Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
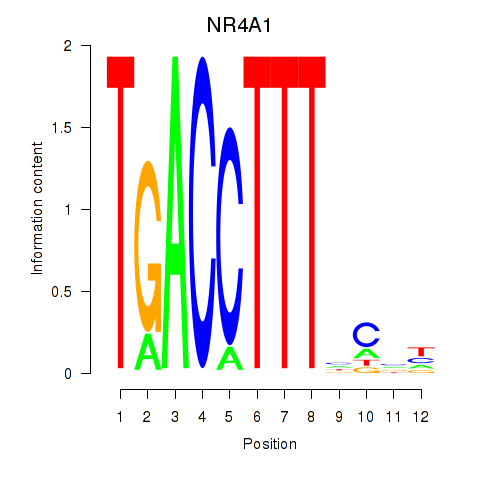
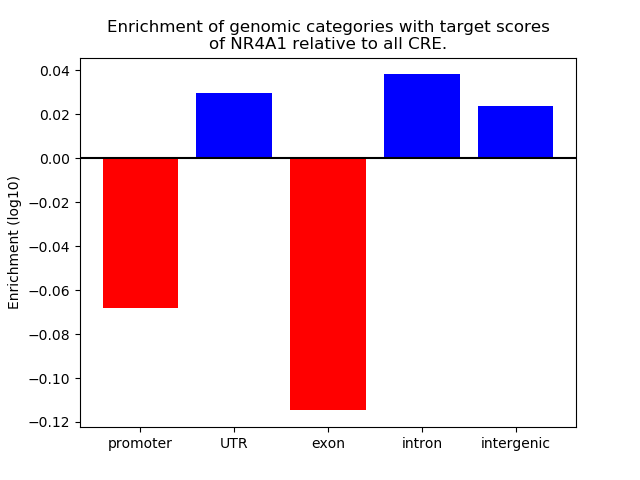
Results for NR4A1
Z-value: 0.99
Transcription factors associated with NR4A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A1
|
ENSG00000123358.15 | nuclear receptor subfamily 4 group A member 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr12_52418688_52418839 | NR4A1 | 2147 | 0.213783 | -0.79 | 1.1e-02 | Click! |
| chr12_52419741_52419907 | NR4A1 | 3208 | 0.165001 | 0.67 | 4.9e-02 | Click! |
| chr12_52438503_52438654 | NR4A1 | 2989 | 0.163142 | 0.62 | 7.5e-02 | Click! |
| chr12_52431070_52431659 | NR4A1 | 135 | 0.937063 | 0.62 | 7.8e-02 | Click! |
| chr12_52418124_52418275 | NR4A1 | 1583 | 0.277251 | -0.61 | 7.9e-02 | Click! |
Activity of the NR4A1 motif across conditions
Conditions sorted by the z-value of the NR4A1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
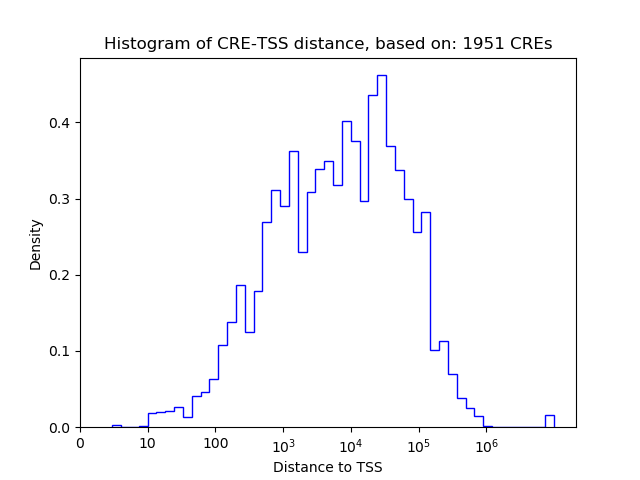
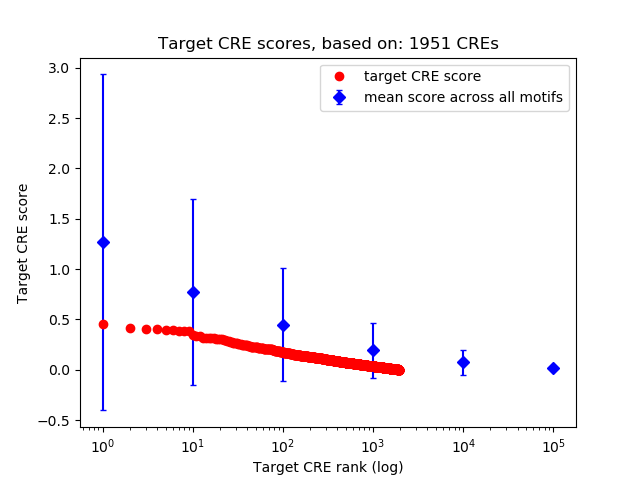
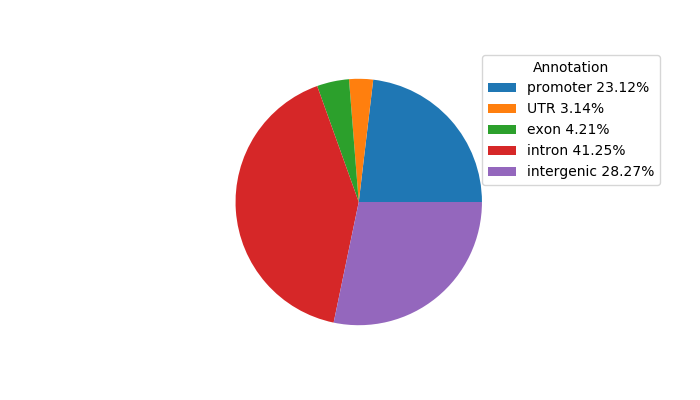
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_158252834_158252985 | 0.45 |
RP11-52J3.3 |
|
1304 |
0.44 |
| chr4_138531037_138531225 | 0.42 |
PCDH18 |
protocadherin 18 |
77483 |
0.13 |
| chr6_81974819_81975050 | 0.41 |
RP1-300G12.2 |
|
269696 |
0.02 |
| chr15_60688043_60688242 | 0.40 |
ANXA2 |
annexin A2 |
1395 |
0.54 |
| chr12_124872214_124872365 | 0.40 |
NCOR2 |
nuclear receptor corepressor 2 |
1081 |
0.65 |
| chr14_55117584_55117850 | 0.39 |
SAMD4A |
sterile alpha motif domain containing 4A |
83080 |
0.09 |
| chr15_62798764_62798915 | 0.39 |
TLN2 |
talin 2 |
54725 |
0.15 |
| chr1_46603624_46603795 | 0.39 |
RP4-533D7.5 |
|
3506 |
0.18 |
| chr22_46471692_46472277 | 0.38 |
FLJ27365 |
hsa-mir-4763 |
4208 |
0.11 |
| chr22_26852060_26852211 | 0.34 |
HPS4 |
Hermansky-Pudlak syndrome 4 |
23212 |
0.12 |
| chr3_16939119_16939271 | 0.34 |
PLCL2 |
phospholipase C-like 2 |
12743 |
0.25 |
| chr12_16759185_16759513 | 0.33 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
116 |
0.98 |
| chr7_66532196_66532424 | 0.32 |
ENSG00000266525 |
. |
47074 |
0.12 |
| chr3_159745160_159745451 | 0.32 |
LINC01100 |
long intergenic non-protein coding RNA 1100 |
11494 |
0.19 |
| chr15_56199751_56199985 | 0.32 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
7984 |
0.25 |
| chr3_125321989_125322196 | 0.32 |
OSBPL11 |
oxysterol binding protein-like 11 |
8158 |
0.22 |
| chr16_72885110_72885261 | 0.32 |
ENSG00000251868 |
. |
29294 |
0.16 |
| chr6_19838575_19839711 | 0.31 |
RP1-167F1.2 |
|
168 |
0.95 |
| chr17_72868164_72868332 | 0.31 |
FDXR |
ferredoxin reductase |
838 |
0.46 |
| chr1_240760678_240760859 | 0.30 |
RP11-467I20.6 |
|
6720 |
0.21 |
| chr18_32995509_32995753 | 0.30 |
ZNF396 |
zinc finger protein 396 |
38330 |
0.16 |
| chr12_106478299_106478450 | 0.30 |
NUAK1 |
NUAK family, SNF1-like kinase, 1 |
558 |
0.81 |
| chr7_72209303_72209642 | 0.30 |
TYW1B |
tRNA-yW synthesizing protein 1 homolog B (S. cerevisiae) |
253 |
0.92 |
| chr3_129322348_129322499 | 0.29 |
PLXND1 |
plexin D1 |
3238 |
0.22 |
| chr2_175863049_175863322 | 0.29 |
CHN1 |
chimerin 1 |
6769 |
0.16 |
| chr14_51897014_51897206 | 0.28 |
FRMD6 |
FERM domain containing 6 |
58745 |
0.13 |
| chr9_130328652_130328977 | 0.27 |
FAM129B |
family with sequence similarity 129, member B |
2553 |
0.26 |
| chr8_99956690_99957063 | 0.27 |
OSR2 |
odd-skipped related transciption factor 2 |
11 |
0.97 |
| chr10_33624842_33625153 | 0.27 |
NRP1 |
neuropilin 1 |
193 |
0.96 |
| chr6_84139999_84140706 | 0.26 |
ME1 |
malic enzyme 1, NADP(+)-dependent, cytosolic |
412 |
0.89 |
| chr1_157980874_157981318 | 0.26 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
14244 |
0.2 |
| chr1_161992740_161992891 | 0.26 |
OLFML2B |
olfactomedin-like 2B |
615 |
0.79 |
| chr15_96883153_96883304 | 0.26 |
ENSG00000222651 |
. |
6738 |
0.16 |
| chr4_180106134_180106285 | 0.25 |
ENSG00000212191 |
. |
499099 |
0.0 |
| chr12_58819523_58819907 | 0.25 |
RP11-362K2.2 |
Protein LOC100506869 |
118192 |
0.06 |
| chr7_90033445_90033883 | 0.25 |
CLDN12 |
claudin 12 |
783 |
0.69 |
| chr17_17804572_17804723 | 0.25 |
TOM1L2 |
target of myb1-like 2 (chicken) |
29269 |
0.14 |
| chr15_48010788_48011252 | 0.25 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
334 |
0.93 |
| chr17_79315981_79316442 | 0.24 |
TMEM105 |
transmembrane protein 105 |
11737 |
0.13 |
| chr5_108790950_108791101 | 0.24 |
PJA2 |
praja ring finger 2, E3 ubiquitin protein ligase |
45330 |
0.2 |
| chr2_189842191_189842342 | 0.23 |
ENSG00000221502 |
. |
552 |
0.78 |
| chr8_119115834_119115985 | 0.23 |
EXT1 |
exostosin glycosyltransferase 1 |
6744 |
0.34 |
| chr1_47489778_47489953 | 0.23 |
CYP4X1 |
cytochrome P450, family 4, subfamily X, polypeptide 1 |
625 |
0.75 |
| chr6_17417832_17418048 | 0.23 |
CAP2 |
CAP, adenylate cyclase-associated protein, 2 (yeast) |
23997 |
0.25 |
| chr7_83819706_83819857 | 0.23 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
4436 |
0.37 |
| chrX_39759988_39760170 | 0.23 |
ENSG00000263972 |
. |
63264 |
0.14 |
| chr17_38439762_38439991 | 0.23 |
CDC6 |
cell division cycle 6 |
4009 |
0.16 |
| chr8_61626221_61626372 | 0.22 |
CHD7 |
chromodomain helicase DNA binding protein 7 |
27628 |
0.23 |
| chr3_11633135_11633286 | 0.22 |
VGLL4 |
vestigial like 4 (Drosophila) |
9345 |
0.2 |
| chr2_238316542_238316753 | 0.22 |
COL6A3 |
collagen, type VI, alpha 3 |
6144 |
0.22 |
| chr14_20903201_20903901 | 0.22 |
KLHL33 |
kelch-like family member 33 |
250 |
0.81 |
| chr1_172105427_172105578 | 0.22 |
ENSG00000207949 |
. |
2545 |
0.27 |
| chr17_13503336_13503489 | 0.22 |
HS3ST3A1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1 |
1832 |
0.45 |
| chr22_24109124_24109275 | 0.22 |
CHCHD10 |
coiled-coil-helix-coiled-coil-helix domain containing 10 |
864 |
0.37 |
| chr6_149068477_149068835 | 0.22 |
UST |
uronyl-2-sulfotransferase |
192 |
0.97 |
| chr4_177714164_177714554 | 0.22 |
VEGFC |
vascular endothelial growth factor C |
478 |
0.89 |
| chr12_54417118_54417269 | 0.21 |
HOXC6 |
homeobox C6 |
4949 |
0.07 |
| chr18_11096558_11096709 | 0.21 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
51954 |
0.18 |
| chr12_16758688_16759086 | 0.21 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
17 |
0.99 |
| chr7_2842191_2842738 | 0.21 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
12428 |
0.25 |
| chr17_5326109_5326345 | 0.21 |
RPAIN |
RPA interacting protein |
2705 |
0.15 |
| chr6_150176715_150177267 | 0.21 |
RP11-350J20.12 |
|
3393 |
0.15 |
| chr4_177707466_177707742 | 0.21 |
VEGFC |
vascular endothelial growth factor C |
6277 |
0.32 |
| chr10_4283577_4283877 | 0.21 |
ENSG00000207124 |
. |
273417 |
0.02 |
| chr19_17797231_17797557 | 0.21 |
UNC13A |
unc-13 homolog A (C. elegans) |
1614 |
0.31 |
| chr8_74333530_74333818 | 0.21 |
RP11-434I12.2 |
|
64978 |
0.13 |
| chrX_31284306_31284787 | 0.21 |
DMD |
dystrophin |
428 |
0.9 |
| chr8_22025466_22025617 | 0.21 |
BMP1 |
bone morphogenetic protein 1 |
2741 |
0.16 |
| chr9_133590287_133590909 | 0.21 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
1231 |
0.49 |
| chr10_93373658_93374073 | 0.20 |
PPP1R3C |
protein phosphatase 1, regulatory subunit 3C |
18946 |
0.27 |
| chr21_17652021_17652264 | 0.20 |
ENSG00000201025 |
. |
4947 |
0.35 |
| chr7_25469565_25469726 | 0.20 |
ENSG00000222101 |
. |
140643 |
0.05 |
| chr9_18476160_18476529 | 0.20 |
ADAMTSL1 |
ADAMTS-like 1 |
2113 |
0.45 |
| chr8_135212673_135212824 | 0.20 |
ZFAT |
zinc finger and AT hook domain containing |
309677 |
0.01 |
| chrX_135230667_135231011 | 0.20 |
FHL1 |
four and a half LIM domains 1 |
102 |
0.97 |
| chr5_135389675_135389826 | 0.20 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
4545 |
0.23 |
| chr20_60931125_60931499 | 0.20 |
RP11-157P1.5 |
|
3243 |
0.15 |
| chr13_52164714_52165080 | 0.20 |
ENSG00000242893 |
. |
3454 |
0.16 |
| chr6_43485479_43485674 | 0.20 |
POLR1C |
polymerase (RNA) I polypeptide C, 30kDa |
726 |
0.33 |
| chr5_58187479_58187710 | 0.19 |
CTD-2176I21.2 |
|
28353 |
0.22 |
| chr6_58148368_58148782 | 0.19 |
ENSG00000212017 |
. |
893645 |
0.0 |
| chr2_227657217_227657687 | 0.19 |
IRS1 |
insulin receptor substrate 1 |
7023 |
0.22 |
| chr6_116571521_116571672 | 0.19 |
TSPYL4 |
TSPY-like 4 |
3665 |
0.18 |
| chr3_143568633_143568784 | 0.19 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
1335 |
0.61 |
| chr7_84119974_84120125 | 0.19 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
1816 |
0.53 |
| chr12_26110310_26110527 | 0.19 |
RASSF8-AS1 |
RASSF8 atnisense RNA 1 |
100 |
0.96 |
| chr2_219149046_219149220 | 0.19 |
TMBIM1 |
transmembrane BAX inhibitor motif containing 1 |
1524 |
0.24 |
| chr12_115889903_115890336 | 0.19 |
RP11-116D17.1 |
HCG2038717; Uncharacterized protein |
89302 |
0.1 |
| chr4_7136849_7137000 | 0.19 |
ENSG00000200867 |
. |
22798 |
0.16 |
| chr7_43651852_43652098 | 0.18 |
STK17A |
serine/threonine kinase 17a |
29311 |
0.15 |
| chr6_26721817_26722019 | 0.18 |
ZNF322 |
zinc finger protein 322 |
61938 |
0.11 |
| chr12_96590087_96590334 | 0.18 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
1818 |
0.37 |
| chr1_228272671_228272822 | 0.18 |
ARF1 |
ADP-ribosylation factor 1 |
1895 |
0.23 |
| chr3_59110595_59110926 | 0.18 |
C3orf67 |
chromosome 3 open reading frame 67 |
74950 |
0.13 |
| chr5_169369055_169369206 | 0.17 |
FAM196B |
family with sequence similarity 196, member B |
38614 |
0.19 |
| chr9_116569857_116570008 | 0.17 |
ENSG00000211556 |
. |
15213 |
0.25 |
| chr12_45628611_45628785 | 0.17 |
ANO6 |
anoctamin 6 |
18795 |
0.27 |
| chr7_90711533_90711739 | 0.17 |
FZD1 |
frizzled family receptor 1 |
182147 |
0.03 |
| chr5_98397075_98397538 | 0.17 |
ENSG00000200351 |
. |
124855 |
0.06 |
| chr22_30022409_30022560 | 0.17 |
NF2 |
neurofibromin 2 (merlin) |
22496 |
0.14 |
| chr15_96879714_96879865 | 0.17 |
ENSG00000222651 |
. |
3299 |
0.19 |
| chr1_43218988_43219139 | 0.17 |
LEPRE1 |
leucine proline-enriched proteoglycan (leprecan) 1 |
1595 |
0.28 |
| chr8_25868117_25868310 | 0.17 |
EBF2 |
early B-cell factor 2 |
30791 |
0.24 |
| chr7_73130944_73131128 | 0.17 |
STX1A |
syntaxin 1A (brain) |
2925 |
0.14 |
| chr16_10678659_10678810 | 0.17 |
EMP2 |
epithelial membrane protein 2 |
4179 |
0.21 |
| chr20_61863942_61864093 | 0.17 |
BIRC7 |
baculoviral IAP repeat containing 7 |
3218 |
0.15 |
| chr6_3749707_3750198 | 0.17 |
RP11-420L9.5 |
|
1393 |
0.41 |
| chr6_26745930_26746188 | 0.17 |
ZNF322 |
zinc finger protein 322 |
86079 |
0.08 |
| chr8_97536368_97536646 | 0.17 |
SDC2 |
syndecan 2 |
30271 |
0.22 |
| chr7_32627060_32627248 | 0.17 |
AVL9 |
AVL9 homolog (S. cerevisiase) |
44287 |
0.18 |
| chr3_197099341_197099661 | 0.17 |
ENSG00000238491 |
. |
33079 |
0.17 |
| chr10_93170267_93170919 | 0.17 |
HECTD2 |
HECT domain containing E3 ubiquitin protein ligase 2 |
491 |
0.89 |
| chr4_157695428_157695747 | 0.17 |
RP11-154F14.2 |
|
66924 |
0.12 |
| chrX_22670943_22671094 | 0.17 |
ENSG00000265819 |
. |
15909 |
0.29 |
| chr10_15390844_15391029 | 0.16 |
FAM171A1 |
family with sequence similarity 171, member A1 |
22122 |
0.25 |
| chr22_44756680_44757093 | 0.16 |
RP1-32I10.10 |
Uncharacterized protein |
4545 |
0.29 |
| chr17_41476988_41477313 | 0.16 |
ARL4D |
ADP-ribosylation factor-like 4D |
823 |
0.46 |
| chr4_105416319_105416884 | 0.16 |
CXXC4 |
CXXC finger protein 4 |
550 |
0.77 |
| chr8_99438878_99439083 | 0.16 |
KCNS2 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
270 |
0.92 |
| chr6_28641788_28642196 | 0.16 |
ENSG00000272278 |
. |
25950 |
0.18 |
| chr2_150187104_150187520 | 0.16 |
LYPD6 |
LY6/PLAUR domain containing 6 |
261 |
0.96 |
| chr6_155654647_155654798 | 0.16 |
TFB1M |
transcription factor B1, mitochondrial |
19104 |
0.23 |
| chr8_495040_495212 | 0.16 |
TDRP |
testis development related protein |
128 |
0.98 |
| chr15_95847267_95847418 | 0.16 |
ENSG00000222076 |
. |
441691 |
0.01 |
| chr22_31479185_31479426 | 0.16 |
RP3-412A9.16 |
|
246 |
0.8 |
| chr2_3699150_3699650 | 0.16 |
ALLC |
allantoicase |
6385 |
0.16 |
| chr2_192111446_192111930 | 0.16 |
MYO1B |
myosin IB |
679 |
0.79 |
| chr15_57669149_57669410 | 0.16 |
CGNL1 |
cingulin-like 1 |
576 |
0.8 |
| chr16_88922465_88922880 | 0.15 |
TRAPPC2L |
trafficking protein particle complex 2-like |
44 |
0.8 |
| chr6_5499328_5499479 | 0.15 |
RP1-232P20.1 |
|
41095 |
0.2 |
| chr8_32406806_32407051 | 0.15 |
NRG1 |
neuregulin 1 |
683 |
0.82 |
| chr7_95401616_95402462 | 0.15 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
123 |
0.98 |
| chr12_12658847_12659363 | 0.15 |
DUSP16 |
dual specificity phosphatase 16 |
14954 |
0.24 |
| chr9_73216401_73216607 | 0.15 |
ENSG00000272232 |
. |
22127 |
0.25 |
| chr10_62587657_62587808 | 0.15 |
CDK1 |
cyclin-dependent kinase 1 |
47821 |
0.16 |
| chr5_58484501_58484817 | 0.15 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
87286 |
0.1 |
| chr2_207905869_207906341 | 0.15 |
ENSG00000253008 |
. |
68692 |
0.1 |
| chr18_52625984_52626182 | 0.15 |
CCDC68 |
coiled-coil domain containing 68 |
656 |
0.76 |
| chr2_208199752_208200019 | 0.15 |
ENSG00000221628 |
. |
65737 |
0.12 |
| chr2_42599068_42599219 | 0.15 |
COX7A2L |
cytochrome c oxidase subunit VIIa polypeptide 2 like |
2993 |
0.32 |
| chr1_144928639_144928790 | 0.15 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
3434 |
0.19 |
| chr5_154229931_154230213 | 0.15 |
FAXDC2 |
fatty acid hydroxylase domain containing 2 |
91 |
0.96 |
| chr7_13056983_13057208 | 0.15 |
ENSG00000222974 |
. |
140786 |
0.05 |
| chr6_99962436_99962683 | 0.15 |
USP45 |
ubiquitin specific peptidase 45 |
693 |
0.65 |
| chr18_32173182_32173613 | 0.15 |
DTNA |
dystrobrevin, alpha |
68 |
0.99 |
| chr4_129439243_129439469 | 0.15 |
PGRMC2 |
progesterone receptor membrane component 2 |
229372 |
0.02 |
| chr22_36138598_36138911 | 0.15 |
APOL5 |
apolipoprotein L, 5 |
24835 |
0.19 |
| chr3_149371875_149372212 | 0.15 |
WWTR1-AS1 |
WWTR1 antisense RNA 1 |
2764 |
0.22 |
| chrX_100334578_100334939 | 0.15 |
TMEM35 |
transmembrane protein 35 |
1049 |
0.47 |
| chr4_108314088_108314239 | 0.15 |
ENSG00000252470 |
. |
42112 |
0.2 |
| chr2_135130087_135130255 | 0.14 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
118341 |
0.06 |
| chr2_234390115_234390266 | 0.14 |
USP40 |
ubiquitin specific peptidase 40 |
7917 |
0.22 |
| chr4_181748441_181748629 | 0.14 |
NA |
NA |
> 106 |
NA |
| chr1_12609904_12610111 | 0.14 |
ENSG00000221340 |
. |
20093 |
0.16 |
| chr5_52333250_52333558 | 0.14 |
CTD-2175A23.1 |
|
47296 |
0.12 |
| chr16_25022284_25022579 | 0.14 |
ARHGAP17 |
Rho GTPase activating protein 17 |
4221 |
0.31 |
| chr14_52584008_52584159 | 0.14 |
NID2 |
nidogen 2 (osteonidogen) |
48371 |
0.15 |
| chr4_170086286_170086437 | 0.14 |
RP11-327O17.2 |
|
36584 |
0.2 |
| chr11_133917712_133918072 | 0.14 |
JAM3 |
junctional adhesion molecule 3 |
20928 |
0.21 |
| chr21_30464791_30465071 | 0.14 |
MAP3K7CL |
MAP3K7 C-terminal like |
356 |
0.83 |
| chr10_128076215_128076453 | 0.14 |
ADAM12 |
ADAM metallopeptidase domain 12 |
690 |
0.79 |
| chr13_110780795_110780946 | 0.14 |
ENSG00000265885 |
. |
30372 |
0.24 |
| chr8_16815000_16815151 | 0.14 |
FGF20 |
fibroblast growth factor 20 |
44274 |
0.17 |
| chr17_6451663_6451961 | 0.14 |
PITPNM3 |
PITPNM family member 3 |
7968 |
0.17 |
| chr15_51039395_51039546 | 0.14 |
SPPL2A |
signal peptide peptidase like 2A |
258 |
0.75 |
| chr5_141703297_141704624 | 0.14 |
SPRY4 |
sprouty homolog 4 (Drosophila) |
213 |
0.88 |
| chr10_36184487_36184638 | 0.14 |
FZD8 |
frizzled family receptor 8 |
254200 |
0.02 |
| chr3_185651354_185651505 | 0.14 |
TRA2B |
transformer 2 beta homolog (Drosophila) |
4372 |
0.23 |
| chr19_18761571_18761809 | 0.14 |
KLHL26 |
kelch-like family member 26 |
13823 |
0.1 |
| chr16_85202371_85202571 | 0.14 |
CTC-786C10.1 |
|
2411 |
0.35 |
| chr11_101278345_101278496 | 0.14 |
ENSG00000263885 |
. |
112216 |
0.07 |
| chr4_81198150_81198317 | 0.14 |
FGF5 |
fibroblast growth factor 5 |
10440 |
0.25 |
| chr2_145417620_145417771 | 0.14 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
139074 |
0.05 |
| chr9_40631681_40631832 | 0.14 |
SPATA31A3 |
SPATA31 subfamily A, member 3 |
68535 |
0.13 |
| chr19_8426733_8426884 | 0.14 |
ANGPTL4 |
angiopoietin-like 4 |
1365 |
0.25 |
| chr9_89996647_89996873 | 0.14 |
DAPK1 |
death-associated protein kinase 1 |
115383 |
0.06 |
| chr2_138721657_138722065 | 0.14 |
HNMT |
histamine N-methyltransferase |
19 |
0.99 |
| chr1_9473851_9474134 | 0.14 |
ENSG00000252956 |
. |
23845 |
0.2 |
| chr19_2639313_2639495 | 0.13 |
CTC-265F19.1 |
|
4447 |
0.15 |
| chr13_42616117_42616369 | 0.13 |
DGKH |
diacylglycerol kinase, eta |
2067 |
0.3 |
| chr18_32290285_32290488 | 0.13 |
DTNA |
dystrobrevin, alpha |
125 |
0.98 |
| chr3_58258156_58258366 | 0.13 |
RPP14 |
ribonuclease P/MRP 14kDa subunit |
33728 |
0.14 |
| chr1_16702300_16702515 | 0.13 |
SZRD1 |
SUZ RNA binding domain containing 1 |
8647 |
0.15 |
| chr2_47260836_47261197 | 0.13 |
AC093732.1 |
|
6647 |
0.2 |
| chr2_44625453_44625606 | 0.13 |
CAMKMT |
calmodulin-lysine N-methyltransferase |
25648 |
0.2 |
| chr17_46561109_46561276 | 0.13 |
ENSG00000206805 |
. |
4733 |
0.14 |
| chr2_24398914_24399065 | 0.13 |
FAM228A |
family with sequence similarity 228, member A |
635 |
0.51 |
| chr2_166430745_166430996 | 0.13 |
CSRNP3 |
cysteine-serine-rich nuclear protein 3 |
251 |
0.96 |
| chr18_60904701_60905063 | 0.13 |
ENSG00000238988 |
. |
42984 |
0.14 |
| chr14_94426547_94426698 | 0.13 |
ASB2 |
ankyrin repeat and SOCS box containing 2 |
2855 |
0.21 |
| chr6_1311351_1312220 | 0.13 |
FOXQ1 |
forkhead box Q1 |
890 |
0.69 |
| chr8_77596836_77596987 | 0.13 |
ZFHX4 |
zinc finger homeobox 4 |
895 |
0.54 |
| chr14_62033353_62033504 | 0.13 |
RP11-47I22.3 |
Uncharacterized protein |
3886 |
0.24 |
| chr18_25754335_25754486 | 0.13 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
3000 |
0.42 |
| chr17_42624321_42624535 | 0.13 |
FZD2 |
frizzled family receptor 2 |
10497 |
0.18 |
| chr11_131758303_131758454 | 0.13 |
AP004372.1 |
|
8624 |
0.25 |
| chr12_66093282_66093481 | 0.13 |
HMGA2 |
high mobility group AT-hook 2 |
124530 |
0.05 |
| chr2_101437362_101437649 | 0.13 |
NPAS2 |
neuronal PAS domain protein 2 |
18 |
0.97 |
| chr10_29824642_29824876 | 0.13 |
ENSG00000207612 |
. |
9267 |
0.19 |
| chr3_75690482_75691208 | 0.13 |
ENSG00000221795 |
. |
10931 |
0.2 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.3 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.1 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0016188 | synaptic vesicle maturation(GO:0016188) |
| 0.0 | 0.1 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.3 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.2 | GO:0099518 | vesicle transport along microtubule(GO:0047496) vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.2 | GO:0002347 | response to tumor cell(GO:0002347) |
| 0.0 | 0.0 | GO:0021801 | cerebral cortex radial glia guided migration(GO:0021801) telencephalon glial cell migration(GO:0022030) |
| 0.0 | 0.0 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.0 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.0 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.0 | GO:0070092 | regulation of glucagon secretion(GO:0070092) |
| 0.0 | 0.0 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.2 | GO:0042745 | circadian sleep/wake cycle(GO:0042745) |
| 0.0 | 0.0 | GO:0061042 | vascular wound healing(GO:0061042) regulation of vascular wound healing(GO:0061043) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.0 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.0 | GO:0051590 | positive regulation of neurotransmitter transport(GO:0051590) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0007520 | myoblast fusion(GO:0007520) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.2 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.0 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.1 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.3 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0010181 | FMN binding(GO:0010181) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.2 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.0 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.0 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.1 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.0 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) |
| 0.0 | 0.0 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.0 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |