Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
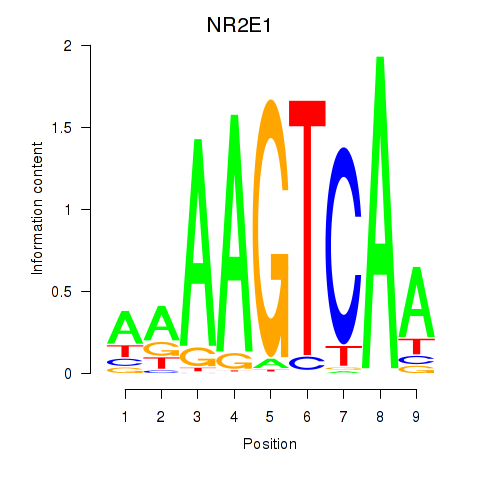
Results for NR2E1
Z-value: 1.47
Transcription factors associated with NR2E1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2E1
|
ENSG00000112333.7 | nuclear receptor subfamily 2 group E member 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_108487390_108487544 | NR2E1 | 205 | 0.724308 | -0.54 | 1.4e-01 | Click! |
| chr6_108490756_108490907 | NR2E1 | 864 | 0.581233 | -0.49 | 1.8e-01 | Click! |
| chr6_108489604_108489769 | NR2E1 | 281 | 0.896148 | -0.48 | 1.9e-01 | Click! |
| chr6_108490135_108490714 | NR2E1 | 457 | 0.803102 | -0.46 | 2.2e-01 | Click! |
| chr6_108487569_108487776 | NR2E1 | 410 | 0.615383 | -0.31 | 4.1e-01 | Click! |
Activity of the NR2E1 motif across conditions
Conditions sorted by the z-value of the NR2E1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
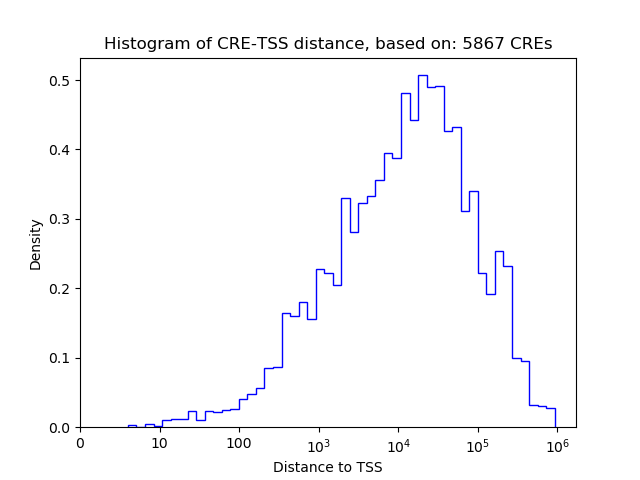
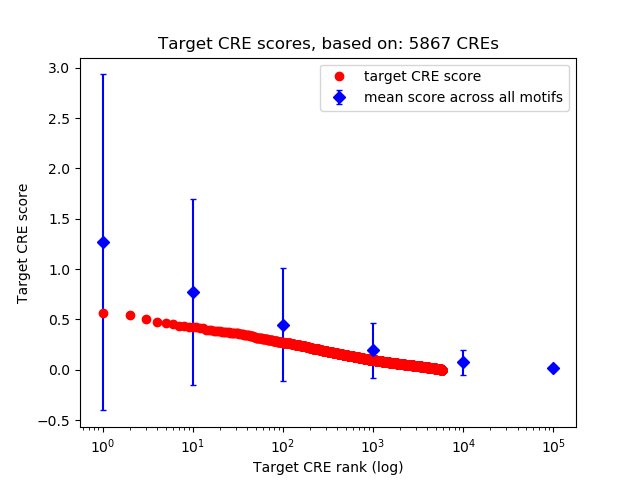
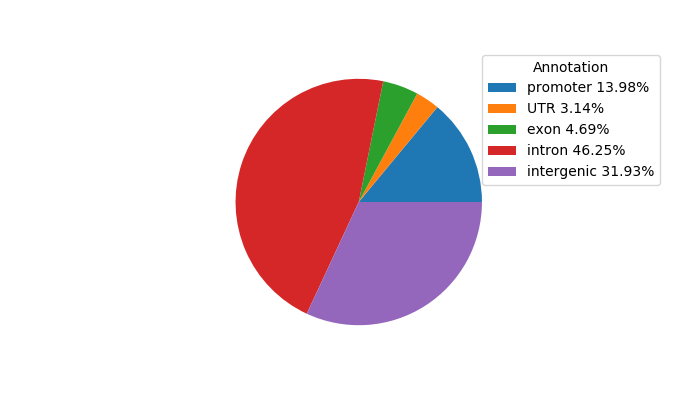
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr4_74455010_74455326 | 0.56 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
30961 |
0.21 |
| chr15_60684685_60685107 | 0.55 |
ANXA2 |
annexin A2 |
1400 |
0.54 |
| chr2_169377724_169378182 | 0.51 |
ENSG00000265694 |
. |
61500 |
0.1 |
| chr11_85962233_85962384 | 0.47 |
EED |
embryonic ectoderm development |
6032 |
0.24 |
| chr9_85677185_85677712 | 0.47 |
RASEF |
RAS and EF-hand domain containing |
595 |
0.84 |
| chr22_33141802_33142090 | 0.46 |
LL22NC01-116C6.1 |
|
37217 |
0.18 |
| chr20_10399686_10399837 | 0.43 |
MKKS |
McKusick-Kaufman syndrome |
12809 |
0.19 |
| chr8_38899485_38899676 | 0.43 |
ENSG00000207199 |
. |
23378 |
0.15 |
| chr4_4494063_4494282 | 0.43 |
STX18-IT1 |
STX18 intronic transcript 1 (non-protein coding) |
10745 |
0.21 |
| chr6_140404065_140404365 | 0.43 |
ENSG00000252107 |
. |
75616 |
0.12 |
| chr8_87354102_87354253 | 0.42 |
WWP1 |
WW domain containing E3 ubiquitin protein ligase 1 |
790 |
0.74 |
| chr5_38758917_38759276 | 0.42 |
RP11-122C5.3 |
|
24586 |
0.23 |
| chr3_149298860_149299373 | 0.42 |
WWTR1 |
WW domain containing transcription regulator 1 |
5077 |
0.23 |
| chr6_85101808_85101959 | 0.40 |
KIAA1009 |
KIAA1009 |
164530 |
0.04 |
| chr1_175946531_175946682 | 0.39 |
ENSG00000252906 |
. |
8930 |
0.26 |
| chr5_24789253_24789404 | 0.39 |
ENSG00000223004 |
. |
22262 |
0.27 |
| chr7_17348882_17349033 | 0.39 |
AC003075.4 |
|
9976 |
0.22 |
| chr6_131360061_131360448 | 0.39 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
23272 |
0.26 |
| chr2_144156290_144156486 | 0.39 |
AC096558.1 |
|
81962 |
0.1 |
| chr2_201351539_201351690 | 0.38 |
SGOL2 |
shugoshin-like 2 (S. pombe) |
23117 |
0.16 |
| chr22_36233928_36234199 | 0.38 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
2202 |
0.43 |
| chr5_158477296_158477548 | 0.38 |
EBF1 |
early B-cell factor 1 |
49279 |
0.14 |
| chr8_108791551_108791785 | 0.38 |
ENSG00000200806 |
. |
105054 |
0.08 |
| chr22_46443774_46444122 | 0.38 |
RP6-109B7.5 |
|
5025 |
0.11 |
| chr9_117879769_117880175 | 0.37 |
TNC |
tenascin C |
506 |
0.85 |
| chr22_24133174_24133325 | 0.37 |
SMARCB1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily b, member 1 |
4051 |
0.11 |
| chr2_168406531_168406682 | 0.37 |
ENSG00000238357 |
. |
81445 |
0.12 |
| chr6_8628285_8628436 | 0.37 |
SLC35B3 |
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
192566 |
0.03 |
| chr7_95545265_95545416 | 0.36 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
111744 |
0.07 |
| chr21_23084240_23084391 | 0.36 |
ENSG00000221210 |
. |
314169 |
0.01 |
| chr16_21289732_21289908 | 0.36 |
CRYM |
crystallin, mu |
26 |
0.98 |
| chr20_34458778_34459038 | 0.36 |
ENSG00000201221 |
. |
16673 |
0.13 |
| chr13_31271674_31272060 | 0.36 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
37778 |
0.19 |
| chr4_177114551_177114709 | 0.35 |
SPATA4 |
spermatogenesis associated 4 |
1398 |
0.46 |
| chr15_36623332_36623712 | 0.35 |
C15orf41 |
chromosome 15 open reading frame 41 |
248290 |
0.02 |
| chr11_37276640_37276791 | 0.35 |
ENSG00000251838 |
. |
447055 |
0.01 |
| chr9_1083553_1083729 | 0.35 |
DMRT2 |
doublesex and mab-3 related transcription factor 2 |
32027 |
0.21 |
| chr5_11961656_11961807 | 0.35 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
57576 |
0.18 |
| chr9_16828717_16828972 | 0.35 |
BNC2 |
basonuclin 2 |
3442 |
0.37 |
| chr10_94632888_94633039 | 0.35 |
EXOC6 |
exocyst complex component 6 |
24686 |
0.24 |
| chr6_54405426_54405699 | 0.34 |
RP11-124I4.2 |
|
170793 |
0.03 |
| chr7_38209906_38210057 | 0.34 |
STARD3NL |
STARD3 N-terminal like |
7843 |
0.32 |
| chr15_95867203_95867354 | 0.34 |
ENSG00000222076 |
. |
421755 |
0.01 |
| chr4_41122726_41122877 | 0.34 |
ENSG00000207198 |
. |
6842 |
0.23 |
| chr2_73963170_73963321 | 0.33 |
TPRKB |
TP53RK binding protein |
1268 |
0.45 |
| chr7_27188769_27189232 | 0.33 |
HOXA-AS3 |
HOXA cluster antisense RNA 3 |
671 |
0.36 |
| chr5_75903118_75903290 | 0.33 |
CTD-2236F14.1 |
|
1591 |
0.31 |
| chr17_14210601_14210865 | 0.33 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
6333 |
0.28 |
| chr3_189743937_189744088 | 0.32 |
ENSG00000265045 |
. |
87711 |
0.08 |
| chr1_6452011_6452162 | 0.32 |
ACOT7 |
acyl-CoA thioesterase 7 |
1335 |
0.27 |
| chr1_170562641_170562838 | 0.32 |
RP11-576I22.2 |
|
60951 |
0.12 |
| chr1_215196200_215196360 | 0.32 |
KCNK2 |
potassium channel, subfamily K, member 2 |
17082 |
0.31 |
| chr2_36689023_36689174 | 0.32 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
51735 |
0.14 |
| chr6_131291045_131291463 | 0.32 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
369 |
0.93 |
| chr5_142136607_142136758 | 0.31 |
ARHGAP26 |
Rho GTPase activating protein 26 |
13267 |
0.24 |
| chr2_153252805_153253009 | 0.31 |
FMNL2 |
formin-like 2 |
61156 |
0.15 |
| chr2_235879385_235879568 | 0.31 |
SH3BP4 |
SH3-domain binding protein 4 |
7853 |
0.34 |
| chr9_138981896_138982047 | 0.31 |
NACC2 |
NACC family member 2, BEN and BTB (POZ) domain containing |
5160 |
0.22 |
| chr3_44771799_44771953 | 0.31 |
ZNF501 |
zinc finger protein 501 |
788 |
0.53 |
| chr5_141703297_141704624 | 0.31 |
SPRY4 |
sprouty homolog 4 (Drosophila) |
213 |
0.88 |
| chr6_13733598_13733910 | 0.31 |
RANBP9 |
RAN binding protein 9 |
21958 |
0.19 |
| chr3_79346408_79346559 | 0.31 |
ENSG00000265193 |
. |
210554 |
0.03 |
| chr13_26477283_26477434 | 0.31 |
AL138815.2 |
Uncharacterized protein |
24679 |
0.17 |
| chr9_128505902_128506053 | 0.31 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
3647 |
0.31 |
| chr4_153194568_153194863 | 0.31 |
ENSG00000244544 |
. |
9118 |
0.23 |
| chr4_157691238_157691428 | 0.30 |
RP11-154F14.2 |
|
71178 |
0.11 |
| chr1_66806954_66807105 | 0.30 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
9157 |
0.31 |
| chr1_100112572_100112850 | 0.30 |
PALMD |
palmdelphin |
962 |
0.68 |
| chr1_185606466_185606617 | 0.30 |
ENSG00000201596 |
. |
3227 |
0.29 |
| chr1_100069801_100069952 | 0.30 |
PALMD |
palmdelphin |
41623 |
0.19 |
| chr1_155897053_155897204 | 0.30 |
ENSG00000252808 |
. |
1251 |
0.26 |
| chr13_24121296_24121482 | 0.30 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
23120 |
0.25 |
| chr8_110632793_110632963 | 0.29 |
SYBU |
syntabulin (syntaxin-interacting) |
12574 |
0.18 |
| chr3_66514197_66514348 | 0.29 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
37084 |
0.22 |
| chr12_114847651_114847803 | 0.29 |
TBX5-AS1 |
TBX5 antisense RNA 1 |
1168 |
0.44 |
| chr12_66590864_66591026 | 0.29 |
IRAK3 |
interleukin-1 receptor-associated kinase 3 |
7908 |
0.16 |
| chr7_143127648_143127805 | 0.29 |
TAS2R60 |
taste receptor, type 2, member 60 |
12820 |
0.12 |
| chr6_75940690_75940841 | 0.29 |
COX7A2 |
cytochrome c oxidase subunit VIIa polypeptide 2 (liver) |
12736 |
0.2 |
| chr7_43739481_43739640 | 0.29 |
COA1 |
cytochrome c oxidase assembly factor 1 homolog (S. cerevisiae) |
29491 |
0.18 |
| chr7_118512519_118512670 | 0.29 |
ENSG00000222226 |
. |
410041 |
0.01 |
| chr17_36861833_36863162 | 0.29 |
CTB-58E17.3 |
|
17 |
0.9 |
| chr3_190802793_190802944 | 0.28 |
OSTN |
osteocrin |
114162 |
0.06 |
| chr7_19144306_19144457 | 0.28 |
AC003986.6 |
|
7716 |
0.17 |
| chr12_78334931_78335200 | 0.28 |
NAV3 |
neuron navigator 3 |
24991 |
0.28 |
| chr1_215178346_215178497 | 0.28 |
KCNK2 |
potassium channel, subfamily K, member 2 |
777 |
0.8 |
| chr14_86002405_86002689 | 0.28 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
5975 |
0.26 |
| chr10_122451080_122451231 | 0.28 |
WDR11-AS1 |
WDR11 antisense RNA 1 |
85241 |
0.09 |
| chr11_91680280_91680431 | 0.28 |
FAT3 |
FAT atypical cadherin 3 |
404907 |
0.01 |
| chr11_6241000_6241151 | 0.28 |
C11orf42 |
chromosome 11 open reading frame 42 |
14279 |
0.1 |
| chr3_134050593_134051228 | 0.28 |
AMOTL2 |
angiomotin like 2 |
39844 |
0.16 |
| chrX_23919479_23919764 | 0.28 |
CXorf58 |
chromosome X open reading frame 58 |
6297 |
0.2 |
| chr21_43353842_43354056 | 0.28 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
7150 |
0.18 |
| chr2_46933523_46933944 | 0.27 |
SOCS5 |
suppressor of cytokine signaling 5 |
7407 |
0.22 |
| chr7_151182359_151182552 | 0.27 |
ENSG00000241959 |
. |
20431 |
0.13 |
| chr15_49083337_49083488 | 0.27 |
RP11-485O10.2 |
|
8025 |
0.18 |
| chr14_70485604_70486019 | 0.27 |
SLC8A3 |
solute carrier family 8 (sodium/calcium exchanger), member 3 |
61086 |
0.14 |
| chr18_30049396_30050442 | 0.27 |
GAREM |
GRB2 associated, regulator of MAPK1 |
476 |
0.87 |
| chr11_12701583_12701887 | 0.27 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
4685 |
0.33 |
| chr3_160192165_160192336 | 0.27 |
ENSG00000238427 |
. |
2046 |
0.26 |
| chr10_54075051_54075395 | 0.27 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
1167 |
0.43 |
| chr10_32201925_32202315 | 0.27 |
ARHGAP12 |
Rho GTPase activating protein 12 |
4318 |
0.31 |
| chr1_32042303_32042778 | 0.27 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
401 |
0.75 |
| chr5_149310354_149310505 | 0.27 |
ENSG00000200334 |
. |
9265 |
0.15 |
| chr19_15362877_15363312 | 0.27 |
EPHX3 |
epoxide hydrolase 3 |
18848 |
0.15 |
| chr2_145653557_145653708 | 0.27 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
375011 |
0.01 |
| chr17_12947974_12948125 | 0.27 |
ELAC2 |
elaC ribonuclease Z 2 |
26545 |
0.19 |
| chr15_35601919_35602070 | 0.27 |
ENSG00000265102 |
. |
62571 |
0.15 |
| chr3_105849899_105850071 | 0.27 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
261589 |
0.02 |
| chr5_146746092_146746307 | 0.27 |
DPYSL3 |
dihydropyrimidinase-like 3 |
34966 |
0.18 |
| chr10_90754998_90755312 | 0.27 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
4008 |
0.17 |
| chr3_93623597_93623748 | 0.27 |
PROS1 |
protein S (alpha) |
53926 |
0.11 |
| chr3_45686186_45686439 | 0.27 |
LIMD1-AS1 |
LIMD1 antisense RNA 1 |
44062 |
0.11 |
| chr12_89210324_89210486 | 0.26 |
ENSG00000252850 |
. |
234989 |
0.02 |
| chr16_69952007_69952311 | 0.26 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
6734 |
0.17 |
| chr2_151333058_151333209 | 0.26 |
RND3 |
Rho family GTPase 3 |
8763 |
0.33 |
| chr8_57086542_57086822 | 0.26 |
PLAG1 |
pleiomorphic adenoma gene 1 |
37156 |
0.12 |
| chr11_12065337_12065964 | 0.26 |
DKK3 |
dickkopf WNT signaling pathway inhibitor 3 |
34334 |
0.19 |
| chr6_108478555_108479189 | 0.26 |
OSTM1 |
osteopetrosis associated transmembrane protein 1 |
8172 |
0.19 |
| chr17_29335129_29336577 | 0.26 |
RP11-848P1.9 |
|
81 |
0.95 |
| chr9_530471_530746 | 0.26 |
RP11-31F19.1 |
|
16709 |
0.13 |
| chr2_157323820_157323971 | 0.26 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
6189 |
0.22 |
| chr17_28037574_28037864 | 0.26 |
RP11-82O19.1 |
|
50402 |
0.08 |
| chr18_28619786_28619937 | 0.26 |
DSC3 |
desmocollin 3 |
2846 |
0.25 |
| chr5_33891538_33892159 | 0.26 |
ADAMTS12 |
ADAM metallopeptidase with thrombospondin type 1 motif, 12 |
198 |
0.94 |
| chr1_182338063_182338214 | 0.26 |
GLUL |
glutamate-ammonia ligase |
22401 |
0.18 |
| chr10_33230803_33231099 | 0.26 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
5772 |
0.31 |
| chr4_123820303_123820454 | 0.26 |
NUDT6 |
nudix (nucleoside diphosphate linked moiety X)-type motif 6 |
14065 |
0.17 |
| chr2_171017422_171017573 | 0.26 |
MYO3B |
myosin IIIB |
17158 |
0.23 |
| chr11_61159610_61160825 | 0.26 |
TMEM216 |
transmembrane protein 216 |
352 |
0.79 |
| chr9_21329043_21329194 | 0.25 |
KLHL9 |
kelch-like family member 9 |
6122 |
0.14 |
| chr4_35309607_35309866 | 0.25 |
ENSG00000206665 |
. |
187784 |
0.03 |
| chr5_36836552_36836703 | 0.25 |
NIPBL |
Nipped-B homolog (Drosophila) |
40234 |
0.2 |
| chr15_70316424_70316624 | 0.25 |
ENSG00000207965 |
. |
55283 |
0.15 |
| chr3_4753555_4753835 | 0.25 |
ENSG00000239126 |
. |
166691 |
0.03 |
| chr3_156800832_156800983 | 0.25 |
ENSG00000222499 |
. |
65776 |
0.09 |
| chr19_43633813_43634101 | 0.25 |
PSG2 |
pregnancy specific beta-1-glycoprotein 2 |
47137 |
0.12 |
| chr10_123334259_123334410 | 0.25 |
FGFR2 |
fibroblast growth factor receptor 2 |
19009 |
0.28 |
| chr2_219582270_219582421 | 0.25 |
TTLL4 |
tubulin tyrosine ligase-like family, member 4 |
6628 |
0.14 |
| chr8_32441306_32441509 | 0.25 |
NRG1 |
neuregulin 1 |
21759 |
0.28 |
| chr2_100101443_100101594 | 0.25 |
REV1 |
REV1, polymerase (DNA directed) |
4917 |
0.24 |
| chr10_27815365_27815516 | 0.25 |
RAB18 |
RAB18, member RAS oncogene family |
21892 |
0.24 |
| chr9_21677527_21678485 | 0.25 |
ENSG00000244230 |
. |
21307 |
0.21 |
| chr15_78171125_78171323 | 0.25 |
CSPG4P13 |
chondroitin sulfate proteoglycan 4 pseudogene 13 |
15802 |
0.15 |
| chr6_106897121_106897272 | 0.25 |
ENSG00000202386 |
. |
60 |
0.97 |
| chr10_34809719_34809870 | 0.25 |
PARD3 |
par-3 family cell polarity regulator |
94135 |
0.09 |
| chr12_13359480_13359859 | 0.25 |
EMP1 |
epithelial membrane protein 1 |
4776 |
0.29 |
| chr6_125932782_125933099 | 0.24 |
RP11-624M8.1 |
|
133234 |
0.05 |
| chr10_115143798_115143949 | 0.24 |
ENSG00000238380 |
. |
30689 |
0.24 |
| chr2_228681607_228681918 | 0.24 |
CCL20 |
chemokine (C-C motif) ligand 20 |
3192 |
0.28 |
| chr11_19472113_19472264 | 0.24 |
ENSG00000200687 |
. |
48660 |
0.12 |
| chr11_86516621_86516772 | 0.24 |
PRSS23 |
protease, serine, 23 |
5111 |
0.32 |
| chr1_59478905_59479109 | 0.24 |
JUN |
jun proto-oncogene |
229222 |
0.02 |
| chr20_18296318_18296469 | 0.24 |
RP4-568F9.3 |
|
638 |
0.6 |
| chr10_36184684_36184984 | 0.24 |
FZD8 |
frizzled family receptor 8 |
254472 |
0.02 |
| chr8_89311514_89311910 | 0.24 |
RP11-586K2.1 |
|
27353 |
0.2 |
| chr5_14448258_14448409 | 0.24 |
TRIO |
trio Rho guanine nucleotide exchange factor |
40224 |
0.21 |
| chr4_54926609_54926760 | 0.24 |
AC110792.1 |
HCG2027126; Uncharacterized protein |
529 |
0.75 |
| chr13_110972233_110972468 | 0.24 |
COL4A2 |
collagen, type IV, alpha 2 |
12736 |
0.21 |
| chr4_173149760_173149911 | 0.24 |
GALNTL6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
176239 |
0.04 |
| chr8_108468386_108468792 | 0.24 |
ANGPT1 |
angiopoietin 1 |
38634 |
0.23 |
| chr12_122884260_122884886 | 0.24 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
12 |
0.98 |
| chr10_32215556_32215707 | 0.24 |
ARHGAP12 |
Rho GTPase activating protein 12 |
2097 |
0.41 |
| chr10_101432551_101432702 | 0.24 |
ENTPD7 |
ectonucleoside triphosphate diphosphohydrolase 7 |
13363 |
0.14 |
| chr1_193376231_193376382 | 0.24 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
220522 |
0.02 |
| chr6_41473165_41473397 | 0.24 |
RP11-328M4.2 |
|
40531 |
0.13 |
| chr6_157358181_157358332 | 0.24 |
RP1-137K2.2 |
|
82584 |
0.1 |
| chr3_191055905_191056279 | 0.24 |
UTS2B |
urotensin 2B |
7767 |
0.23 |
| chr2_145753937_145754126 | 0.24 |
ENSG00000253036 |
. |
338607 |
0.01 |
| chr15_49726228_49726382 | 0.24 |
FGF7 |
fibroblast growth factor 7 |
10848 |
0.24 |
| chr13_49928496_49928647 | 0.23 |
CAB39L |
calcium binding protein 39-like |
3308 |
0.31 |
| chr3_137486919_137487668 | 0.23 |
SOX14 |
SRY (sex determining region Y)-box 14 |
3714 |
0.37 |
| chr20_46121886_46122388 | 0.23 |
ENSG00000201742 |
. |
308 |
0.88 |
| chr2_46946794_46946945 | 0.23 |
SOCS5 |
suppressor of cytokine signaling 5 |
20543 |
0.19 |
| chr21_33317813_33317964 | 0.23 |
HUNK |
hormonally up-regulated Neu-associated kinase |
21024 |
0.24 |
| chr5_75902860_75903011 | 0.23 |
CTD-2236F14.1 |
|
1860 |
0.28 |
| chr5_158520357_158520666 | 0.23 |
EBF1 |
early B-cell factor 1 |
6190 |
0.27 |
| chr12_86229562_86230264 | 0.23 |
RASSF9 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 9 |
435 |
0.89 |
| chr21_29172171_29172322 | 0.23 |
ADAMTS5 |
ADAM metallopeptidase with thrombospondin type 1 motif, 5 |
833414 |
0.0 |
| chr5_137358347_137358498 | 0.23 |
RP11-325L7.1 |
|
10041 |
0.14 |
| chr22_46422730_46422990 | 0.23 |
RP6-109B7.5 |
|
26113 |
0.09 |
| chr1_61561178_61561329 | 0.23 |
NFIA |
nuclear factor I/A |
7169 |
0.21 |
| chr16_11303461_11303612 | 0.23 |
RMI2 |
RecQ mediated genome instability 2 |
39970 |
0.09 |
| chr5_9533993_9534144 | 0.23 |
CTD-2201E9.2 |
|
10778 |
0.18 |
| chr12_115119174_115119325 | 0.23 |
TBX3 |
T-box 3 |
2146 |
0.36 |
| chr3_192634406_192635568 | 0.23 |
MB21D2 |
Mab-21 domain containing 2 |
963 |
0.71 |
| chr3_187984581_187985052 | 0.23 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
27164 |
0.24 |
| chr6_148886821_148887018 | 0.23 |
ENSG00000223322 |
. |
41543 |
0.21 |
| chr2_224703111_224703262 | 0.23 |
AP1S3 |
adaptor-related protein complex 1, sigma 3 subunit |
442 |
0.89 |
| chr12_102093877_102094028 | 0.23 |
CHPT1 |
choline phosphotransferase 1 |
2502 |
0.21 |
| chr12_1428214_1428403 | 0.23 |
RP5-951N9.2 |
|
66691 |
0.11 |
| chr22_37583582_37583762 | 0.23 |
C1QTNF6 |
C1q and tumor necrosis factor related protein 6 |
654 |
0.59 |
| chrX_45365887_45366208 | 0.22 |
RP11-342D14.1 |
|
186344 |
0.03 |
| chr14_36786542_36786693 | 0.22 |
MBIP |
MAP3K12 binding inhibitory protein 1 |
2459 |
0.33 |
| chr8_32137811_32137962 | 0.22 |
ENSG00000200246 |
. |
23874 |
0.22 |
| chr7_130761600_130761776 | 0.22 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
11545 |
0.21 |
| chr4_17813550_17813701 | 0.22 |
NCAPG |
non-SMC condensin I complex, subunit G |
1100 |
0.4 |
| chr1_59246622_59247052 | 0.22 |
JUN |
jun proto-oncogene |
2948 |
0.28 |
| chr20_34459115_34459914 | 0.22 |
ENSG00000201221 |
. |
16067 |
0.13 |
| chr7_34027760_34027911 | 0.22 |
BMPER |
BMP binding endothelial regulator |
82690 |
0.11 |
| chr3_7340877_7341028 | 0.22 |
GRM7 |
glutamate receptor, metabotropic 7 |
103785 |
0.08 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060750 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) |
| 0.1 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.1 | GO:0060523 | prostate epithelial cord elongation(GO:0060523) |
| 0.1 | 0.5 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.1 | 0.2 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.1 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.1 | 0.2 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.1 | 0.2 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.1 | 0.2 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.1 | 0.4 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.1 | 0.2 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.2 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.4 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.0 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0072194 | kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.4 | GO:0046718 | viral entry into host cell(GO:0046718) |
| 0.0 | 0.1 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0060044 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.2 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.2 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.1 | GO:0045978 | negative regulation of nucleoside metabolic process(GO:0045978) negative regulation of ATP metabolic process(GO:1903579) |
| 0.0 | 0.2 | GO:0048843 | negative regulation of axon extension involved in axon guidance(GO:0048843) |
| 0.0 | 0.1 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0051877 | pigment granule aggregation in cell center(GO:0051877) |
| 0.0 | 0.4 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.2 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0061525 | hindgut morphogenesis(GO:0007442) hindgut development(GO:0061525) |
| 0.0 | 0.1 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.1 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.0 | 0.4 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0047484 | regulation of response to osmotic stress(GO:0047484) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.0 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.0 | GO:0032278 | positive regulation of gonadotropin secretion(GO:0032278) positive regulation of follicle-stimulating hormone secretion(GO:0046881) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.6 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0021955 | central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0022030 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.1 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0010664 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0048865 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0050922 | negative regulation of chemotaxis(GO:0050922) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0045542 | positive regulation of cholesterol biosynthetic process(GO:0045542) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.1 | GO:0050686 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) platelet activating factor metabolic process(GO:0046469) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.0 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.1 | GO:0051657 | maintenance of organelle location(GO:0051657) |
| 0.0 | 0.0 | GO:0002248 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.0 | GO:0045605 | negative regulation of epidermal cell differentiation(GO:0045605) |
| 0.0 | 0.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.1 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.0 | GO:0008614 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.1 | GO:1904893 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.4 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.2 | GO:0071456 | cellular response to decreased oxygen levels(GO:0036294) cellular response to oxygen levels(GO:0071453) cellular response to hypoxia(GO:0071456) |
| 0.0 | 0.0 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.0 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0031116 | positive regulation of microtubule polymerization(GO:0031116) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.1 | GO:0010324 | phagocytosis, engulfment(GO:0006911) membrane invagination(GO:0010324) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0060745 | mammary gland branching involved in pregnancy(GO:0060745) |
| 0.0 | 0.1 | GO:0007379 | segment specification(GO:0007379) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.0 | GO:1902339 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0042574 | retinal metabolic process(GO:0042574) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0014034 | neural crest formation(GO:0014029) neural crest cell fate commitment(GO:0014034) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.2 | GO:0008210 | estrogen metabolic process(GO:0008210) |
| 0.0 | 0.0 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.0 | GO:0043551 | regulation of phosphatidylinositol 3-kinase activity(GO:0043551) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.2 | GO:0007076 | mitotic chromosome condensation(GO:0007076) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.2 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.1 | GO:0070207 | protein homotrimerization(GO:0070207) |
| 0.0 | 0.1 | GO:0039692 | single stranded viral RNA replication via double stranded DNA intermediate(GO:0039692) viral RNA genome replication(GO:0039694) RNA replication(GO:0039703) |
| 0.0 | 0.1 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.0 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0046636 | negative regulation of alpha-beta T cell activation(GO:0046636) |
| 0.0 | 0.0 | GO:0080111 | DNA demethylation(GO:0080111) |
| 0.0 | 0.0 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0001710 | mesodermal cell fate commitment(GO:0001710) |
| 0.0 | 0.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.0 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.0 | 0.0 | GO:0007619 | courtship behavior(GO:0007619) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.4 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.2 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0016011 | dystroglycan complex(GO:0016011) sarcoglycan complex(GO:0016012) |
| 0.0 | 0.1 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.1 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.4 | GO:0032587 | ruffle membrane(GO:0032587) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0005638 | lamin filament(GO:0005638) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.3 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.3 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.4 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.3 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0031781 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.2 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0047115 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.0 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.2 | GO:1990782 | receptor tyrosine kinase binding(GO:0030971) protein tyrosine kinase binding(GO:1990782) |
| 0.0 | 0.0 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.0 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.0 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.6 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.8 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.6 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.2 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME PLC BETA MEDIATED EVENTS | Genes involved in PLC beta mediated events |
| 0.0 | 0.4 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.5 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 1.0 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.1 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |