Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
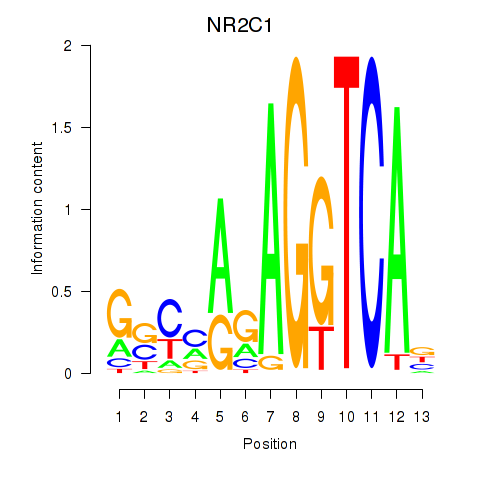
Results for NR2C1
Z-value: 0.73
Transcription factors associated with NR2C1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2C1
|
ENSG00000120798.12 | nuclear receptor subfamily 2 group C member 1 |
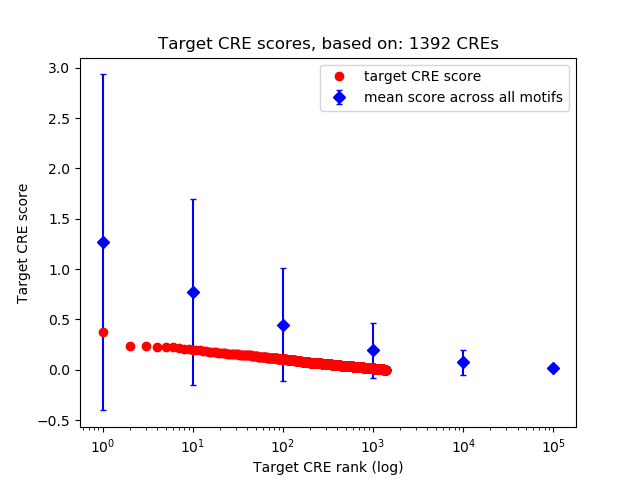
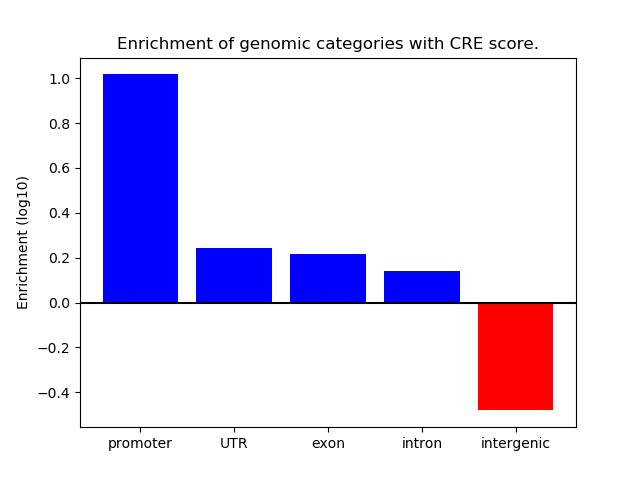
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_95466827_95467360 | NR2C1 | 220 | 0.944692 | 0.16 | 6.9e-01 | Click! |
| chr12_95466323_95466760 | NR2C1 | 252 | 0.933981 | 0.15 | 7.0e-01 | Click! |
Activity of the NR2C1 motif across conditions
Conditions sorted by the z-value of the NR2C1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
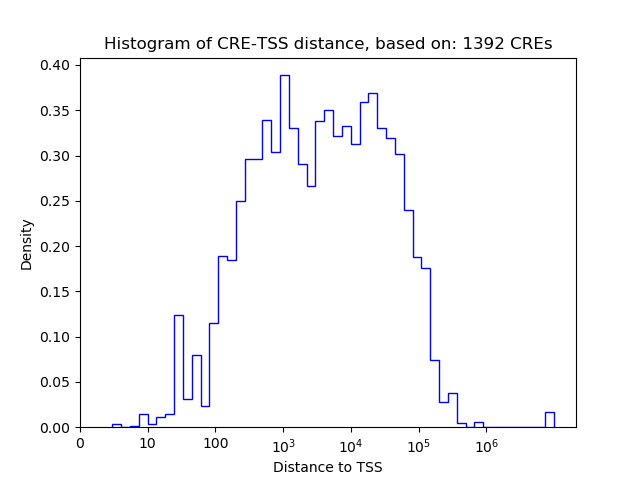
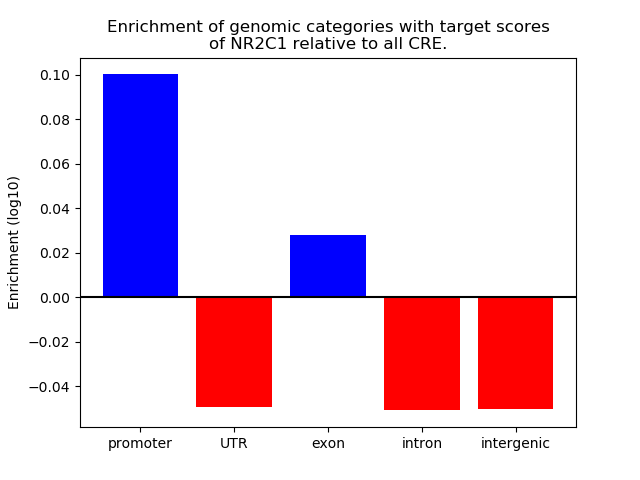
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_7254194_7254453 | 0.38 |
ACAP1 |
ArfGAP with coiled-coil, ankyrin repeat and PH domains 1 |
139 |
0.83 |
| chr4_138452050_138452321 | 0.24 |
PCDH18 |
protocadherin 18 |
1380 |
0.62 |
| chr12_52701821_52702383 | 0.24 |
RP11-845M18.6 |
|
53 |
0.94 |
| chr20_48601776_48601927 | 0.23 |
SNAI1 |
snail family zinc finger 1 |
2315 |
0.25 |
| chr1_5998344_5998611 | 0.22 |
ENSG00000266687 |
. |
44493 |
0.12 |
| chr17_64300095_64300347 | 0.22 |
PRKCA |
protein kinase C, alpha |
1277 |
0.48 |
| chr16_70669285_70669554 | 0.21 |
IL34 |
interleukin 34 |
11049 |
0.15 |
| chr3_194930898_194931049 | 0.21 |
ENSG00000206600 |
. |
4543 |
0.2 |
| chr5_98397075_98397538 | 0.21 |
ENSG00000200351 |
. |
124855 |
0.06 |
| chr1_233750665_233750953 | 0.20 |
KCNK1 |
potassium channel, subfamily K, member 1 |
1059 |
0.6 |
| chr15_86163825_86163989 | 0.19 |
RP11-815J21.3 |
|
7013 |
0.15 |
| chr11_63647208_63647359 | 0.19 |
ENSG00000202089 |
. |
2874 |
0.19 |
| chr3_170138450_170138601 | 0.19 |
CLDN11 |
claudin 11 |
1670 |
0.46 |
| chr15_74232964_74233156 | 0.19 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
12471 |
0.14 |
| chr3_112358713_112358917 | 0.18 |
CCDC80 |
coiled-coil domain containing 80 |
1301 |
0.54 |
| chr2_74606600_74607395 | 0.18 |
DCTN1 |
dynactin 1 |
413 |
0.73 |
| chr9_132428871_132429094 | 0.18 |
PRRX2 |
paired related homeobox 2 |
1062 |
0.42 |
| chr9_35688355_35688710 | 0.17 |
TPM2 |
tropomyosin 2 (beta) |
1368 |
0.2 |
| chr10_10866573_10866724 | 0.17 |
CELF2 |
CUGBP, Elav-like family member 2 |
180611 |
0.03 |
| chr3_14514213_14514497 | 0.17 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
605 |
0.8 |
| chr17_48287259_48287746 | 0.17 |
COL1A1 |
collagen, type I, alpha 1 |
8509 |
0.12 |
| chr22_31091709_31091860 | 0.17 |
OSBP2 |
oxysterol binding protein 2 |
908 |
0.52 |
| chr2_217555203_217555354 | 0.16 |
AC007563.5 |
|
3909 |
0.2 |
| chr15_76005245_76005482 | 0.16 |
CSPG4 |
chondroitin sulfate proteoglycan 4 |
174 |
0.9 |
| chr22_45899514_45899973 | 0.16 |
FBLN1 |
fibulin 1 |
817 |
0.67 |
| chr5_141931313_141931519 | 0.16 |
ENSG00000252831 |
. |
17506 |
0.21 |
| chr14_105156045_105157032 | 0.16 |
INF2 |
inverted formin, FH2 and WH2 domain containing |
564 |
0.57 |
| chr22_33199313_33199464 | 0.15 |
TIMP3 |
TIMP metallopeptidase inhibitor 3 |
1701 |
0.44 |
| chr12_125002282_125002834 | 0.15 |
NCOR2 |
nuclear receptor corepressor 2 |
282 |
0.95 |
| chr6_158903222_158903414 | 0.15 |
ENSG00000243373 |
. |
44489 |
0.14 |
| chr6_683619_684080 | 0.15 |
EXOC2 |
exocyst complex component 2 |
9262 |
0.26 |
| chr8_145027702_145027989 | 0.15 |
PLEC |
plectin |
243 |
0.86 |
| chr17_42621446_42621636 | 0.15 |
FZD2 |
frizzled family receptor 2 |
13384 |
0.17 |
| chr10_13908254_13908427 | 0.15 |
FRMD4A |
FERM domain containing 4A |
7428 |
0.26 |
| chr5_148787363_148787711 | 0.15 |
ENSG00000208035 |
. |
20944 |
0.11 |
| chr10_71234623_71234840 | 0.15 |
TSPAN15 |
tetraspanin 15 |
8893 |
0.22 |
| chr5_54932725_54932876 | 0.15 |
SLC38A9 |
solute carrier family 38, member 9 |
55648 |
0.13 |
| chr6_148829582_148830289 | 0.15 |
ENSG00000223322 |
. |
15441 |
0.29 |
| chr19_49371899_49372050 | 0.15 |
PLEKHA4 |
pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 |
85 |
0.93 |
| chr7_77827156_77827307 | 0.14 |
RP5-1185I7.1 |
|
207494 |
0.03 |
| chr8_145024226_145024570 | 0.14 |
PLEC |
plectin |
646 |
0.55 |
| chr6_33383816_33383967 | 0.14 |
PHF1 |
PHD finger protein 1 |
1049 |
0.32 |
| chr6_133561688_133561839 | 0.14 |
EYA4 |
eyes absent homolog 4 (Drosophila) |
27 |
0.99 |
| chr2_132431108_132431555 | 0.14 |
C2orf27A |
chromosome 2 open reading frame 27A |
48617 |
0.15 |
| chr2_219646380_219646634 | 0.14 |
CYP27A1 |
cytochrome P450, family 27, subfamily A, polypeptide 1 |
28 |
0.97 |
| chr6_56621224_56621427 | 0.14 |
DST |
dystonin |
29352 |
0.23 |
| chr8_82771414_82771608 | 0.14 |
SNX16 |
sorting nexin 16 |
16410 |
0.24 |
| chr19_50031295_50031537 | 0.14 |
RCN3 |
reticulocalbin 3, EF-hand calcium binding domain |
131 |
0.88 |
| chr3_23754408_23754630 | 0.14 |
ENSG00000238672 |
. |
54583 |
0.12 |
| chr3_119421863_119422475 | 0.14 |
MAATS1 |
MYCBP-associated, testis expressed 1 |
289 |
0.89 |
| chr18_52968140_52968291 | 0.13 |
TCF4 |
transcription factor 4 |
1642 |
0.52 |
| chr5_115151532_115151821 | 0.13 |
CDO1 |
cysteine dioxygenase type 1 |
975 |
0.53 |
| chr5_134582010_134582210 | 0.13 |
C5orf66 |
chromosome 5 open reading frame 66 |
91519 |
0.08 |
| chr20_33585796_33585947 | 0.13 |
MYH7B |
myosin, heavy chain 7B, cardiac muscle, beta |
3177 |
0.19 |
| chr20_62600161_62600928 | 0.13 |
ZNF512B |
zinc finger protein 512B |
674 |
0.45 |
| chr15_66710166_66710343 | 0.13 |
MAP2K1 |
mitogen-activated protein kinase kinase 1 |
31099 |
0.09 |
| chr19_50031610_50031873 | 0.13 |
RCN3 |
reticulocalbin 3, EF-hand calcium binding domain |
142 |
0.87 |
| chr7_130575854_130576216 | 0.13 |
ENSG00000226380 |
. |
13737 |
0.26 |
| chr17_15164310_15164681 | 0.13 |
RP11-849N15.1 |
|
163 |
0.92 |
| chr2_198537960_198538125 | 0.13 |
RFTN2 |
raftlin family member 2 |
2677 |
0.27 |
| chr1_42744774_42744925 | 0.13 |
FOXJ3 |
forkhead box J3 |
591 |
0.8 |
| chr10_69915983_69916134 | 0.13 |
ENSG00000222371 |
. |
1778 |
0.38 |
| chr19_36821987_36822637 | 0.12 |
ENSG00000222730 |
. |
961 |
0.5 |
| chr3_9691165_9692185 | 0.12 |
MTMR14 |
myotubularin related protein 14 |
479 |
0.8 |
| chr17_13503336_13503489 | 0.12 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
1832 |
0.45 |
| chr22_44893289_44894077 | 0.12 |
LDOC1L |
leucine zipper, down-regulated in cancer 1-like |
495 |
0.88 |
| chr1_116610133_116610284 | 0.12 |
MAB21L3 |
mab-21-like 3 (C. elegans) |
44168 |
0.15 |
| chr13_36729008_36729209 | 0.12 |
DCLK1 |
doublecortin-like kinase 1 |
23665 |
0.21 |
| chr6_116832868_116833176 | 0.12 |
FAM26E |
family with sequence similarity 26, member E |
213 |
0.73 |
| chr6_6737313_6737878 | 0.12 |
LY86-AS1 |
LY86 antisense RNA 1 |
114591 |
0.06 |
| chr5_321961_322616 | 0.12 |
AHRR |
aryl-hydrocarbon receptor repressor |
397 |
0.84 |
| chr16_31464634_31465197 | 0.12 |
ARMC5 |
armadillo repeat containing 5 |
4486 |
0.1 |
| chr19_980574_980725 | 0.12 |
WDR18 |
WD repeat domain 18 |
3682 |
0.1 |
| chr8_105601818_105602057 | 0.12 |
LRP12 |
low density lipoprotein receptor-related protein 12 |
685 |
0.56 |
| chr11_131758303_131758454 | 0.12 |
AP004372.1 |
|
8624 |
0.25 |
| chr20_62601411_62601964 | 0.12 |
ZNF512B |
zinc finger protein 512B |
469 |
0.61 |
| chr7_121950011_121950419 | 0.12 |
FEZF1 |
FEZ family zinc finger 1 |
530 |
0.77 |
| chr10_121413293_121413467 | 0.12 |
BAG3 |
BCL2-associated athanogene 3 |
2498 |
0.33 |
| chr6_149450767_149450918 | 0.12 |
ENSG00000263481 |
. |
47088 |
0.15 |
| chrY_1463557_1463741 | 0.11 |
NA |
NA |
> 106 |
NA |
| chr7_75595517_75595668 | 0.11 |
POR |
P450 (cytochrome) oxidoreductase |
1569 |
0.35 |
| chr3_190251616_190251894 | 0.11 |
IL1RAP |
interleukin 1 receptor accessory protein |
19864 |
0.24 |
| chr1_223900690_223901533 | 0.11 |
CAPN2 |
calpain 2, (m/II) large subunit |
1077 |
0.55 |
| chr7_135442774_135442995 | 0.11 |
FAM180A |
family with sequence similarity 180, member A |
9290 |
0.17 |
| chr5_135390084_135390235 | 0.11 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
4379 |
0.23 |
| chr5_155409653_155409804 | 0.11 |
SGCD |
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
112374 |
0.07 |
| chr10_30238566_30238767 | 0.11 |
KIAA1462 |
KIAA1462 |
109787 |
0.07 |
| chr18_43901758_43901909 | 0.11 |
RNF165 |
ring finger protein 165 |
4939 |
0.31 |
| chr22_25348343_25348574 | 0.11 |
KIAA1671 |
KIAA1671 |
239 |
0.93 |
| chr3_157160417_157160653 | 0.11 |
PTX3 |
pentraxin 3, long |
5957 |
0.27 |
| chr2_174918153_174918304 | 0.11 |
SP3 |
Sp3 transcription factor |
87798 |
0.09 |
| chr3_127175696_127176137 | 0.11 |
TPRA1 |
transmembrane protein, adipocyte asscociated 1 |
123364 |
0.05 |
| chr12_118124375_118124764 | 0.11 |
KSR2 |
kinase suppressor of ras 2 |
12373 |
0.3 |
| chr1_156087932_156088663 | 0.11 |
LMNA |
lamin A/C |
3784 |
0.13 |
| chr13_20135605_20135812 | 0.11 |
TPTE2 |
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
24805 |
0.21 |
| chr7_116963548_116963909 | 0.11 |
WNT2 |
wingless-type MMTV integration site family member 2 |
385 |
0.89 |
| chr6_125698814_125699135 | 0.11 |
RP11-735G4.1 |
|
3504 |
0.35 |
| chr17_6923216_6923383 | 0.11 |
MIR497HG |
mir-497-195 cluster host gene (non-protein coding) |
321 |
0.56 |
| chr1_201472955_201473310 | 0.11 |
CSRP1 |
cysteine and glycine-rich protein 1 |
2835 |
0.21 |
| chrX_1513557_1513776 | 0.11 |
SLC25A6 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 |
2049 |
0.26 |
| chr8_144302737_144302888 | 0.11 |
GPIHBP1 |
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
7744 |
0.14 |
| chr1_236046770_236047092 | 0.11 |
LYST |
lysosomal trafficking regulator |
59 |
0.97 |
| chr5_141667234_141667739 | 0.11 |
SPRY4 |
sprouty homolog 4 (Drosophila) |
36261 |
0.18 |
| chr15_63673071_63673237 | 0.10 |
CA12 |
carbonic anhydrase XII |
880 |
0.7 |
| chr3_124762287_124762438 | 0.10 |
HEG1 |
heart development protein with EGF-like domains 1 |
12440 |
0.18 |
| chr8_27492286_27492503 | 0.10 |
SCARA3 |
scavenger receptor class A, member 3 |
696 |
0.66 |
| chr11_44642855_44643154 | 0.10 |
CD82 |
CD82 molecule |
3184 |
0.25 |
| chr22_46471692_46472277 | 0.10 |
FLJ27365 |
hsa-mir-4763 |
4208 |
0.11 |
| chr18_60904701_60905063 | 0.10 |
ENSG00000238988 |
. |
42984 |
0.14 |
| chr6_115106861_115107012 | 0.10 |
ENSG00000251882 |
. |
81204 |
0.12 |
| chr3_126702505_126703684 | 0.10 |
PLXNA1 |
plexin A1 |
4343 |
0.34 |
| chr16_51184442_51184635 | 0.10 |
SALL1 |
spalt-like transcription factor 1 |
30 |
0.97 |
| chr19_11313913_11314064 | 0.10 |
CTC-510F12.2 |
|
316 |
0.77 |
| chr7_47576453_47576928 | 0.10 |
TNS3 |
tensin 3 |
2185 |
0.45 |
| chr19_36523254_36523431 | 0.10 |
CLIP3 |
CAP-GLY domain containing linker protein 3 |
231 |
0.85 |
| chr7_48134906_48135164 | 0.10 |
UPP1 |
uridine phosphorylase 1 |
6062 |
0.24 |
| chr19_46141740_46141935 | 0.10 |
EML2 |
echinoderm microtubule associated protein like 2 |
294 |
0.6 |
| chr20_35170676_35170845 | 0.10 |
MYL9 |
myosin, light chain 9, regulatory |
860 |
0.53 |
| chr1_23720565_23720716 | 0.10 |
TCEA3 |
transcription elongation factor A (SII), 3 |
6728 |
0.14 |
| chr4_184797459_184798247 | 0.10 |
STOX2 |
storkhead box 2 |
28656 |
0.22 |
| chr4_8632079_8632376 | 0.10 |
CPZ |
carboxypeptidase Z |
28530 |
0.2 |
| chr22_37960000_37960286 | 0.10 |
CDC42EP1 |
CDC42 effector protein (Rho GTPase binding) 1 |
270 |
0.86 |
| chr14_53465156_53465307 | 0.10 |
RP11-368P15.3 |
|
38242 |
0.17 |
| chr10_76951664_76951987 | 0.10 |
ENSG00000263626 |
. |
7109 |
0.19 |
| chr1_17914943_17915229 | 0.10 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
175 |
0.97 |
| chr20_49431977_49432128 | 0.10 |
BCAS4 |
breast carcinoma amplified sequence 4 |
20445 |
0.17 |
| chr9_118916796_118916947 | 0.10 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
788 |
0.73 |
| chr5_77189257_77189408 | 0.10 |
TBCA |
tubulin folding cofactor A |
24728 |
0.26 |
| chr19_13106826_13107198 | 0.10 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
360 |
0.73 |
| chr14_102975190_102975782 | 0.10 |
ANKRD9 |
ankyrin repeat domain 9 |
86 |
0.97 |
| chr17_772969_773237 | 0.09 |
NXN |
nucleoredoxin |
5752 |
0.17 |
| chr13_114543321_114543472 | 0.09 |
GAS6 |
growth arrest-specific 6 |
4379 |
0.26 |
| chr10_128593271_128593431 | 0.09 |
DOCK1 |
dedicator of cytokinesis 1 |
627 |
0.78 |
| chr17_21355804_21356348 | 0.09 |
KCNJ12 |
potassium inwardly-rectifying channel, subfamily J, member 12 |
47628 |
0.15 |
| chr6_91085638_91085845 | 0.09 |
ENSG00000266760 |
. |
63280 |
0.12 |
| chr3_141163249_141163400 | 0.09 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
12759 |
0.22 |
| chr1_95392942_95393358 | 0.09 |
RP4-639F20.1 |
|
28 |
0.82 |
| chr4_8594642_8594896 | 0.09 |
CPZ |
carboxypeptidase Z |
286 |
0.92 |
| chr11_126229473_126229631 | 0.09 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
3683 |
0.19 |
| chr17_10031368_10031819 | 0.09 |
GAS7 |
growth arrest-specific 7 |
13723 |
0.19 |
| chr11_118491742_118491923 | 0.09 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
13474 |
0.11 |
| chr18_53665373_53665862 | 0.09 |
ENSG00000201816 |
. |
81208 |
0.12 |
| chr13_102108148_102108308 | 0.09 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
3210 |
0.35 |
| chr15_74222319_74222668 | 0.09 |
LOXL1-AS1 |
LOXL1 antisense RNA 1 |
1904 |
0.26 |
| chr4_38036867_38037018 | 0.09 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
7533 |
0.29 |
| chr1_157938633_157938826 | 0.09 |
KIRREL |
kin of IRRE like (Drosophila) |
24334 |
0.18 |
| chr15_25936715_25936881 | 0.09 |
ATP10A |
ATPase, class V, type 10A |
3174 |
0.29 |
| chr1_170632786_170633084 | 0.09 |
PRRX1 |
paired related homeobox 1 |
112 |
0.98 |
| chr16_72911078_72911415 | 0.09 |
ENSG00000251868 |
. |
55355 |
0.12 |
| chr19_46110497_46110665 | 0.09 |
OPA3 |
optic atrophy 3 (autosomal recessive, with chorea and spastic paraplegia) |
5111 |
0.1 |
| chr19_11306068_11306280 | 0.09 |
KANK2 |
KN motif and ankyrin repeat domains 2 |
187 |
0.9 |
| chr9_130601009_130601344 | 0.09 |
ENSG00000222455 |
. |
2325 |
0.13 |
| chr22_38809140_38809341 | 0.09 |
RP3-434P1.6 |
|
11053 |
0.12 |
| chr17_41477567_41477895 | 0.09 |
ARL4D |
ADP-ribosylation factor-like 4D |
1404 |
0.28 |
| chr12_116820769_116821023 | 0.08 |
ENSG00000264037 |
. |
45227 |
0.17 |
| chr4_114215036_114215187 | 0.08 |
ANK2 |
ankyrin 2, neuronal |
973 |
0.69 |
| chr7_151424638_151424789 | 0.08 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
8636 |
0.25 |
| chr3_99356620_99357126 | 0.08 |
COL8A1 |
collagen, type VIII, alpha 1 |
446 |
0.89 |
| chrX_68357146_68357332 | 0.08 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
25298 |
0.27 |
| chr3_131220949_131221185 | 0.08 |
MRPL3 |
mitochondrial ribosomal protein L3 |
714 |
0.66 |
| chr9_132330432_132330663 | 0.08 |
RP11-492E3.2 |
|
7141 |
0.16 |
| chr15_68572266_68572681 | 0.08 |
FEM1B |
fem-1 homolog b (C. elegans) |
176 |
0.95 |
| chr16_34542907_34543058 | 0.08 |
ENSG00000221532 |
. |
32119 |
0.22 |
| chr7_45928070_45928578 | 0.08 |
IGFBP1 |
insulin-like growth factor binding protein 1 |
228 |
0.93 |
| chr12_21927918_21928225 | 0.08 |
KCNJ8 |
potassium inwardly-rectifying channel, subfamily J, member 8 |
316 |
0.88 |
| chr2_62638521_62638672 | 0.08 |
ENSG00000241625 |
. |
79717 |
0.09 |
| chr3_64672065_64672629 | 0.08 |
ADAMTS9 |
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
1008 |
0.5 |
| chr16_72972618_72972769 | 0.08 |
ENSG00000221799 |
. |
46063 |
0.15 |
| chr3_14494001_14494152 | 0.08 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
19674 |
0.22 |
| chr22_24114508_24114741 | 0.08 |
MMP11 |
matrix metallopeptidase 11 (stromelysin 3) |
382 |
0.71 |
| chr3_161133992_161134265 | 0.08 |
SPTSSB |
serine palmitoyltransferase, small subunit B |
43460 |
0.19 |
| chr14_23288815_23289060 | 0.08 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
7 |
0.95 |
| chr20_52390008_52390159 | 0.08 |
ENSG00000238468 |
. |
104786 |
0.07 |
| chr3_194930191_194930342 | 0.08 |
ENSG00000206600 |
. |
5250 |
0.19 |
| chr8_128752538_128752942 | 0.08 |
MYC |
v-myc avian myelocytomatosis viral oncogene homolog |
4263 |
0.31 |
| chr5_172753069_172753220 | 0.08 |
STC2 |
stanniocalcin 2 |
656 |
0.71 |
| chr14_29229093_29229354 | 0.08 |
RP11-966I7.1 |
|
5246 |
0.18 |
| chr5_142305337_142305488 | 0.08 |
ARHGAP26 |
Rho GTPase activating protein 26 |
18525 |
0.25 |
| chr8_13424609_13424760 | 0.08 |
C8orf48 |
chromosome 8 open reading frame 48 |
332 |
0.9 |
| chr3_101499681_101500261 | 0.08 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
579 |
0.73 |
| chr6_116382044_116382245 | 0.08 |
FRK |
fyn-related kinase |
223 |
0.94 |
| chr11_113545454_113545605 | 0.08 |
TMPRSS5 |
transmembrane protease, serine 5 |
31495 |
0.18 |
| chr9_21552799_21553028 | 0.08 |
MIR31HG |
MIR31 host gene (non-protein coding) |
6755 |
0.18 |
| chr1_94073851_94074066 | 0.08 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
5696 |
0.23 |
| chr5_71462052_71462317 | 0.08 |
ENSG00000264099 |
. |
3110 |
0.3 |
| chr3_112351475_112351838 | 0.08 |
CCDC80 |
coiled-coil domain containing 80 |
5288 |
0.27 |
| chr6_30594742_30595248 | 0.08 |
ATAT1 |
alpha tubulin acetyltransferase 1 |
332 |
0.73 |
| chr19_45889271_45889481 | 0.08 |
PPP1R13L |
protein phosphatase 1, regulatory subunit 13 like |
500 |
0.68 |
| chr3_99219380_99219557 | 0.08 |
ENSG00000266030 |
. |
133735 |
0.05 |
| chr8_99376729_99376932 | 0.08 |
KCNS2 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
62420 |
0.11 |
| chr1_8087615_8088020 | 0.08 |
ERRFI1 |
ERBB receptor feedback inhibitor 1 |
1449 |
0.45 |
| chr22_51080446_51080597 | 0.07 |
ARSA |
arylsulfatase A |
13914 |
0.1 |
| chr9_112542645_112542955 | 0.07 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
31 |
0.37 |
| chr8_116675050_116675578 | 0.07 |
TRPS1 |
trichorhinophalangeal syndrome I |
1409 |
0.6 |
| chr22_50737983_50738362 | 0.07 |
PLXNB2 |
plexin B2 |
1719 |
0.19 |
| chr2_154335613_154335771 | 0.07 |
RPRM |
reprimo, TP53 dependent G2 arrest mediator candidate |
370 |
0.89 |
| chr17_47653834_47654054 | 0.07 |
NXPH3 |
neurexophilin 3 |
460 |
0.67 |
| chr11_45355125_45355276 | 0.07 |
SYT13 |
synaptotagmin XIII |
47330 |
0.16 |
| chr21_28662522_28662673 | 0.07 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
323765 |
0.01 |
| chr6_131571316_131571502 | 0.07 |
AKAP7 |
A kinase (PRKA) anchor protein 7 |
110 |
0.98 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0060424 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0001554 | luteolysis(GO:0001554) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.2 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0006911 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.0 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.0 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |