17_76123369_76123520 | 0.23 |
TMC6 |
transmembrane channel-like 6 |
343 |
0.8 |
| chr16_23540202_23540353 | 0.23 |
GGA2 |
golgi-associated, gamma adaptin ear containing, ARF binding protein 2 |
6961 |
0.14 |
| chr8_144408226_144408377 | 0.23 |
TOP1MT |
topoisomerase (DNA) I, mitochondrial |
5680 |
0.1 |
| chr19_17622860_17623539 | 0.23 |
PGLS |
6-phosphogluconolactonase |
525 |
0.61 |
| chr22_46402476_46402694 | 0.23 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
29576 |
0.1 |
| chr6_10520951_10521149 | 0.22 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
534 |
0.77 |
| chr14_24740164_24740379 | 0.22 |
RABGGTA |
Rab geranylgeranyltransferase, alpha subunit |
534 |
0.5 |
| chr3_50648522_50648700 | 0.22 |
CISH |
cytokine inducible SH2-containing protein |
592 |
0.48 |
| chr11_86748898_86749631 | 0.22 |
TMEM135 |
transmembrane protein 135 |
182 |
0.97 |
| chr2_201500251_201500402 | 0.22 |
AOX1 |
aldehyde oxidase 1 |
24434 |
0.2 |
| chr19_54827917_54828068 | 0.21 |
LILRA5 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
3583 |
0.12 |
| chr1_9690042_9690512 | 0.21 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
21513 |
0.15 |
| chr11_64611077_64611228 | 0.21 |
CDC42BPG |
CDC42 binding protein kinase gamma (DMPK-like) |
889 |
0.43 |
| chr2_69029555_69029831 | 0.21 |
ARHGAP25 |
Rho GTPase activating protein 25 |
4670 |
0.25 |
| chr12_120675375_120675841 | 0.21 |
PXN |
paxillin |
10961 |
0.12 |
| chr6_41612221_41612372 | 0.21 |
MDFI |
MyoD family inhibitor |
6031 |
0.16 |
| chr17_1547338_1547608 | 0.21 |
SCARF1 |
scavenger receptor class F, member 1 |
1565 |
0.23 |
| chr4_108910417_108910832 | 0.21 |
HADH |
hydroxyacyl-CoA dehydrogenase |
246 |
0.92 |
| chr4_75550401_75550552 | 0.21 |
AC142293.3 |
|
35812 |
0.18 |
| chr20_58633559_58633836 | 0.21 |
C20orf197 |
chromosome 20 open reading frame 197 |
2717 |
0.35 |
| chr3_37598700_37598851 | 0.21 |
ENSG00000239105 |
. |
15929 |
0.25 |
| chr11_44592282_44592433 | 0.21 |
CD82 |
CD82 molecule |
2271 |
0.34 |
| chr9_95716734_95716885 | 0.20 |
FGD3 |
FYVE, RhoGEF and PH domain containing 3 |
7076 |
0.22 |
| chr19_55770559_55770710 | 0.20 |
PPP6R1 |
protein phosphatase 6, regulatory subunit 1 |
271 |
0.8 |
| chr1_2805652_2805803 | 0.20 |
TTC34 |
tetratricopeptide repeat domain 34 |
87441 |
0.08 |
| chr20_33878344_33878495 | 0.20 |
FAM83C |
family with sequence similarity 83, member C |
1785 |
0.2 |
| chr6_15246387_15247495 | 0.20 |
JARID2 |
jumonji, AT rich interactive domain 2 |
414 |
0.82 |
| chr6_120039243_120039394 | 0.20 |
ENSG00000265725 |
. |
297007 |
0.01 |
| chr14_102976422_102976710 | 0.20 |
ANKRD9 |
ankyrin repeat domain 9 |
430 |
0.81 |
| chr19_52154679_52154830 | 0.20 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
4603 |
0.12 |
| chr8_144951605_144951756 | 0.20 |
EPPK1 |
epiplakin 1 |
952 |
0.39 |
| chr3_48955621_48956131 | 0.19 |
ARIH2 |
ariadne RBR E3 ubiquitin protein ligase 2 |
378 |
0.68 |
| chr16_85649875_85650315 | 0.19 |
GSE1 |
Gse1 coiled-coil protein |
3137 |
0.26 |
| chr8_19797296_19797595 | 0.19 |
LPL |
lipoprotein lipase |
963 |
0.69 |
| chr3_15482302_15482608 | 0.19 |
EAF1-AS1 |
EAF1 antisense RNA 1 |
354 |
0.48 |
| chr4_79580897_79581048 | 0.19 |
ENSG00000238816 |
. |
19718 |
0.2 |
| chr3_151608599_151608750 | 0.19 |
SUCNR1 |
succinate receptor 1 |
17243 |
0.21 |
| chr20_39676850_39677439 | 0.19 |
TOP1 |
topoisomerase (DNA) I |
19686 |
0.2 |
| chr17_35716056_35716207 | 0.19 |
ACACA |
acetyl-CoA carboxylase alpha |
72 |
0.97 |
| chr2_225455716_225455867 | 0.18 |
CUL3 |
cullin 3 |
5681 |
0.34 |
| chr11_58342434_58343027 | 0.18 |
LPXN |
leupaxin |
604 |
0.68 |
| chr16_1451750_1451901 | 0.18 |
LA16c-312E8.2 |
|
7010 |
0.08 |
| chr2_74198290_74198479 | 0.18 |
ENSG00000201876 |
. |
3051 |
0.21 |
| chr10_102133099_102133250 | 0.18 |
ENSG00000212325 |
. |
25029 |
0.12 |
| chr19_42302290_42302441 | 0.18 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
1280 |
0.33 |
| chr15_81591495_81592151 | 0.18 |
IL16 |
interleukin 16 |
66 |
0.98 |
| chr18_21296917_21297068 | 0.18 |
LAMA3 |
laminin, alpha 3 |
27344 |
0.2 |
| chr16_85591287_85591449 | 0.18 |
GSE1 |
Gse1 coiled-coil protein |
53647 |
0.13 |
| chr8_72648716_72648867 | 0.18 |
ENSG00000200191 |
. |
79069 |
0.09 |
| chr22_39493202_39493353 | 0.18 |
APOBEC3H |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
3 |
0.97 |
| chr10_72147214_72147395 | 0.17 |
LRRC20 |
leucine rich repeat containing 20 |
4922 |
0.2 |
| chr12_120972611_120972974 | 0.17 |
RNF10 |
ring finger protein 10 |
177 |
0.79 |
| chr12_123210436_123210665 | 0.17 |
HCAR1 |
hydroxycarboxylic acid receptor 1 |
4579 |
0.15 |
| chr4_140967052_140967203 | 0.17 |
RP11-392B6.1 |
|
82042 |
0.1 |
| chrX_153979641_153979978 | 0.17 |
GAB3 |
GRB2-associated binding protein 3 |
457 |
0.73 |
| chr19_43058209_43058360 | 0.17 |
CEACAM1 |
carcinoembryonic antigen-related cell adhesion molecule 1 (biliary glycoprotein) |
7102 |
0.19 |
| chrX_38663358_38663509 | 0.17 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
297 |
0.64 |
| chr1_150533351_150533580 | 0.17 |
ADAMTSL4-AS1 |
ADAMTSL4 antisense RNA 1 |
504 |
0.58 |
| chr2_85197275_85197757 | 0.17 |
KCMF1 |
potassium channel modulatory factor 1 |
700 |
0.74 |
| chr17_57297960_57298694 | 0.17 |
GDPD1 |
glycerophosphodiester phosphodiesterase domain containing 1 |
404 |
0.75 |
| chr6_79787718_79787933 | 0.17 |
PHIP |
pleckstrin homology domain interacting protein |
128 |
0.98 |
| chr8_144699859_144700339 | 0.17 |
TSTA3 |
tissue specific transplantation antigen P35B |
119 |
0.9 |
| chr6_157097564_157098248 | 0.17 |
ARID1B |
AT rich interactive domain 1B (SWI1-like) |
1157 |
0.47 |
| chr9_113799034_113800104 | 0.17 |
LPAR1 |
lysophosphatidic acid receptor 1 |
754 |
0.74 |
| chr20_10600368_10600683 | 0.17 |
JAG1 |
jagged 1 |
42629 |
0.18 |
| chr5_134722208_134722661 | 0.17 |
H2AFY |
H2A histone family, member Y |
12467 |
0.16 |
| chr21_46302756_46302907 | 0.17 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
9079 |
0.12 |
| chr4_71704020_71704773 | 0.16 |
GRSF1 |
G-rich RNA sequence binding factor 1 |
322 |
0.85 |
| chr1_27159845_27160009 | 0.16 |
ZDHHC18 |
zinc finger, DHHC-type containing 18 |
1416 |
0.28 |
| chr20_62184252_62184657 | 0.16 |
C20orf195 |
chromosome 20 open reading frame 195 |
81 |
0.94 |
| chr8_125861063_125861214 | 0.16 |
ENSG00000263735 |
. |
26838 |
0.22 |
| chr13_30267815_30267966 | 0.16 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
98065 |
0.08 |
| chr11_3886844_3887200 | 0.16 |
STIM1 |
stromal interaction molecule 1 |
1311 |
0.29 |
| chr2_224701437_224701763 | 0.16 |
AP1S3 |
adaptor-related protein complex 1, sigma 3 subunit |
601 |
0.83 |
| chr12_105076123_105076407 | 0.16 |
ENSG00000264295 |
. |
90854 |
0.08 |
| chr1_150949314_150950272 | 0.16 |
CERS2 |
ceramide synthase 2 |
2314 |
0.14 |
| chr1_202829892_202830043 | 0.16 |
RP11-480I12.5 |
|
1740 |
0.26 |
| chr14_65175690_65176293 | 0.16 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
4676 |
0.25 |
| chr17_80056849_80057346 | 0.16 |
FASN |
fatty acid synthase |
889 |
0.36 |
| chr13_67732158_67732309 | 0.16 |
PCDH9 |
protocadherin 9 |
70339 |
0.13 |
| chr7_55121925_55122165 | 0.15 |
EGFR |
epidermal growth factor receptor |
35234 |
0.22 |
| chr8_38691975_38692126 | 0.15 |
RP11-723D22.3 |
|
11907 |
0.16 |
| chr5_74964815_74965486 | 0.15 |
ENSG00000207333 |
. |
40274 |
0.14 |
| chr19_38806035_38806435 | 0.15 |
YIF1B |
Yip1 interacting factor homolog B (S. cerevisiae) |
173 |
0.87 |
| chr2_109204886_109205231 | 0.15 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
136 |
0.97 |
| chr12_82554330_82554481 | 0.15 |
ENSG00000221260 |
. |
104317 |
0.08 |
| chr7_36247494_36247909 | 0.15 |
AC007327.5 |
|
23015 |
0.2 |
| chr2_158299843_158300108 | 0.15 |
CYTIP |
cytohesin 1 interacting protein |
679 |
0.68 |
| chr3_172240144_172241280 | 0.15 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
553 |
0.83 |
| chr18_74716892_74717079 | 0.15 |
MBP |
myelin basic protein |
5093 |
0.27 |
| chr3_16883621_16883873 | 0.15 |
PLCL2 |
phospholipase C-like 2 |
42705 |
0.19 |
| chr8_23510263_23510414 | 0.14 |
NKX3-1 |
NK3 homeobox 1 |
30064 |
0.14 |
| chr8_48642101_48642252 | 0.14 |
SPIDR |
scaffolding protein involved in DNA repair |
2361 |
0.28 |
| chr16_57831764_57831915 | 0.14 |
KIFC3 |
kinesin family member C3 |
90 |
0.96 |
| chr1_73682669_73682820 | 0.14 |
ENSG00000251825 |
. |
25206 |
0.28 |
| chr22_22173562_22173713 | 0.14 |
ENSG00000200985 |
. |
26914 |
0.13 |
| chr2_75076571_75077006 | 0.14 |
HK2 |
hexokinase 2 |
14491 |
0.24 |
| chr17_41622724_41622875 | 0.14 |
ETV4 |
ets variant 4 |
240 |
0.78 |
| chr12_9912992_9913534 | 0.14 |
CD69 |
CD69 molecule |
234 |
0.92 |
| chr18_43146663_43146829 | 0.14 |
SLC14A2 |
solute carrier family 14 (urea transporter), member 2 |
48020 |
0.13 |
| chr1_95261259_95261410 | 0.14 |
SLC44A3 |
solute carrier family 44, member 3 |
24564 |
0.19 |
| chr16_2566413_2567218 | 0.14 |
ATP6V0C |
ATPase, H+ transporting, lysosomal 16kDa, V0 subunit c |
2185 |
0.1 |
| chr22_23579910_23580061 | 0.14 |
BCR |
breakpoint cluster region |
16162 |
0.16 |
| chr10_104405800_104406167 | 0.14 |
TRIM8 |
tripartite motif containing 8 |
1339 |
0.42 |
| chr11_43965819_43966142 | 0.14 |
C11orf96 |
chromosome 11 open reading frame 96 |
1925 |
0.3 |
| chr6_72077062_72077213 | 0.14 |
ENSG00000199094 |
. |
9597 |
0.21 |
| chr12_27485485_27485755 | 0.13 |
ARNTL2 |
aryl hydrocarbon receptor nuclear translocator-like 2 |
167 |
0.96 |
| chr1_169679248_169679951 | 0.13 |
SELL |
selectin L |
1240 |
0.48 |
| chr1_11902762_11902970 | 0.13 |
NPPA-AS1 |
NPPA antisense RNA 1 |
1052 |
0.37 |
| chr21_40179962_40180113 | 0.13 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
1523 |
0.53 |
| chr1_3540485_3541321 | 0.13 |
RP11-46F15.2 |
|
404 |
0.59 |
| chr5_80215972_80216123 | 0.13 |
CTC-459I6.1 |
|
32752 |
0.19 |
| chr1_230410321_230410472 | 0.13 |
RP5-956O18.2 |
|
6167 |
0.24 |
| chr12_11804572_11805680 | 0.13 |
ETV6 |
ets variant 6 |
2338 |
0.4 |
| chr17_38486750_38487092 | 0.13 |
RARA |
retinoic acid receptor, alpha |
10719 |
0.11 |
| chr11_73093161_73093312 | 0.13 |
RELT |
RELT tumor necrosis factor receptor |
5523 |
0.16 |
| chr10_116127396_116127547 | 0.13 |
AFAP1L2 |
actin filament associated protein 1-like 2 |
36768 |
0.17 |
| chr10_3510829_3511012 | 0.13 |
RP11-184A2.3 |
|
282339 |
0.01 |
| chr14_24808275_24808901 | 0.13 |
RIPK3 |
receptor-interacting serine-threonine kinase 3 |
663 |
0.41 |
| chr17_56405277_56405428 | 0.13 |
BZRAP1 |
benzodiazepine receptor (peripheral) associated protein 1 |
93 |
0.91 |
| chr10_129863856_129864007 | 0.13 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
18097 |
0.25 |
| chr4_130014442_130014634 | 0.13 |
SCLT1 |
sodium channel and clathrin linker 1 |
6 |
0.54 |
| chr22_45730655_45730806 | 0.13 |
FAM118A |
family with sequence similarity 118, member A |
5205 |
0.22 |
| chr4_184063920_184064196 | 0.13 |
ENSG00000252702 |
. |
26512 |
0.16 |
| chr18_34409568_34409805 | 0.13 |
KIAA1328 |
KIAA1328 |
483 |
0.5 |
| chr6_11093389_11093895 | 0.13 |
SMIM13 |
small integral membrane protein 13 |
624 |
0.67 |
| chr15_70391403_70391876 | 0.12 |
TLE3 |
transducin-like enhancer of split 3 (E(sp1) homolog, Drosophila) |
1124 |
0.58 |
| chr2_73297200_73297351 | 0.12 |
SFXN5 |
sideroflexin 5 |
1527 |
0.42 |
| chr10_79637658_79637809 | 0.12 |
AL391421.1 |
Uncharacterized protein; cDNA FLJ43696 fis, clone TBAES2007964 |
11100 |
0.18 |
| chr22_40415035_40415186 | 0.12 |
FAM83F |
family with sequence similarity 83, member F |
9418 |
0.17 |
| chr11_441089_441413 | 0.12 |
ENSG00000243562 |
. |
558 |
0.44 |
| chr3_197677297_197678136 | 0.12 |
RPL35A |
ribosomal protein L35a |
288 |
0.79 |
| chr2_220462950_220463200 | 0.12 |
STK11IP |
serine/threonine kinase 11 interacting protein |
479 |
0.64 |
| chr19_12896715_12896961 | 0.12 |
ENSG00000263800 |
. |
1104 |
0.23 |
| chr11_111749891_111750174 | 0.12 |
C11orf1 |
chromosome 11 open reading frame 1 |
84 |
0.55 |
| chr11_46639581_46640063 | 0.12 |
HARBI1 |
harbinger transposase derived 1 |
363 |
0.56 |
| chr3_57530329_57530480 | 0.12 |
DNAH12 |
dynein, axonemal, heavy chain 12 |
333 |
0.79 |
| chr9_138967584_138968045 | 0.12 |
NACC2 |
NACC family member 2, BEN and BTB (POZ) domain containing |
19317 |
0.19 |
| chr17_55335025_55335422 | 0.12 |
MSI2 |
musashi RNA-binding protein 2 |
844 |
0.72 |
| chr11_3860853_3861244 | 0.12 |
RHOG |
ras homolog family member G |
1024 |
0.35 |
| chr16_3018938_3019181 | 0.12 |
PAQR4 |
progestin and adipoQ receptor family member IV |
187 |
0.85 |
| chr19_58919335_58919902 | 0.12 |
ZNF584 |
zinc finger protein 584 |
374 |
0.67 |
| chr17_17727205_17727405 | 0.12 |
SREBF1 |
sterol regulatory element binding transcription factor 1 |
373 |
0.81 |
| chr22_40574055_40575183 | 0.12 |
TNRC6B |
trinucleotide repeat containing 6B |
673 |
0.79 |
| chr9_114424483_114425187 | 0.12 |
GNG10 |
guanine nucleotide binding protein (G protein), gamma 10 |
1220 |
0.43 |
| chr3_41244711_41244955 | 0.12 |
CTNNB1 |
catenin (cadherin-associated protein), beta 1, 88kDa |
3231 |
0.41 |
| chr6_159103866_159104017 | 0.12 |
SYTL3 |
synaptotagmin-like 3 |
19746 |
0.18 |
| chr2_238876520_238877346 | 0.12 |
UBE2F |
ubiquitin-conjugating enzyme E2F (putative) |
502 |
0.82 |
| chr20_37359167_37359318 | 0.11 |
SLC32A1 |
solute carrier family 32 (GABA vesicular transporter), member 1 |
6137 |
0.18 |
| chr18_9488845_9488996 | 0.11 |
RALBP1 |
ralA binding protein 1 |
12787 |
0.15 |
| chr12_93984725_93985802 | 0.11 |
SOCS2 |
suppressor of cytokine signaling 2 |
16429 |
0.18 |
| chr2_95943344_95943495 | 0.11 |
PROM2 |
prominin 2 |
3174 |
0.28 |
| chr12_39298878_39299365 | 0.11 |
CPNE8 |
copine VIII |
312 |
0.84 |
| chr1_202923356_202923584 | 0.11 |
ADIPOR1 |
adiponectin receptor 1 |
3943 |
0.14 |
| chr3_50648196_50648499 | 0.11 |
CISH |
cytokine inducible SH2-containing protein |
856 |
0.4 |
| chr1_40849833_40850457 | 0.11 |
SMAP2 |
small ArfGAP2 |
9825 |
0.18 |
| chr14_45603973_45604448 | 0.11 |
FKBP3 |
FK506 binding protein 3, 25kDa |
312 |
0.77 |
| chr20_2489860_2490111 | 0.11 |
ZNF343 |
zinc finger protein 343 |
207 |
0.92 |
| chr12_49246930_49247081 | 0.11 |
DDX23 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 |
1059 |
0.33 |
| chr17_42173254_42173470 | 0.11 |
HDAC5 |
histone deacetylase 5 |
2807 |
0.14 |
| chr4_2802491_2802761 | 0.11 |
SH3BP2 |
SH3-domain binding protein 2 |
1901 |
0.35 |
| chr12_56122984_56124090 | 0.11 |
CD63 |
CD63 molecule |
46 |
0.89 |
| chr1_1072756_1072964 | 0.11 |
C1orf159 |
chromosome 1 open reading frame 159 |
21119 |
0.08 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.1 | 0.2 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.1 | 0.3 | GO:0033015 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.1 | 0.2 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.2 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.1 | GO:0034371 | chylomicron remodeling(GO:0034371) |
| 0.0 | 0.2 | GO:0010626 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0032049 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.2 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.4 | GO:0043666 | regulation of phosphoprotein phosphatase activity(GO:0043666) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.1 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.3 | GO:0046854 | phosphatidylinositol phosphorylation(GO:0046854) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.2 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.3 | GO:0035336 | long-chain fatty-acyl-CoA metabolic process(GO:0035336) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0071425 | hematopoietic stem cell proliferation(GO:0071425) |
| 0.0 | 0.1 | GO:0045116 | protein neddylation(GO:0045116) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.0 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.1 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.0 | 0.0 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.0 | 0.1 | GO:0000712 | resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.0 | GO:0045943 | positive regulation of transcription from RNA polymerase I promoter(GO:0045943) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0042769 | DNA damage response, detection of DNA damage(GO:0042769) |
| 0.0 | 0.0 | GO:0032971 | regulation of muscle filament sliding(GO:0032971) regulation of actin filament-based movement(GO:1903115) |
| 0.0 | 0.0 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.0 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.0 | GO:0014045 | establishment of endothelial blood-brain barrier(GO:0014045) central nervous system vasculogenesis(GO:0022009) |
| 0.0 | 0.2 | GO:0018208 | peptidyl-proline modification(GO:0018208) |
| 0.0 | 0.0 | GO:0045292 | mRNA cis splicing, via spliceosome(GO:0045292) |
| 0.0 | 0.1 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.1 | GO:0075713 | establishment of viral latency(GO:0019043) establishment of integrated proviral latency(GO:0075713) |
| 0.0 | 0.0 | GO:0014067 | negative regulation of phosphatidylinositol 3-kinase signaling(GO:0014067) |
| 0.0 | 0.0 | GO:0032431 | activation of phospholipase A2 activity(GO:0032431) |
| 0.0 | 0.0 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.0 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.0 | GO:0014009 | glial cell proliferation(GO:0014009) |
| 0.0 | 0.1 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0070822 | Sin3 complex(GO:0016580) Sin3-type complex(GO:0070822) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.0 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.2 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.4 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.2 | GO:0004661 | protein geranylgeranyltransferase activity(GO:0004661) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.2 | GO:0001228 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0000828 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.0 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.3 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0005527 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.1 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.1 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.0 | GO:1901474 | azole transporter activity(GO:0045118) azole transmembrane transporter activity(GO:1901474) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.0 | 0.0 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.3 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.2 | REACTOME GROWTH HORMONE RECEPTOR SIGNALING | Genes involved in Growth hormone receptor signaling |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS | Genes involved in Synthesis of bile acids and bile salts |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |