Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
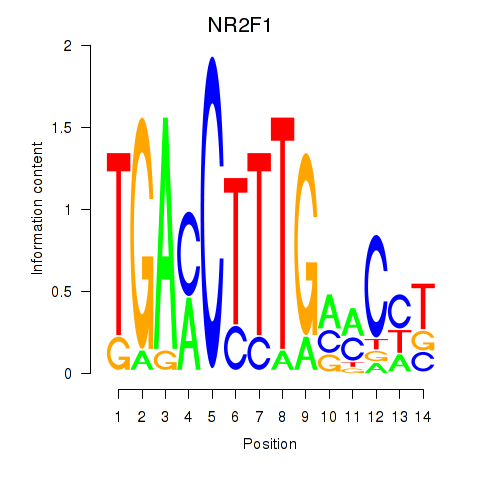
Results for NR2F1
Z-value: 1.05
Transcription factors associated with NR2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2F1
|
ENSG00000175745.7 | nuclear receptor subfamily 2 group F member 1 |
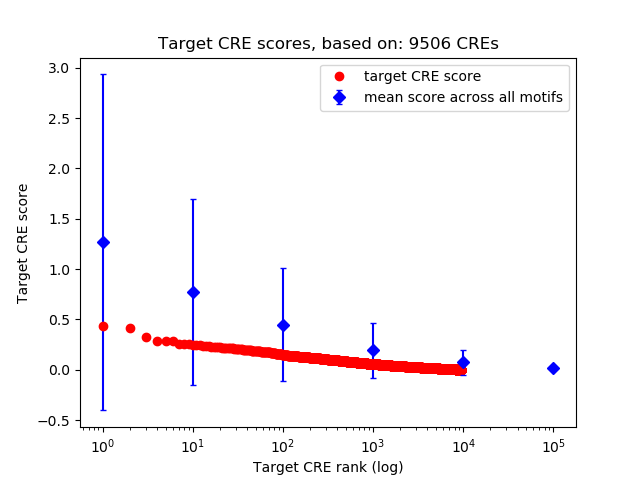
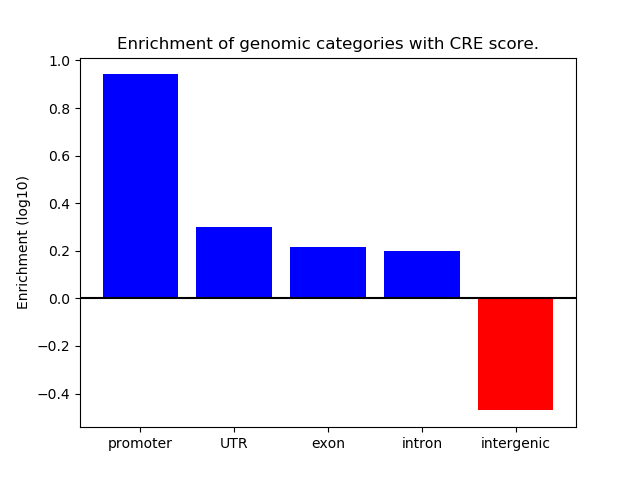
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr5_92919252_92919403 | NR2F1 | 284 | 0.899493 | -0.19 | 6.2e-01 | Click! |
| chr5_92918344_92919004 | NR2F1 | 369 | 0.842576 | -0.09 | 8.2e-01 | Click! |
| chr5_92919468_92919846 | NR2F1 | 614 | 0.692294 | 0.02 | 9.6e-01 | Click! |
| chr5_92918170_92918321 | NR2F1 | 798 | 0.536761 | -0.01 | 9.8e-01 | Click! |
Activity of the NR2F1 motif across conditions
Conditions sorted by the z-value of the NR2F1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
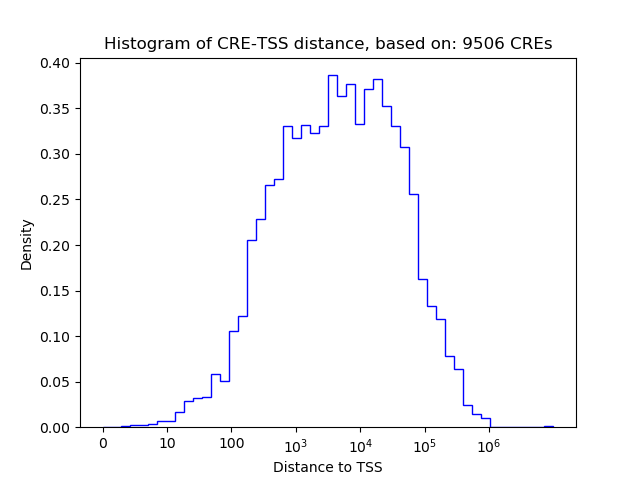
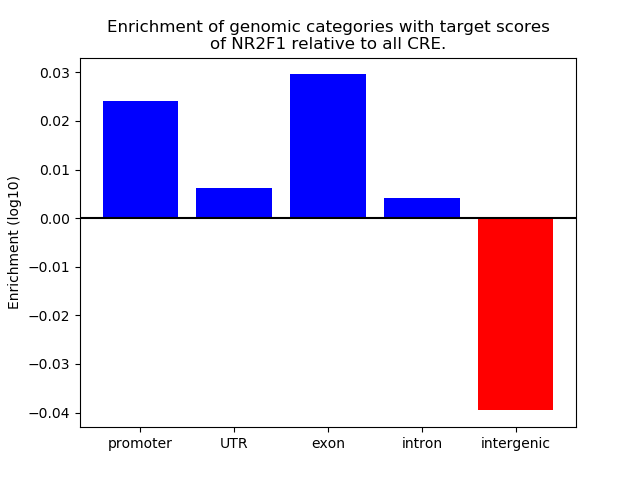
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr10_112123532_112123887 | 0.43 |
SMNDC1 |
survival motor neuron domain containing 1 |
59000 |
0.12 |
| chr22_39639623_39639926 | 0.41 |
PDGFB |
platelet-derived growth factor beta polypeptide |
753 |
0.58 |
| chr11_65324607_65325239 | 0.32 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
313 |
0.76 |
| chr10_99441863_99442014 | 0.28 |
AVPI1 |
arginine vasopressin-induced 1 |
5142 |
0.16 |
| chr12_56327336_56327517 | 0.28 |
WIBG |
within bgcn homolog (Drosophila) |
1024 |
0.3 |
| chr13_114543321_114543472 | 0.28 |
GAS6 |
growth arrest-specific 6 |
4379 |
0.26 |
| chr1_215550174_215550325 | 0.26 |
KCTD3 |
potassium channel tetramerization domain containing 3 |
190486 |
0.03 |
| chr7_150435640_150435791 | 0.26 |
GIMAP5 |
GTPase, IMAP family member 5 |
1279 |
0.41 |
| chr19_17797231_17797557 | 0.25 |
UNC13A |
unc-13 homolog A (C. elegans) |
1614 |
0.31 |
| chr12_6711834_6712315 | 0.25 |
CHD4 |
chromodomain helicase DNA binding protein 4 |
3772 |
0.1 |
| chr12_24691177_24691328 | 0.24 |
ENSG00000240481 |
. |
168470 |
0.04 |
| chr9_130740055_130740787 | 0.24 |
FAM102A |
family with sequence similarity 102, member A |
2371 |
0.18 |
| chr2_42599068_42599219 | 0.24 |
COX7A2L |
cytochrome c oxidase subunit VIIa polypeptide 2 like |
2993 |
0.32 |
| chr22_45065479_45065630 | 0.24 |
PRR5 |
proline rich 5 (renal) |
961 |
0.61 |
| chr10_77873536_77873687 | 0.23 |
ENSG00000221232 |
. |
13458 |
0.28 |
| chr6_170493106_170493257 | 0.23 |
RP11-302L19.1 |
|
15440 |
0.24 |
| chr11_64073389_64074392 | 0.23 |
ESRRA |
estrogen-related receptor alpha |
28 |
0.92 |
| chrX_56829451_56830094 | 0.22 |
ENSG00000204272 |
. |
74080 |
0.12 |
| chr6_151701170_151702048 | 0.22 |
ENSG00000252615 |
. |
2375 |
0.22 |
| chr12_663051_663343 | 0.22 |
B4GALNT3 |
beta-1,4-N-acetyl-galactosaminyl transferase 3 |
10898 |
0.16 |
| chr14_22997122_22997273 | 0.22 |
TRAJ15 |
T cell receptor alpha joining 15 |
1383 |
0.28 |
| chr19_18304075_18305119 | 0.22 |
MPV17L2 |
MPV17 mitochondrial membrane protein-like 2 |
605 |
0.57 |
| chr20_62369505_62369786 | 0.22 |
RP4-583P15.14 |
|
22 |
0.92 |
| chr4_143323082_143323233 | 0.22 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
29255 |
0.27 |
| chr19_1466791_1467069 | 0.22 |
ENSG00000267317 |
. |
4166 |
0.08 |
| chr16_28997666_28997863 | 0.21 |
LAT |
linker for activation of T cells |
231 |
0.83 |
| chr22_31033767_31033918 | 0.21 |
SLC35E4 |
solute carrier family 35, member E4 |
1332 |
0.32 |
| chr3_132377366_132377517 | 0.21 |
UBA5 |
ubiquitin-like modifier activating enzyme 5 |
869 |
0.47 |
| chr4_7972052_7972961 | 0.21 |
AFAP1 |
actin filament associated protein 1 |
30853 |
0.14 |
| chr3_97591824_97591975 | 0.21 |
CRYBG3 |
beta-gamma crystallin domain containing 3 |
3920 |
0.3 |
| chr12_8190824_8190975 | 0.21 |
FOXJ2 |
forkhead box J2 |
4943 |
0.17 |
| chr5_67564084_67564251 | 0.21 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
11901 |
0.28 |
| chr20_30873595_30873746 | 0.21 |
ENSG00000264308 |
. |
8079 |
0.16 |
| chr2_19073151_19073433 | 0.21 |
NT5C1B |
5'-nucleotidase, cytosolic IB |
302454 |
0.01 |
| chr11_101987791_101987942 | 0.20 |
YAP1 |
Yes-associated protein 1 |
4621 |
0.19 |
| chr19_48222929_48223134 | 0.20 |
EHD2 |
EH-domain containing 2 |
6352 |
0.14 |
| chr11_122722660_122722811 | 0.20 |
CRTAM |
cytotoxic and regulatory T cell molecule |
10276 |
0.21 |
| chr11_118788335_118788931 | 0.20 |
BCL9L |
B-cell CLL/lymphoma 9-like |
980 |
0.33 |
| chr3_124770412_124771279 | 0.20 |
HEG1 |
heart development protein with EGF-like domains 1 |
3957 |
0.22 |
| chr1_25944952_25945229 | 0.20 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
749 |
0.68 |
| chr8_145010856_145011411 | 0.20 |
PLEC |
plectin |
2625 |
0.16 |
| chr11_71750284_71750541 | 0.20 |
NUMA1 |
nuclear mitotic apparatus protein 1 |
459 |
0.68 |
| chr9_136238609_136239100 | 0.19 |
SURF4 |
surfeit 4 |
4060 |
0.08 |
| chr14_52687906_52688082 | 0.19 |
PTGDR |
prostaglandin D2 receptor (DP) |
46437 |
0.16 |
| chr3_194117300_194117501 | 0.19 |
GP5 |
glycoprotein V (platelet) |
1683 |
0.34 |
| chr1_17914943_17915229 | 0.19 |
ARHGEF10L |
Rho guanine nucleotide exchange factor (GEF) 10-like |
175 |
0.97 |
| chr10_43761610_43761802 | 0.19 |
RASGEF1A |
RasGEF domain family, member 1A |
661 |
0.77 |
| chr7_25013883_25014034 | 0.19 |
OSBPL3 |
oxysterol binding protein-like 3 |
5802 |
0.3 |
| chr4_188524057_188524208 | 0.19 |
ZFP42 |
ZFP42 zinc finger protein |
392793 |
0.01 |
| chr17_77022963_77023114 | 0.19 |
C1QTNF1-AS1 |
C1QTNF1 antisense RNA 1 |
645 |
0.56 |
| chr12_47615454_47615605 | 0.18 |
PCED1B |
PC-esterase domain containing 1B |
1755 |
0.4 |
| chr12_54132562_54132781 | 0.18 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
11142 |
0.17 |
| chr3_171175410_171175561 | 0.18 |
TNIK |
TRAF2 and NCK interacting kinase |
2367 |
0.33 |
| chr15_31622749_31622975 | 0.18 |
KLF13 |
Kruppel-like factor 13 |
3804 |
0.36 |
| chr5_9762754_9762905 | 0.18 |
ENSG00000222054 |
. |
1810 |
0.44 |
| chr1_12675825_12676469 | 0.18 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
1590 |
0.37 |
| chr19_18898403_18898596 | 0.18 |
COMP |
cartilage oligomeric matrix protein |
3615 |
0.19 |
| chr2_158296405_158296556 | 0.18 |
CYTIP |
cytohesin 1 interacting protein |
554 |
0.73 |
| chr3_124762287_124762438 | 0.18 |
HEG1 |
heart development protein with EGF-like domains 1 |
12440 |
0.18 |
| chr9_132428871_132429094 | 0.18 |
PRRX2 |
paired related homeobox 2 |
1062 |
0.42 |
| chr6_119669093_119669825 | 0.18 |
MAN1A1 |
mannosidase, alpha, class 1A, member 1 |
1442 |
0.51 |
| chr14_103560765_103560916 | 0.18 |
RP11-736N17.8 |
|
220 |
0.92 |
| chr5_35813662_35813813 | 0.18 |
CTD-2113L7.1 |
|
13383 |
0.2 |
| chr18_3411935_3412439 | 0.18 |
TGIF1 |
TGFB-induced factor homeobox 1 |
115 |
0.97 |
| chr11_118212224_118212431 | 0.18 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
675 |
0.54 |
| chr10_6094124_6094387 | 0.17 |
IL2RA |
interleukin 2 receptor, alpha |
9998 |
0.15 |
| chr10_8099034_8099185 | 0.17 |
GATA3 |
GATA binding protein 3 |
2340 |
0.45 |
| chr14_98443402_98443682 | 0.17 |
C14orf64 |
chromosome 14 open reading frame 64 |
841 |
0.77 |
| chr9_130738670_130738821 | 0.17 |
FAM102A |
family with sequence similarity 102, member A |
4047 |
0.14 |
| chr5_172246747_172246898 | 0.17 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
14456 |
0.16 |
| chr6_170189987_170191002 | 0.17 |
RP1-266L20.9 |
|
19985 |
0.14 |
| chr16_10713819_10713970 | 0.17 |
EMP2 |
epithelial membrane protein 2 |
39339 |
0.13 |
| chr14_99729436_99729607 | 0.17 |
AL109767.1 |
|
236 |
0.94 |
| chr6_26614195_26614680 | 0.17 |
ABT1 |
activator of basal transcription 1 |
17257 |
0.17 |
| chr6_143247533_143248138 | 0.17 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
18503 |
0.25 |
| chr5_133446419_133446570 | 0.17 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
3908 |
0.27 |
| chr19_1470719_1471158 | 0.16 |
C19orf25 |
chromosome 19 open reading frame 25 |
7958 |
0.07 |
| chr20_17831337_17831488 | 0.16 |
ENSG00000221220 |
. |
8185 |
0.22 |
| chr12_45269258_45270040 | 0.16 |
NELL2 |
NEL-like 2 (chicken) |
54 |
0.98 |
| chr9_137967884_137968035 | 0.16 |
OLFM1 |
olfactomedin 1 |
447 |
0.88 |
| chr4_38080893_38081044 | 0.16 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
33476 |
0.22 |
| chr2_44844069_44844220 | 0.16 |
ENSG00000252896 |
. |
165125 |
0.04 |
| chr16_87520451_87520724 | 0.16 |
ZCCHC14 |
zinc finger, CCHC domain containing 14 |
5064 |
0.23 |
| chr10_33405368_33405626 | 0.16 |
ENSG00000263576 |
. |
17933 |
0.22 |
| chr3_64207181_64207332 | 0.16 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
3875 |
0.26 |
| chr1_189407105_189407256 | 0.16 |
ENSG00000252553 |
. |
228206 |
0.02 |
| chr14_105953334_105954468 | 0.16 |
CRIP1 |
cysteine-rich protein 1 (intestinal) |
647 |
0.53 |
| chr4_89518835_89518986 | 0.16 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
4381 |
0.2 |
| chr11_66090538_66090689 | 0.16 |
CD248 |
CD248 molecule, endosialin |
6098 |
0.07 |
| chr18_56105832_56106078 | 0.16 |
ENSG00000207778 |
. |
12351 |
0.17 |
| chr22_47428996_47429147 | 0.15 |
ENSG00000221672 |
. |
185268 |
0.03 |
| chr9_79018493_79018995 | 0.15 |
RFK |
riboflavin kinase |
9323 |
0.24 |
| chr11_16118588_16118739 | 0.15 |
ENSG00000221556 |
. |
27199 |
0.24 |
| chr3_53033553_53033704 | 0.15 |
SFMBT1 |
Scm-like with four mbt domains 1 |
45653 |
0.12 |
| chr2_181971569_181971726 | 0.15 |
UBE2E3 |
ubiquitin-conjugating enzyme E2E 3 |
124897 |
0.06 |
| chr10_134351637_134352716 | 0.15 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
533 |
0.8 |
| chr2_230195712_230195863 | 0.15 |
PID1 |
phosphotyrosine interaction domain containing 1 |
59786 |
0.15 |
| chr8_66751756_66751954 | 0.15 |
PDE7A |
phosphodiesterase 7A |
872 |
0.73 |
| chr8_111917370_111917521 | 0.15 |
KCNV1 |
potassium channel, subfamily V, member 1 |
929369 |
0.0 |
| chr14_76447085_76447352 | 0.15 |
TGFB3 |
transforming growth factor, beta 3 |
118 |
0.97 |
| chr3_48630851_48631002 | 0.15 |
COL7A1 |
collagen, type VII, alpha 1 |
1667 |
0.2 |
| chr19_48221689_48221840 | 0.15 |
EHD2 |
EH-domain containing 2 |
5085 |
0.15 |
| chr3_69888019_69888170 | 0.15 |
MITF |
microphthalmia-associated transcription factor |
27269 |
0.22 |
| chr4_157720406_157720557 | 0.15 |
RP11-154F14.2 |
|
42030 |
0.17 |
| chr4_87763331_87763482 | 0.15 |
SLC10A6 |
solute carrier family 10 (sodium/bile acid cotransporter), member 6 |
7010 |
0.26 |
| chr21_30257999_30258150 | 0.15 |
N6AMT1 |
N-6 adenine-specific DNA methyltransferase 1 (putative) |
381 |
0.9 |
| chr8_132052704_132052867 | 0.15 |
ADCY8 |
adenylate cyclase 8 (brain) |
206 |
0.97 |
| chr6_56010689_56010840 | 0.15 |
COL21A1 |
collagen, type XXI, alpha 1 |
20971 |
0.28 |
| chr20_821653_822360 | 0.15 |
FAM110A |
family with sequence similarity 110, member A |
3279 |
0.3 |
| chr11_63304045_63305245 | 0.14 |
RARRES3 |
retinoic acid receptor responder (tazarotene induced) 3 |
352 |
0.84 |
| chr12_111177324_111178139 | 0.14 |
PPP1CC |
protein phosphatase 1, catalytic subunit, gamma isozyme |
2913 |
0.28 |
| chr22_41798411_41798579 | 0.14 |
TEF |
thyrotrophic embryonic factor |
19556 |
0.13 |
| chr11_128336036_128336187 | 0.14 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
39178 |
0.19 |
| chr7_75622595_75622746 | 0.14 |
POR |
P450 (cytochrome) oxidoreductase |
11441 |
0.16 |
| chr13_76334947_76335146 | 0.14 |
LMO7 |
LIM domain 7 |
249 |
0.95 |
| chr12_94441024_94441175 | 0.14 |
ENSG00000223126 |
. |
38276 |
0.18 |
| chr2_55361422_55361625 | 0.14 |
RTN4 |
reticulon 4 |
21766 |
0.16 |
| chr11_46268403_46268554 | 0.14 |
CTD-2589M5.4 |
|
27620 |
0.16 |
| chr3_138952873_138953024 | 0.14 |
MRPS22 |
mitochondrial ribosomal protein S22 |
109913 |
0.06 |
| chr7_129546585_129546878 | 0.14 |
UBE2H |
ubiquitin-conjugating enzyme E2H |
44436 |
0.1 |
| chr11_46370356_46370605 | 0.14 |
DGKZ |
diacylglycerol kinase, zeta |
1342 |
0.36 |
| chr1_207090442_207090593 | 0.14 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
4695 |
0.16 |
| chr19_2503722_2503936 | 0.14 |
ENSG00000252962 |
. |
690 |
0.64 |
| chr13_43406909_43407060 | 0.14 |
FAM216B |
family with sequence similarity 216, member B |
51233 |
0.18 |
| chr1_28385802_28385953 | 0.14 |
EYA3 |
eyes absent homolog 3 (Drosophila) |
1279 |
0.41 |
| chr18_52968140_52968291 | 0.14 |
TCF4 |
transcription factor 4 |
1642 |
0.52 |
| chr1_156091493_156091688 | 0.14 |
LMNA |
lamin A/C |
4361 |
0.13 |
| chr7_157405294_157405606 | 0.14 |
AC005481.5 |
Uncharacterized protein |
1265 |
0.52 |
| chr17_15151294_15151445 | 0.14 |
ENSG00000265110 |
. |
3644 |
0.18 |
| chr9_21611364_21611515 | 0.14 |
MIR31HG |
MIR31 host gene (non-protein coding) |
51771 |
0.12 |
| chr4_150608043_150608194 | 0.14 |
ENSG00000202331 |
. |
220594 |
0.02 |
| chr2_231090336_231091061 | 0.14 |
SP140 |
SP140 nuclear body protein |
219 |
0.54 |
| chr15_83348879_83349094 | 0.14 |
AP3B2 |
adaptor-related protein complex 3, beta 2 subunit |
19006 |
0.12 |
| chr3_150014037_150014188 | 0.14 |
TSC22D2 |
TSC22 domain family, member 2 |
112010 |
0.06 |
| chr3_195839047_195839198 | 0.14 |
TFRC |
transferrin receptor |
30062 |
0.14 |
| chr3_156561383_156561569 | 0.14 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
17328 |
0.27 |
| chr11_85538689_85538889 | 0.13 |
SYTL2 |
synaptotagmin-like 2 |
16605 |
0.17 |
| chrY_14617944_14618095 | 0.13 |
GYG2P1 |
glycogenin 2 pseudogene 1 |
85898 |
0.1 |
| chr5_37303368_37303519 | 0.13 |
ENSG00000251880 |
. |
23794 |
0.18 |
| chr3_53184243_53184394 | 0.13 |
PRKCD |
protein kinase C, delta |
5707 |
0.19 |
| chr3_30655384_30655643 | 0.13 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
7420 |
0.31 |
| chr1_113050352_113050503 | 0.13 |
WNT2B |
wingless-type MMTV integration site family, member 2B |
973 |
0.56 |
| chr17_46618647_46619025 | 0.13 |
HOXB-AS1 |
HOXB cluster antisense RNA 1 |
2077 |
0.14 |
| chr3_131220949_131221185 | 0.13 |
MRPL3 |
mitochondrial ribosomal protein L3 |
714 |
0.66 |
| chr2_131582424_131582575 | 0.13 |
AC133785.1 |
|
12068 |
0.16 |
| chr7_44185996_44186184 | 0.13 |
GCK |
glucokinase (hexokinase 4) |
347 |
0.78 |
| chr13_34061518_34061669 | 0.13 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
136826 |
0.05 |
| chr1_228073244_228073456 | 0.13 |
ENSG00000264483 |
. |
56034 |
0.1 |
| chr3_86997346_86997497 | 0.13 |
VGLL3 |
vestigial like 3 (Drosophila) |
42431 |
0.22 |
| chr10_32215556_32215707 | 0.13 |
ARHGAP12 |
Rho GTPase activating protein 12 |
2097 |
0.41 |
| chr3_15403025_15403378 | 0.13 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
20326 |
0.13 |
| chr1_236222466_236222646 | 0.13 |
ENSG00000252822 |
. |
1421 |
0.44 |
| chr17_27892819_27893926 | 0.13 |
ABHD15 |
abhydrolase domain containing 15 |
783 |
0.31 |
| chr15_96725215_96725403 | 0.13 |
NR2F2-AS1 |
NR2F2 antisense RNA 1 |
40975 |
0.19 |
| chr2_110872434_110873339 | 0.13 |
MALL |
mal, T-cell differentiation protein-like |
713 |
0.63 |
| chr19_43969380_43969688 | 0.13 |
LYPD3 |
LY6/PLAUR domain containing 3 |
278 |
0.87 |
| chr11_62169601_62169752 | 0.13 |
SCGB1A1 |
secretoglobin, family 1A, member 1 (uteroglobin) |
2899 |
0.16 |
| chr1_45273651_45274047 | 0.13 |
BTBD19 |
BTB (POZ) domain containing 19 |
305 |
0.68 |
| chr14_22849167_22849401 | 0.13 |
ENSG00000251002 |
. |
52435 |
0.09 |
| chr6_143227809_143227960 | 0.13 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
38454 |
0.19 |
| chr1_208000240_208000391 | 0.13 |
ENSG00000203709 |
. |
24447 |
0.21 |
| chr5_118503926_118504077 | 0.13 |
ENSG00000264536 |
. |
13669 |
0.18 |
| chr18_53542896_53543047 | 0.13 |
ENSG00000201816 |
. |
203854 |
0.03 |
| chr9_34253878_34254029 | 0.13 |
ENSG00000222426 |
. |
28715 |
0.11 |
| chr1_156087932_156088663 | 0.13 |
LMNA |
lamin A/C |
3784 |
0.13 |
| chr16_9192642_9192817 | 0.13 |
RP11-473I1.9 |
|
5976 |
0.16 |
| chr17_64297613_64298278 | 0.13 |
PRKCA |
protein kinase C, alpha |
999 |
0.57 |
| chr10_134754880_134755031 | 0.13 |
TTC40 |
tetratricopeptide repeat domain 40 |
1134 |
0.61 |
| chr7_116140807_116141026 | 0.13 |
CAV2 |
caveolin 2 |
1082 |
0.38 |
| chr5_45163064_45163215 | 0.13 |
MRPS30 |
mitochondrial ribosomal protein S30 |
354112 |
0.01 |
| chr19_5293472_5293908 | 0.13 |
PTPRS |
protein tyrosine phosphatase, receptor type, S |
427 |
0.88 |
| chr12_88966591_88966742 | 0.13 |
KITLG |
KIT ligand |
7572 |
0.25 |
| chr11_102702875_102703026 | 0.13 |
MMP3 |
matrix metallopeptidase 3 (stromelysin 1, progelatinase) |
6491 |
0.19 |
| chr1_111212990_111213454 | 0.13 |
KCNA3 |
potassium voltage-gated channel, shaker-related subfamily, member 3 |
4433 |
0.21 |
| chr12_39821314_39821465 | 0.13 |
KIF21A |
kinesin family member 21A |
15383 |
0.23 |
| chr1_156676036_156676613 | 0.13 |
CRABP2 |
cellular retinoic acid binding protein 2 |
716 |
0.47 |
| chr2_175496712_175497047 | 0.13 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
2428 |
0.33 |
| chr19_1512246_1512438 | 0.13 |
ADAMTSL5 |
ADAMTS-like 5 |
663 |
0.43 |
| chr15_73925957_73926274 | 0.13 |
NPTN |
neuroplastin |
360 |
0.91 |
| chr18_71409183_71409334 | 0.13 |
ENSG00000241866 |
. |
2912 |
0.42 |
| chr11_108541562_108541713 | 0.13 |
DDX10 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 10 |
5788 |
0.3 |
| chrX_101410162_101410938 | 0.13 |
BEX5 |
brain expressed, X-linked 5 |
212 |
0.92 |
| chr11_128382129_128382364 | 0.13 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
6957 |
0.25 |
| chr16_18033135_18033286 | 0.13 |
NPIPA8 |
nuclear pore complex interacting protein family, member A8 |
384882 |
0.01 |
| chr6_158406704_158406855 | 0.13 |
SYNJ2 |
synaptojanin 2 |
3709 |
0.23 |
| chr6_130070208_130070770 | 0.13 |
ENSG00000211996 |
. |
1607 |
0.48 |
| chr10_82214562_82215407 | 0.12 |
TSPAN14 |
tetraspanin 14 |
873 |
0.63 |
| chr7_104660604_104660779 | 0.12 |
KMT2E |
lysine (K)-specific methyltransferase 2E |
5783 |
0.17 |
| chr12_13056708_13056859 | 0.12 |
GPRC5A |
G protein-coupled receptor, family C, group 5, member A |
5220 |
0.15 |
| chr16_80308693_80308866 | 0.12 |
DYNLRB2 |
dynein, light chain, roadblock-type 2 |
265852 |
0.02 |
| chr3_9691165_9692185 | 0.12 |
MTMR14 |
myotubularin related protein 14 |
479 |
0.8 |
| chr20_61064540_61064691 | 0.12 |
GATA5 |
GATA binding protein 5 |
13589 |
0.16 |
| chr5_101952794_101952945 | 0.12 |
RP11-58B2.1 |
|
111896 |
0.07 |
| chr6_149805475_149805626 | 0.12 |
ZC3H12D |
zinc finger CCCH-type containing 12D |
576 |
0.72 |
| chr8_144815037_144815188 | 0.12 |
ENSG00000265660 |
. |
211 |
0.79 |
| chr4_39355902_39356119 | 0.12 |
RFC1 |
replication factor C (activator 1) 1, 145kDa |
11959 |
0.16 |
| chr1_205473812_205474567 | 0.12 |
CDK18 |
cyclin-dependent kinase 18 |
259 |
0.91 |
| chr10_30003940_30004091 | 0.12 |
ENSG00000222092 |
. |
3171 |
0.3 |
| chr17_75663137_75663644 | 0.12 |
SEPT9 |
septin 9 |
185395 |
0.03 |
| chr1_208418253_208418520 | 0.12 |
PLXNA2 |
plexin A2 |
721 |
0.82 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.1 | 0.1 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 | 0.2 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0034287 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.2 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0061072 | iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:0032928 | regulation of superoxide anion generation(GO:0032928) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.1 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.1 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.1 | GO:0070670 | response to interleukin-4(GO:0070670) |
| 0.0 | 0.0 | GO:0007262 | STAT protein import into nucleus(GO:0007262) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.1 | GO:0061364 | apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.1 | GO:0031579 | membrane raft organization(GO:0031579) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.1 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.1 | GO:0034122 | negative regulation of toll-like receptor signaling pathway(GO:0034122) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.0 | 0.0 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.0 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.0 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.0 | 0.0 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.0 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.2 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.0 | GO:0045110 | intermediate filament bundle assembly(GO:0045110) |
| 0.0 | 0.0 | GO:0046877 | regulation of saliva secretion(GO:0046877) |
| 0.0 | 0.0 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.0 | GO:0021819 | layer formation in cerebral cortex(GO:0021819) |
| 0.0 | 0.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.0 | GO:0097205 | regulation of glomerular filtration(GO:0003093) glomerular filtration(GO:0003094) renal filtration(GO:0097205) regulation of renal system process(GO:0098801) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.0 | GO:0060065 | uterus development(GO:0060065) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0016081 | synaptic vesicle docking(GO:0016081) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0016103 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0090026 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.0 | GO:0071364 | cellular response to epidermal growth factor stimulus(GO:0071364) |
| 0.0 | 0.0 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.0 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.1 | GO:0035089 | establishment of apical/basal cell polarity(GO:0035089) establishment of monopolar cell polarity(GO:0061162) establishment or maintenance of monopolar cell polarity(GO:0061339) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.2 | GO:0046677 | response to antibiotic(GO:0046677) |
| 0.0 | 0.0 | GO:0006925 | inflammatory cell apoptotic process(GO:0006925) |
| 0.0 | 0.0 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.0 | GO:0048714 | positive regulation of oligodendrocyte differentiation(GO:0048714) |
| 0.0 | 0.0 | GO:0071277 | cellular response to calcium ion(GO:0071277) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.2 | GO:0035115 | embryonic forelimb morphogenesis(GO:0035115) |
| 0.0 | 0.0 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.0 | GO:0060664 | epithelial cell proliferation involved in salivary gland morphogenesis(GO:0060664) |
| 0.0 | 0.1 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.0 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0002093 | auditory receptor cell morphogenesis(GO:0002093) auditory receptor cell stereocilium organization(GO:0060088) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.0 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.0 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.0 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.0 | GO:0031904 | endosome lumen(GO:0031904) |
| 0.0 | 0.1 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.0 | GO:0016939 | kinesin II complex(GO:0016939) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.1 | GO:0052658 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.2 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.1 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0031779 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0000977 | transcription regulatory region sequence-specific DNA binding(GO:0000976) RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.2 | GO:0052866 | phosphatidylinositol phosphate phosphatase activity(GO:0052866) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.0 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0008967 | phosphoglycolate phosphatase activity(GO:0008967) |
| 0.0 | 0.1 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.0 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.0 | GO:1990939 | ATP-dependent microtubule motor activity, plus-end-directed(GO:0008574) ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.0 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.0 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.2 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.1 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.3 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.2 | REACTOME RECRUITMENT OF NUMA TO MITOTIC CENTROSOMES | Genes involved in Recruitment of NuMA to mitotic centrosomes |
| 0.0 | 0.2 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.1 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.1 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |