Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
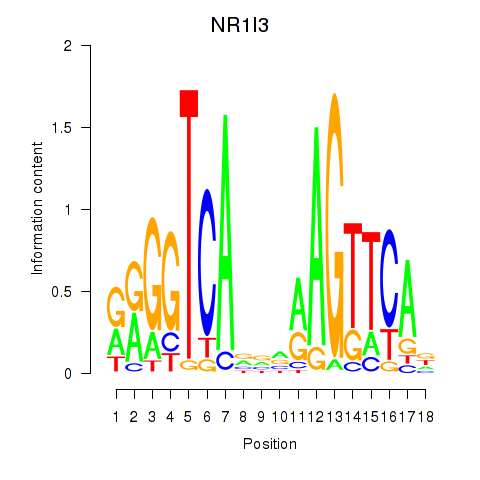
Results for NR1I3
Z-value: 0.66
Transcription factors associated with NR1I3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR1I3
|
ENSG00000143257.7 | nuclear receptor subfamily 1 group I member 3 |
Activity of the NR1I3 motif across conditions
Conditions sorted by the z-value of the NR1I3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
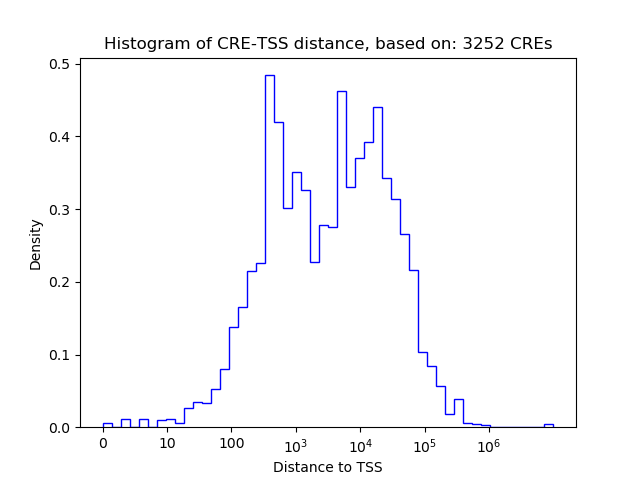
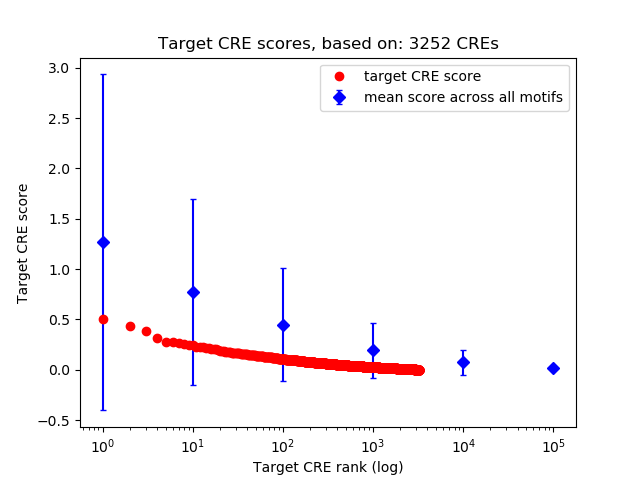
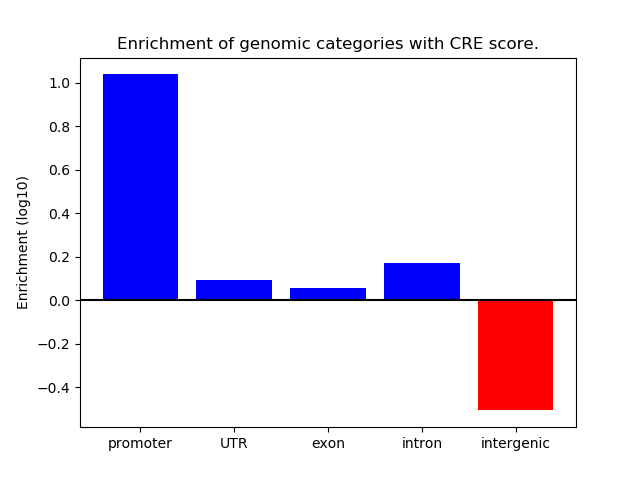
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_36398415_36398818 | 0.51 |
TYROBP |
TYRO protein tyrosine kinase binding protein |
533 |
0.58 |
| chr19_6773186_6773436 | 0.44 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
342 |
0.82 |
| chr10_134369040_134369355 | 0.39 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
17554 |
0.21 |
| chr7_106506867_106507018 | 0.31 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
1018 |
0.65 |
| chr17_1548578_1548729 | 0.28 |
SCARF1 |
scavenger receptor class F, member 1 |
385 |
0.74 |
| chr16_29674660_29674967 | 0.28 |
QPRT |
quinolinate phosphoribosyltransferase |
213 |
0.51 |
| chr1_161186326_161186719 | 0.27 |
FCER1G |
Fc fragment of IgE, high affinity I, receptor for; gamma polypeptide |
1435 |
0.17 |
| chr8_142099867_142100113 | 0.26 |
DENND3 |
DENN/MADD domain containing 3 |
27387 |
0.18 |
| chr1_42230520_42230906 | 0.25 |
ENSG00000264896 |
. |
5901 |
0.3 |
| chr7_43804203_43804455 | 0.24 |
BLVRA |
biliverdin reductase A |
539 |
0.79 |
| chr7_116655483_116655634 | 0.23 |
ST7 |
suppression of tumorigenicity 7 |
568 |
0.75 |
| chr16_53552847_53552998 | 0.23 |
AKTIP |
AKT interacting protein |
14599 |
0.21 |
| chr1_169662937_169663139 | 0.23 |
SELL |
selectin L |
17801 |
0.18 |
| chr9_140541651_140541974 | 0.21 |
EHMT1 |
euchromatic histone-lysine N-methyltransferase 1 |
28358 |
0.12 |
| chr7_17246351_17246502 | 0.21 |
AC003075.4 |
|
74501 |
0.11 |
| chr20_4792810_4792961 | 0.21 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
2884 |
0.28 |
| chr1_109705357_109705508 | 0.21 |
ENSG00000238310 |
. |
45038 |
0.08 |
| chr6_133089264_133089460 | 0.20 |
VNN2 |
vanin 2 |
4764 |
0.14 |
| chr9_136727962_136728162 | 0.19 |
SARDH |
sarcosine dehydrogenase |
122985 |
0.05 |
| chr8_116889935_116890086 | 0.19 |
TRPS1 |
trichorhinophalangeal syndrome I |
68111 |
0.14 |
| chr16_50747073_50747224 | 0.19 |
RP11-327F22.6 |
|
393 |
0.68 |
| chr13_96205566_96205866 | 0.19 |
CLDN10 |
claudin 10 |
737 |
0.72 |
| chr2_27109742_27109997 | 0.18 |
DPYSL5 |
dihydropyrimidinase-like 5 |
38493 |
0.13 |
| chr8_90868157_90868315 | 0.18 |
ENSG00000207359 |
. |
44816 |
0.15 |
| chr17_7920233_7920425 | 0.18 |
GUCY2D |
guanylate cyclase 2D, membrane (retina-specific) |
14417 |
0.1 |
| chr8_24151584_24151953 | 0.17 |
ADAM28 |
ADAM metallopeptidase domain 28 |
144 |
0.97 |
| chr11_47380887_47381038 | 0.17 |
MYBPC3 |
myosin binding protein C, cardiac |
6709 |
0.11 |
| chr1_44985301_44985452 | 0.17 |
ENSG00000263381 |
. |
25789 |
0.18 |
| chr12_9889522_9889673 | 0.16 |
CLECL1 |
C-type lectin-like 1 |
3702 |
0.19 |
| chr20_47402410_47402561 | 0.16 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
41935 |
0.17 |
| chr7_101460813_101461304 | 0.16 |
CUX1 |
cut-like homeobox 1 |
138 |
0.97 |
| chr4_6911822_6912351 | 0.16 |
TBC1D14 |
TBC1 domain family, member 14 |
111 |
0.97 |
| chr20_5634032_5634183 | 0.16 |
GPCPD1 |
glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) |
42435 |
0.18 |
| chr21_27942979_27943130 | 0.16 |
CYYR1 |
cysteine/tyrosine-rich 1 |
2441 |
0.43 |
| chr15_70017287_70017438 | 0.16 |
ENSG00000238870 |
. |
5799 |
0.29 |
| chr2_208999817_208999981 | 0.16 |
CRYGC |
crystallin, gamma C |
5345 |
0.12 |
| chr17_28053735_28053927 | 0.16 |
RP11-82O19.1 |
|
34290 |
0.11 |
| chrX_71299756_71300007 | 0.15 |
RGAG4 |
retrotransposon gag domain containing 4 |
51797 |
0.11 |
| chr22_31625332_31625483 | 0.15 |
ENSG00000202019 |
. |
751 |
0.5 |
| chr11_58912766_58913207 | 0.15 |
FAM111A |
family with sequence similarity 111, member A |
450 |
0.8 |
| chr6_37017654_37018643 | 0.15 |
COX6A1P2 |
cytochrome c oxidase subunit VIa polypeptide 1 pseudogene 2 |
5541 |
0.22 |
| chr12_112215247_112215398 | 0.15 |
ALDH2 |
aldehyde dehydrogenase 2 family (mitochondrial) |
10596 |
0.18 |
| chr16_87993472_87993623 | 0.15 |
BANP |
BTG3 associated nuclear protein |
70 |
0.98 |
| chr10_11214521_11214797 | 0.15 |
RP3-323N1.2 |
|
1320 |
0.48 |
| chr11_128585758_128586117 | 0.15 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
20019 |
0.17 |
| chr13_110307530_110307681 | 0.15 |
LINC00676 |
long intergenic non-protein coding RNA 676 |
73024 |
0.13 |
| chr19_16698681_16699367 | 0.15 |
CTD-3222D19.5 |
|
228 |
0.74 |
| chr12_104963796_104963969 | 0.14 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
18744 |
0.22 |
| chr18_9488845_9488996 | 0.14 |
RALBP1 |
ralA binding protein 1 |
12787 |
0.15 |
| chrX_48542477_48542733 | 0.14 |
WAS |
Wiskott-Aldrich syndrome |
437 |
0.75 |
| chr5_14173969_14174187 | 0.14 |
TRIO |
trio Rho guanine nucleotide exchange factor |
9829 |
0.32 |
| chr15_91383344_91383495 | 0.14 |
CTD-3094K11.1 |
|
465 |
0.73 |
| chr5_147184374_147184525 | 0.14 |
JAKMIP2 |
janus kinase and microtubule interacting protein 2 |
22111 |
0.17 |
| chr5_10445338_10445751 | 0.14 |
ROPN1L |
rhophilin associated tail protein 1-like |
3556 |
0.18 |
| chr10_73506779_73506930 | 0.14 |
C10orf105 |
chromosome 10 open reading frame 105 |
9273 |
0.2 |
| chr14_103585199_103585350 | 0.14 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
4524 |
0.19 |
| chr3_29185770_29185921 | 0.13 |
ENSG00000238470 |
. |
133112 |
0.05 |
| chr8_134532733_134532928 | 0.13 |
ST3GAL1 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 1 |
21204 |
0.27 |
| chr17_80057367_80057586 | 0.13 |
FASN |
fatty acid synthase |
1268 |
0.24 |
| chr15_70827379_70827556 | 0.13 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
167153 |
0.04 |
| chr20_19638965_19639116 | 0.13 |
ENSG00000232675 |
. |
35811 |
0.18 |
| chr1_153363293_153363489 | 0.13 |
S100A8 |
S100 calcium binding protein A8 |
61 |
0.95 |
| chr16_90169651_90169802 | 0.13 |
PRDM7 |
PR domain containing 7 |
27388 |
0.14 |
| chr17_2700217_2700494 | 0.13 |
RAP1GAP2 |
RAP1 GTPase activating protein 2 |
579 |
0.74 |
| chr9_137335451_137335655 | 0.13 |
RXRA |
retinoid X receptor, alpha |
37125 |
0.19 |
| chr17_80833603_80833820 | 0.13 |
TBCD |
tubulin folding cofactor D |
8317 |
0.18 |
| chr6_159464766_159465080 | 0.13 |
TAGAP |
T-cell activation RhoGTPase activating protein |
1127 |
0.51 |
| chrX_53342784_53342935 | 0.13 |
IQSEC2 |
IQ motif and Sec7 domain 2 |
7663 |
0.18 |
| chrX_77582442_77582995 | 0.13 |
CYSLTR1 |
cysteinyl leukotriene receptor 1 |
262 |
0.96 |
| chr19_48833533_48834100 | 0.13 |
EMP3 |
epithelial membrane protein 3 |
4951 |
0.12 |
| chr2_152148385_152148674 | 0.12 |
NMI |
N-myc (and STAT) interactor |
1958 |
0.35 |
| chr13_41588793_41589349 | 0.12 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
4379 |
0.23 |
| chr6_2733092_2733243 | 0.12 |
MYLK4 |
myosin light chain kinase family, member 4 |
17987 |
0.2 |
| chr12_96792273_96792988 | 0.12 |
CDK17 |
cyclin-dependent kinase 17 |
512 |
0.8 |
| chr4_16271608_16271759 | 0.12 |
TAPT1-AS1 |
TAPT1 antisense RNA 1 (head to head) |
43015 |
0.15 |
| chr16_16170769_16170920 | 0.12 |
ABCC1 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 1 |
67131 |
0.11 |
| chr15_75160410_75160789 | 0.12 |
SCAMP2 |
secretory carrier membrane protein 2 |
4782 |
0.12 |
| chr17_48907044_48907195 | 0.12 |
WFIKKN2 |
WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 |
4892 |
0.17 |
| chr10_77189804_77189955 | 0.12 |
RP11-399K21.10 |
|
1467 |
0.43 |
| chr17_28048860_28049057 | 0.12 |
RP11-82O19.1 |
|
39163 |
0.1 |
| chr1_38469725_38469960 | 0.12 |
FHL3 |
four and a half LIM domains 3 |
1335 |
0.32 |
| chr11_118095313_118095652 | 0.12 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
254 |
0.89 |
| chr11_47615011_47615162 | 0.12 |
C1QTNF4 |
C1q and tumor necrosis factor related protein 4 |
1125 |
0.28 |
| chr21_46302756_46302907 | 0.12 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
9079 |
0.12 |
| chr8_95917651_95917917 | 0.11 |
ENSG00000238791 |
. |
563 |
0.66 |
| chrX_153979641_153979978 | 0.11 |
GAB3 |
GRB2-associated binding protein 3 |
457 |
0.73 |
| chr2_44397166_44397731 | 0.11 |
PPM1B |
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
1399 |
0.32 |
| chr4_37603077_37603228 | 0.11 |
RP11-36B15.1 |
|
12655 |
0.21 |
| chr18_19284100_19284279 | 0.11 |
ABHD3 |
abhydrolase domain containing 3 |
403 |
0.75 |
| chr22_39493202_39493353 | 0.11 |
APOBEC3H |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3H |
3 |
0.97 |
| chr20_57582980_57583192 | 0.11 |
CTSZ |
cathepsin Z |
784 |
0.56 |
| chr9_88357119_88357540 | 0.11 |
AGTPBP1 |
ATP/GTP binding protein 1 |
385 |
0.93 |
| chr14_91869059_91869463 | 0.11 |
CCDC88C |
coiled-coil domain containing 88C |
14429 |
0.23 |
| chr1_12149159_12149310 | 0.11 |
ENSG00000201135 |
. |
11193 |
0.15 |
| chr22_38199332_38199483 | 0.11 |
H1F0 |
H1 histone family, member 0 |
1707 |
0.2 |
| chr17_56744023_56744174 | 0.11 |
ENSG00000199426 |
. |
40 |
0.96 |
| chr2_216871280_216871528 | 0.11 |
MREG |
melanoregulin |
6942 |
0.25 |
| chr18_47012500_47013343 | 0.11 |
C18orf32 |
chromosome 18 open reading frame 32 |
62 |
0.8 |
| chr12_51319016_51319581 | 0.11 |
METTL7A |
methyltransferase like 7A |
764 |
0.59 |
| chr8_121761934_121762232 | 0.11 |
RP11-713M15.1 |
|
11410 |
0.25 |
| chr16_55471445_55471596 | 0.11 |
MMP2 |
matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) |
41222 |
0.16 |
| chr3_16978815_16978966 | 0.11 |
ENSG00000264818 |
. |
4202 |
0.26 |
| chr1_12227882_12228546 | 0.10 |
TNFRSF1B |
tumor necrosis factor receptor superfamily, member 1B |
1154 |
0.44 |
| chr21_46078204_46078355 | 0.10 |
KRTAP12-3 |
keratin associated protein 12-3 |
430 |
0.64 |
| chr5_58055562_58056142 | 0.10 |
RP11-479O16.1 |
|
24754 |
0.24 |
| chr4_15704055_15704284 | 0.10 |
BST1 |
bone marrow stromal cell antigen 1 |
404 |
0.85 |
| chr2_64834665_64834935 | 0.10 |
ENSG00000252414 |
. |
28774 |
0.17 |
| chr3_185903259_185903649 | 0.10 |
ETV5 |
ets variant 5 |
75347 |
0.1 |
| chr4_120983549_120983700 | 0.10 |
RP11-679C8.2 |
|
4489 |
0.25 |
| chr11_61124561_61124712 | 0.10 |
CYB561A3 |
cytochrome b561 family, member A3 |
115 |
0.94 |
| chr3_69140694_69141185 | 0.10 |
ARL6IP5 |
ADP-ribosylation-like factor 6 interacting protein 5 |
6767 |
0.15 |
| chr13_74276583_74277049 | 0.10 |
KLF12 |
Kruppel-like factor 12 |
292370 |
0.01 |
| chr20_47445241_47445454 | 0.10 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
927 |
0.69 |
| chr9_35775939_35776090 | 0.10 |
NPR2 |
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
16137 |
0.07 |
| chr6_15268964_15269231 | 0.10 |
JARID2 |
jumonji, AT rich interactive domain 2 |
19954 |
0.17 |
| chr8_142129282_142129557 | 0.10 |
DENND3 |
DENN/MADD domain containing 3 |
2042 |
0.34 |
| chr17_62707569_62707720 | 0.10 |
RP13-104F24.1 |
|
47077 |
0.12 |
| chr3_58339519_58339685 | 0.10 |
PXK |
PX domain containing serine/threonine kinase |
20569 |
0.17 |
| chr1_198608071_198608226 | 0.10 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
1 |
0.98 |
| chr6_135644010_135644161 | 0.10 |
RP3-388E23.2 |
|
21379 |
0.2 |
| chr6_43612507_43612686 | 0.10 |
RSPH9 |
radial spoke head 9 homolog (Chlamydomonas) |
187 |
0.91 |
| chr9_141108330_141108481 | 0.10 |
TUBBP5 |
tubulin, beta pseudogene 5 |
38908 |
0.18 |
| chr6_35696426_35696695 | 0.10 |
FKBP5 |
FK506 binding protein 5 |
200 |
0.9 |
| chr17_54759510_54759661 | 0.10 |
NOG |
noggin |
88525 |
0.08 |
| chr10_103608684_103608835 | 0.10 |
KCNIP2 |
Kv channel interacting protein 2 |
5082 |
0.18 |
| chr2_137140100_137140251 | 0.10 |
ENSG00000251976 |
. |
7727 |
0.32 |
| chr10_49731303_49731459 | 0.10 |
ARHGAP22 |
Rho GTPase activating protein 22 |
900 |
0.64 |
| chr12_94601752_94602458 | 0.10 |
RP11-74K11.2 |
|
22285 |
0.18 |
| chr5_139971225_139971376 | 0.10 |
ENSG00000200235 |
. |
12048 |
0.08 |
| chr20_5126928_5127079 | 0.10 |
CDS2 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
19392 |
0.12 |
| chr6_120039243_120039394 | 0.10 |
ENSG00000265725 |
. |
297007 |
0.01 |
| chr2_71299748_71299899 | 0.09 |
NAGK |
N-acetylglucosamine kinase |
844 |
0.45 |
| chr3_49841438_49841709 | 0.09 |
FAM212A |
family with sequence similarity 212, member A |
886 |
0.35 |
| chr10_51566009_51566382 | 0.09 |
NCOA4 |
nuclear receptor coactivator 4 |
965 |
0.54 |
| chr14_77500493_77500704 | 0.09 |
IRF2BPL |
interferon regulatory factor 2 binding protein-like |
5564 |
0.22 |
| chr20_58633559_58633836 | 0.09 |
C20orf197 |
chromosome 20 open reading frame 197 |
2717 |
0.35 |
| chr10_14596775_14596926 | 0.09 |
FAM107B |
family with sequence similarity 107, member B |
622 |
0.8 |
| chr1_228457559_228457839 | 0.09 |
RP5-1139B12.2 |
|
4399 |
0.14 |
| chr20_44543417_44543568 | 0.09 |
PLTP |
phospholipid transfer protein |
2698 |
0.13 |
| chr2_73297200_73297351 | 0.09 |
SFXN5 |
sideroflexin 5 |
1527 |
0.42 |
| chr5_153652567_153652718 | 0.09 |
ENSG00000221070 |
. |
67044 |
0.1 |
| chr17_38128484_38128998 | 0.09 |
GSDMA |
gasdermin A |
6965 |
0.12 |
| chr9_71160716_71160867 | 0.09 |
TMEM252 |
transmembrane protein 252 |
5008 |
0.3 |
| chr15_80274344_80274495 | 0.09 |
BCL2A1 |
BCL2-related protein A1 |
10631 |
0.19 |
| chr19_45250325_45250702 | 0.09 |
BCL3 |
B-cell CLL/lymphoma 3 |
1291 |
0.3 |
| chr3_4516286_4516499 | 0.09 |
SUMF1 |
sulfatase modifying factor 1 |
7427 |
0.21 |
| chr1_203186842_203186993 | 0.09 |
CHIT1 |
chitinase 1 (chitotriosidase) |
11882 |
0.15 |
| chr4_113068308_113068459 | 0.09 |
C4orf32 |
chromosome 4 open reading frame 32 |
1830 |
0.44 |
| chr1_198901485_198902338 | 0.09 |
ENSG00000207759 |
. |
73629 |
0.11 |
| chr20_826611_826818 | 0.09 |
FAM110A |
family with sequence similarity 110, member A |
947 |
0.64 |
| chrX_49047229_49047448 | 0.09 |
PRICKLE3 |
prickle homolog 3 (Drosophila) |
4493 |
0.09 |
| chr19_7413904_7414409 | 0.09 |
CTB-133G6.1 |
|
308 |
0.88 |
| chr9_131550054_131550269 | 0.09 |
TBC1D13 |
TBC1 domain family, member 13 |
533 |
0.64 |
| chr12_48220962_48221113 | 0.09 |
HDAC7 |
histone deacetylase 7 |
5878 |
0.17 |
| chr11_46331268_46331419 | 0.09 |
CREB3L1 |
cAMP responsive element binding protein 3-like 1 |
1365 |
0.39 |
| chr6_20174159_20174424 | 0.09 |
RP11-239H6.2 |
|
38027 |
0.18 |
| chr16_53133173_53134292 | 0.09 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
660 |
0.77 |
| chr12_29311473_29311703 | 0.09 |
FAR2 |
fatty acyl CoA reductase 2 |
9455 |
0.26 |
| chr12_118796178_118796667 | 0.09 |
TAOK3 |
TAO kinase 3 |
488 |
0.85 |
| chr3_195823637_195823788 | 0.09 |
TFRC |
transferrin receptor |
14652 |
0.17 |
| chr15_92404681_92404832 | 0.09 |
SLCO3A1 |
solute carrier organic anion transporter family, member 3A1 |
7405 |
0.27 |
| chr1_183549128_183549279 | 0.09 |
NCF2 |
neutrophil cytosolic factor 2 |
10513 |
0.2 |
| chr17_56405277_56405428 | 0.09 |
BZRAP1 |
benzodiazepine receptor (peripheral) associated protein 1 |
93 |
0.91 |
| chr14_95650210_95650361 | 0.08 |
CTD-2240H23.2 |
|
1906 |
0.28 |
| chr1_230281012_230281163 | 0.08 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
78069 |
0.1 |
| chr2_145139373_145139524 | 0.08 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
48145 |
0.16 |
| chr6_96969080_96969553 | 0.08 |
UFL1-AS1 |
UFL1 antisense RNA 1 |
229 |
0.65 |
| chr20_61202559_61202764 | 0.08 |
ENSG00000207764 |
. |
40542 |
0.09 |
| chr6_109621799_109621950 | 0.08 |
ENSG00000201023 |
. |
4823 |
0.21 |
| chr2_178217813_178218079 | 0.08 |
ENSG00000238295 |
. |
7257 |
0.15 |
| chr12_57107519_57107670 | 0.08 |
NACA |
nascent polypeptide-associated complex alpha subunit |
5790 |
0.14 |
| chr16_22200588_22200739 | 0.08 |
EEF2K |
eukaryotic elongation factor-2 kinase |
16940 |
0.16 |
| chr10_51574545_51574696 | 0.08 |
NCOA4 |
nuclear receptor coactivator 4 |
1320 |
0.42 |
| chr1_150536239_150536708 | 0.08 |
ADAMTSL4-AS1 |
ADAMTSL4 antisense RNA 1 |
2504 |
0.12 |
| chr11_44592282_44592433 | 0.08 |
CD82 |
CD82 molecule |
2271 |
0.34 |
| chr12_32636420_32636776 | 0.08 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
2308 |
0.33 |
| chr12_116983809_116983960 | 0.08 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
13302 |
0.27 |
| chr6_31508773_31509384 | 0.08 |
ENSG00000265236 |
. |
123 |
0.54 |
| chr2_2689870_2690021 | 0.08 |
ENSG00000263570 |
. |
331157 |
0.01 |
| chr20_39407503_39407654 | 0.08 |
ENSG00000238908 |
. |
75291 |
0.11 |
| chr1_186587084_186587465 | 0.08 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
62285 |
0.14 |
| chr7_2648316_2648467 | 0.08 |
IQCE |
IQ motif containing E |
11858 |
0.16 |
| chr11_57194286_57194587 | 0.08 |
SLC43A3 |
solute carrier family 43, member 3 |
18 |
0.95 |
| chr14_73931285_73931436 | 0.08 |
ENSG00000251393 |
. |
2231 |
0.24 |
| chr5_81148309_81148643 | 0.08 |
SSBP2 |
single-stranded DNA binding protein 2 |
101404 |
0.07 |
| chr19_42302514_42302665 | 0.08 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
1504 |
0.28 |
| chr1_14951057_14951208 | 0.08 |
KAZN |
kazrin, periplakin interacting protein |
25919 |
0.28 |
| chrX_45626469_45627939 | 0.08 |
ENSG00000207725 |
. |
20674 |
0.22 |
| chr6_26155918_26156313 | 0.08 |
HIST1H1E |
histone cluster 1, H1e |
444 |
0.58 |
| chr21_40182599_40182907 | 0.08 |
ETS2 |
v-ets avian erythroblastosis virus E26 oncogene homolog 2 |
1193 |
0.62 |
| chr9_134001509_134001851 | 0.08 |
NUP214 |
nucleoporin 214kDa |
213 |
0.82 |
| chr1_161008159_161008677 | 0.08 |
TSTD1 |
thiosulfate sulfurtransferase (rhodanese)-like domain containing 1 |
101 |
0.93 |
| chr1_198905629_198906537 | 0.08 |
ENSG00000207759 |
. |
77801 |
0.11 |
| chr12_120675375_120675841 | 0.08 |
PXN |
paxillin |
10961 |
0.12 |
| chr17_9990899_9991143 | 0.08 |
ENSG00000221486 |
. |
16814 |
0.19 |
| chr2_70144352_70144560 | 0.08 |
MXD1 |
MAX dimerization protein 1 |
2136 |
0.21 |
| chr1_182138764_182138915 | 0.08 |
ZNF648 |
zinc finger protein 648 |
107992 |
0.07 |
| chr14_65175690_65176293 | 0.08 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
4676 |
0.25 |
| chr14_102976422_102976710 | 0.08 |
ANKRD9 |
ankyrin repeat domain 9 |
430 |
0.81 |
| chr9_93569356_93569567 | 0.08 |
SYK |
spleen tyrosine kinase |
5252 |
0.35 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048680 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.3 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.2 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.1 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.2 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.1 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) |
| 0.0 | 0.0 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.1 | GO:0050901 | leukocyte tethering or rolling(GO:0050901) leukocyte adhesion to vascular endothelial cell(GO:0061756) |
| 0.0 | 0.1 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.1 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.0 | 0.2 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.0 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.0 | GO:0045332 | lipid translocation(GO:0034204) phospholipid translocation(GO:0045332) |
| 0.0 | 0.0 | GO:0032780 | negative regulation of ATPase activity(GO:0032780) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.0 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.2 | GO:0016628 | oxidoreductase activity, acting on the CH-CH group of donors, NAD or NADP as acceptor(GO:0016628) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.0 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.2 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.0 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.0 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME PD1 SIGNALING | Genes involved in PD-1 signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.4 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |