Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
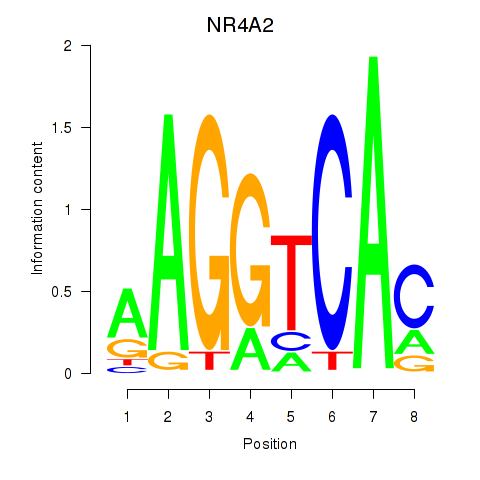
Results for NR4A2
Z-value: 0.40
Transcription factors associated with NR4A2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR4A2
|
ENSG00000153234.9 | nuclear receptor subfamily 4 group A member 2 |
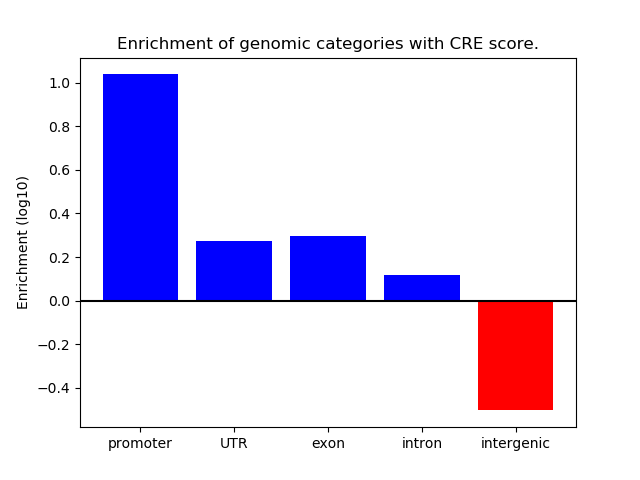
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
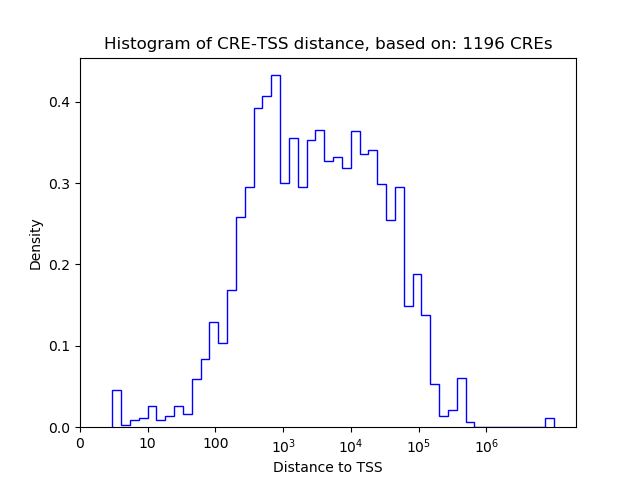
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_157178968_157179119 | NR4A2 | 7590 | 0.280844 | -0.81 | 8.0e-03 | Click! |
| chr2_156698821_156698972 | NR4A2 | 487737 | 0.005077 | -0.71 | 3.3e-02 | Click! |
| chr2_157176521_157176855 | NR4A2 | 9945 | 0.273363 | -0.68 | 4.5e-02 | Click! |
| chr2_157177580_157177731 | NR4A2 | 8978 | 0.275974 | -0.67 | 4.7e-02 | Click! |
| chr2_157186149_157186300 | NR4A2 | 409 | 0.898176 | -0.66 | 5.5e-02 | Click! |
Activity of the NR4A2 motif across conditions
Conditions sorted by the z-value of the NR4A2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
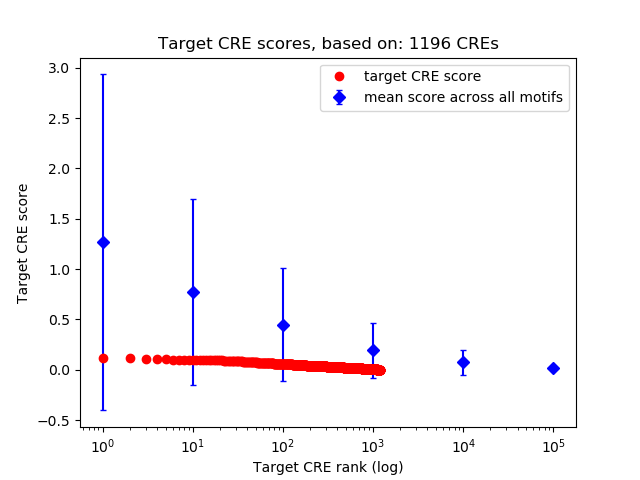
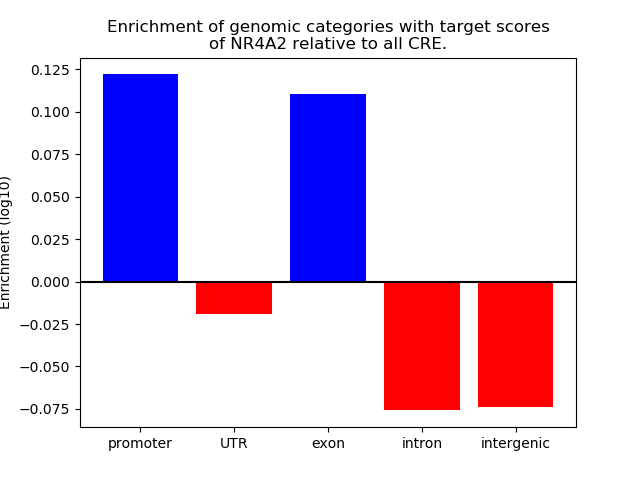
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_32718292_32718517 | 0.12 |
LCK |
lymphocyte-specific protein tyrosine kinase |
1529 |
0.2 |
| chr1_6524907_6525205 | 0.11 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
634 |
0.58 |
| chr11_82828119_82828301 | 0.11 |
PCF11 |
PCF11 cleavage and polyadenylation factor subunit |
39820 |
0.12 |
| chr14_100531999_100532513 | 0.11 |
EVL |
Enah/Vasp-like |
498 |
0.76 |
| chr11_121324494_121324645 | 0.11 |
RP11-730K11.1 |
|
847 |
0.59 |
| chr9_134127628_134128133 | 0.10 |
FAM78A |
family with sequence similarity 78, member A |
18000 |
0.15 |
| chr1_198616062_198616213 | 0.10 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
7845 |
0.25 |
| chr11_117860130_117860281 | 0.10 |
IL10RA |
interleukin 10 receptor, alpha |
3096 |
0.25 |
| chr5_175085682_175085863 | 0.10 |
HRH2 |
histamine receptor H2 |
739 |
0.7 |
| chr7_50349443_50349694 | 0.10 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
1250 |
0.61 |
| chr7_106506867_106507018 | 0.10 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
1018 |
0.65 |
| chr7_8171794_8172008 | 0.10 |
AC006042.6 |
|
18246 |
0.2 |
| chrX_128916701_128916852 | 0.10 |
SASH3 |
SAM and SH3 domain containing 3 |
2816 |
0.27 |
| chr13_30510013_30510639 | 0.10 |
LINC00572 |
long intergenic non-protein coding RNA 572 |
9538 |
0.3 |
| chr11_64511299_64511931 | 0.09 |
RASGRP2 |
RAS guanyl releasing protein 2 (calcium and DAG-regulated) |
2 |
0.96 |
| chr12_46611391_46611598 | 0.09 |
SLC38A1 |
solute carrier family 38, member 1 |
49990 |
0.18 |
| chrX_46432402_46432561 | 0.09 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
738 |
0.71 |
| chr14_22966248_22966484 | 0.09 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
10195 |
0.1 |
| chr1_198611164_198611406 | 0.09 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
2993 |
0.32 |
| chr19_39108076_39108634 | 0.09 |
MAP4K1 |
mitogen-activated protein kinase kinase kinase kinase 1 |
209 |
0.87 |
| chr8_66843424_66843611 | 0.09 |
PDE7A |
phosphodiesterase 7A |
89330 |
0.09 |
| chr3_121379556_121379755 | 0.09 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
90 |
0.96 |
| chr19_54880943_54881230 | 0.09 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
972 |
0.39 |
| chr7_142424483_142424634 | 0.09 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
32761 |
0.16 |
| chr14_102281143_102281546 | 0.09 |
CTD-2017C7.2 |
|
4686 |
0.17 |
| chr5_142177952_142178103 | 0.09 |
ARHGAP26 |
Rho GTPase activating protein 26 |
25745 |
0.22 |
| chr11_65185943_65186437 | 0.09 |
ENSG00000245532 |
. |
25739 |
0.09 |
| chr10_30316643_30316960 | 0.09 |
KIAA1462 |
KIAA1462 |
31652 |
0.24 |
| chr1_198617358_198617617 | 0.09 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
9195 |
0.24 |
| chr21_35884173_35884324 | 0.09 |
KCNE1 |
potassium voltage-gated channel, Isk-related family, member 1 |
325 |
0.89 |
| chr22_44391651_44392034 | 0.09 |
PARVB |
parvin, beta |
3249 |
0.27 |
| chr14_22974214_22974379 | 0.09 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
18125 |
0.09 |
| chr8_27237778_27238009 | 0.08 |
PTK2B |
protein tyrosine kinase 2 beta |
275 |
0.93 |
| chr17_38483011_38483310 | 0.08 |
RARA |
retinoic acid receptor, alpha |
8623 |
0.11 |
| chr17_38018344_38018495 | 0.08 |
IKZF3 |
IKAROS family zinc finger 3 (Aiolos) |
1960 |
0.26 |
| chr14_22946001_22946152 | 0.08 |
TRAJ60 |
T cell receptor alpha joining 60 (pseudogene) |
780 |
0.46 |
| chr15_75082239_75082553 | 0.08 |
ENSG00000264386 |
. |
1298 |
0.27 |
| chr17_28085184_28085335 | 0.08 |
RP11-82O19.1 |
|
2862 |
0.22 |
| chr14_98443402_98443682 | 0.08 |
C14orf64 |
chromosome 14 open reading frame 64 |
841 |
0.77 |
| chr7_38389126_38389277 | 0.08 |
AMPH |
amphiphysin |
113512 |
0.07 |
| chr1_150536239_150536708 | 0.08 |
ADAMTSL4-AS1 |
ADAMTSL4 antisense RNA 1 |
2504 |
0.12 |
| chr20_39768553_39768784 | 0.08 |
RP1-1J6.2 |
|
2025 |
0.31 |
| chrX_128910765_128910976 | 0.08 |
SASH3 |
SAM and SH3 domain containing 3 |
3090 |
0.26 |
| chr7_45016674_45016991 | 0.08 |
MYO1G |
myosin IG |
1865 |
0.25 |
| chr17_76755792_76756034 | 0.08 |
CYTH1 |
cytohesin 1 |
22441 |
0.17 |
| chrX_48772131_48772362 | 0.08 |
PIM2 |
pim-2 oncogene |
766 |
0.45 |
| chr16_88767040_88767399 | 0.08 |
RNF166 |
ring finger protein 166 |
208 |
0.83 |
| chr19_3478950_3479105 | 0.08 |
C19orf77 |
chromosome 19 open reading frame 77 |
59 |
0.95 |
| chr1_167602601_167602772 | 0.08 |
RP3-455J7.4 |
|
2775 |
0.25 |
| chr16_81030456_81031156 | 0.07 |
CMC2 |
C-x(9)-C motif containing 2 |
1458 |
0.31 |
| chr7_38389675_38390189 | 0.07 |
AMPH |
amphiphysin |
112781 |
0.07 |
| chr20_61551174_61551325 | 0.07 |
DIDO1 |
death inducer-obliterator 1 |
5491 |
0.16 |
| chr1_90074397_90074573 | 0.07 |
RP11-413E1.4 |
|
20799 |
0.15 |
| chr6_144472562_144472798 | 0.07 |
STX11 |
syntaxin 11 |
1017 |
0.64 |
| chr10_82259532_82259683 | 0.07 |
RP11-137H2.4 |
|
36091 |
0.14 |
| chr10_52273037_52273188 | 0.07 |
RP11-512N4.2 |
|
40538 |
0.15 |
| chr16_28504630_28504781 | 0.07 |
CLN3 |
ceroid-lipofuscinosis, neuronal 3 |
611 |
0.54 |
| chr9_95857151_95857367 | 0.07 |
RP11-274J16.5 |
|
552 |
0.64 |
| chr12_56732992_56733270 | 0.07 |
IL23A |
interleukin 23, alpha subunit p19 |
468 |
0.6 |
| chr5_39206788_39206954 | 0.07 |
FYB |
FYN binding protein |
3742 |
0.34 |
| chr1_28974520_28974765 | 0.07 |
ENSG00000270103 |
. |
470 |
0.72 |
| chr7_38384658_38384902 | 0.07 |
AMPH |
amphiphysin |
117933 |
0.06 |
| chr10_130834433_130834622 | 0.07 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
430921 |
0.01 |
| chr1_247573351_247573502 | 0.07 |
NLRP3 |
NLR family, pyrin domain containing 3 |
6032 |
0.19 |
| chr14_55799512_55799838 | 0.07 |
RP11-665C16.5 |
|
6544 |
0.25 |
| chr16_17107310_17107606 | 0.07 |
CTD-2576D5.4 |
|
120903 |
0.07 |
| chr6_149415953_149416198 | 0.06 |
RP11-162J8.3 |
|
62366 |
0.12 |
| chr6_14755159_14755442 | 0.06 |
ENSG00000206960 |
. |
108534 |
0.08 |
| chr13_43564402_43564714 | 0.06 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
809 |
0.73 |
| chr6_2999439_2999644 | 0.06 |
RP1-90J20.8 |
|
173 |
0.82 |
| chr3_60066609_60066760 | 0.06 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
109101 |
0.08 |
| chr18_60980797_60980948 | 0.06 |
RP11-28F1.2 |
|
443 |
0.81 |
| chr4_78722448_78722599 | 0.06 |
CNOT6L |
CCR4-NOT transcription complex, subunit 6-like |
17694 |
0.25 |
| chr13_100308687_100308874 | 0.06 |
ENSG00000263615 |
. |
13467 |
0.18 |
| chr12_117536177_117536841 | 0.06 |
TESC |
tescalcin |
742 |
0.76 |
| chr10_14702052_14702247 | 0.06 |
ENSG00000201766 |
. |
3344 |
0.25 |
| chr8_6131851_6132002 | 0.06 |
RP11-115C21.2 |
|
132137 |
0.05 |
| chr2_99347754_99348710 | 0.06 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
643 |
0.77 |
| chr7_142344591_142344772 | 0.06 |
MTRNR2L6 |
MT-RNR2-like 6 |
29423 |
0.21 |
| chr6_57146781_57146932 | 0.06 |
PRIM2 |
primase, DNA, polypeptide 2 (58kDa) |
35566 |
0.17 |
| chr17_66288424_66288865 | 0.06 |
ARSG |
arylsulfatase G |
985 |
0.41 |
| chr12_104854827_104855220 | 0.06 |
CHST11 |
carbohydrate (chondroitin 4) sulfotransferase 11 |
4244 |
0.32 |
| chrX_3617821_3618286 | 0.06 |
PRKX |
protein kinase, X-linked |
13596 |
0.21 |
| chr12_96502184_96502412 | 0.06 |
ENSG00000266889 |
. |
6271 |
0.22 |
| chr9_117159748_117160008 | 0.06 |
AKNA |
AT-hook transcription factor |
3193 |
0.27 |
| chr5_137416537_137416688 | 0.06 |
WNT8A |
wingless-type MMTV integration site family, member 8A |
2969 |
0.16 |
| chr15_64888931_64889431 | 0.06 |
ENSG00000207223 |
. |
55906 |
0.09 |
| chr18_9779691_9779842 | 0.06 |
ENSG00000242651 |
. |
50858 |
0.12 |
| chr6_34360722_34361201 | 0.06 |
NUDT3 |
nudix (nucleoside diphosphate linked moiety X)-type motif 3 |
510 |
0.78 |
| chr6_25137772_25138250 | 0.06 |
ENSG00000222373 |
. |
54630 |
0.11 |
| chr9_135036030_135036181 | 0.06 |
NTNG2 |
netrin G2 |
1229 |
0.56 |
| chr11_6399629_6399806 | 0.06 |
SMPD1 |
sphingomyelin phosphodiesterase 1, acid lysosomal |
11938 |
0.14 |
| chr13_41187397_41187548 | 0.06 |
FOXO1 |
forkhead box O1 |
53262 |
0.14 |
| chr10_88296004_88296305 | 0.06 |
RP11-77P6.2 |
|
14452 |
0.18 |
| chr7_127749136_127749293 | 0.06 |
ENSG00000207588 |
. |
27301 |
0.2 |
| chr20_57232821_57232972 | 0.06 |
STX16 |
syntaxin 16 |
5833 |
0.18 |
| chr13_33000514_33000892 | 0.06 |
N4BP2L1 |
NEDD4 binding protein 2-like 1 |
1448 |
0.44 |
| chrX_69522152_69522303 | 0.06 |
KIF4A |
kinesin family member 4A |
12287 |
0.11 |
| chr7_139929395_139929688 | 0.06 |
ENSG00000199283 |
. |
12129 |
0.17 |
| chr5_1594076_1595069 | 0.06 |
SDHAP3 |
succinate dehydrogenase complex, subunit A, flavoprotein pseudogene 3 |
1168 |
0.53 |
| chr17_78619420_78619571 | 0.06 |
RPTOR |
regulatory associated protein of MTOR, complex 1 |
100227 |
0.07 |
| chrX_47489439_47489603 | 0.06 |
CFP |
complement factor properdin |
157 |
0.92 |
| chr10_130833753_130834277 | 0.06 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
431433 |
0.01 |
| chr20_37434777_37435281 | 0.06 |
PPP1R16B |
protein phosphatase 1, regulatory subunit 16B |
662 |
0.71 |
| chr4_185776582_185776733 | 0.06 |
ENSG00000266698 |
. |
4393 |
0.22 |
| chr1_40867584_40867735 | 0.06 |
SMAP2 |
small ArfGAP2 |
5152 |
0.19 |
| chr6_57680311_57680462 | 0.06 |
ENSG00000212017 |
. |
425456 |
0.01 |
| chr12_14540053_14540282 | 0.06 |
ATF7IP |
activating transcription factor 7 interacting protein |
2169 |
0.39 |
| chr8_53853126_53853277 | 0.06 |
NPBWR1 |
neuropeptides B/W receptor 1 |
2210 |
0.4 |
| chr2_204606154_204606338 | 0.05 |
ENSG00000211573 |
. |
23275 |
0.19 |
| chr12_62658631_62659028 | 0.05 |
USP15 |
ubiquitin specific peptidase 15 |
4620 |
0.23 |
| chr10_99280593_99280744 | 0.05 |
UBTD1 |
ubiquitin domain containing 1 |
22043 |
0.1 |
| chr11_128380502_128380688 | 0.05 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
5306 |
0.26 |
| chr14_78107992_78108340 | 0.05 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
25050 |
0.16 |
| chr17_53346307_53346458 | 0.05 |
HLF |
hepatic leukemia factor |
1205 |
0.54 |
| chr10_73521558_73521709 | 0.05 |
C10orf54 |
chromosome 10 open reading frame 54 |
4246 |
0.22 |
| chr2_86079401_86079552 | 0.05 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
15301 |
0.17 |
| chr22_39240481_39240642 | 0.05 |
NPTXR |
neuronal pentraxin receptor |
574 |
0.67 |
| chr6_42332393_42332544 | 0.05 |
ENSG00000221252 |
. |
47666 |
0.14 |
| chr22_39417668_39418190 | 0.05 |
APOBEC3D |
apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3D |
472 |
0.69 |
| chr2_198651086_198651566 | 0.05 |
BOLL |
boule-like RNA-binding protein |
28 |
0.98 |
| chr14_77422597_77423323 | 0.05 |
ENSG00000266553 |
. |
11148 |
0.21 |
| chr13_113622985_113623455 | 0.05 |
MCF2L-AS1 |
MCF2L antisense RNA 1 |
82 |
0.79 |
| chr3_72788363_72788610 | 0.05 |
ENSG00000222838 |
. |
47524 |
0.16 |
| chrY_7143368_7143656 | 0.05 |
PRKY |
protein kinase, Y-linked, pseudogene |
1158 |
0.57 |
| chr5_175960940_175961324 | 0.05 |
RNF44 |
ring finger protein 44 |
3289 |
0.16 |
| chr16_745521_745735 | 0.05 |
FBXL16 |
F-box and leucine-rich repeat protein 16 |
397 |
0.57 |
| chr14_69247883_69248049 | 0.05 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
9994 |
0.22 |
| chr8_53081485_53081636 | 0.05 |
RP11-26M5.3 |
|
18180 |
0.22 |
| chr10_31287025_31287176 | 0.05 |
ZNF438 |
zinc finger protein 438 |
1346 |
0.57 |
| chr12_47615454_47615605 | 0.05 |
PCED1B |
PC-esterase domain containing 1B |
1755 |
0.4 |
| chr19_42705324_42705475 | 0.05 |
ENSG00000265122 |
. |
8169 |
0.09 |
| chr7_142494910_142495350 | 0.05 |
PRSS3P2 |
protease, serine, 3 pseudogene 2 |
13999 |
0.17 |
| chr7_150066210_150066658 | 0.05 |
REPIN1 |
replication initiator 1 |
474 |
0.45 |
| chr1_226900274_226900494 | 0.05 |
ITPKB |
inositol-trisphosphate 3-kinase B |
24775 |
0.18 |
| chr1_19933664_19933939 | 0.05 |
MINOS1-NBL1 |
MINOS1-NBL1 readthrough |
10240 |
0.13 |
| chr6_29894399_29894823 | 0.05 |
HLA-A |
major histocompatibility complex, class I, A |
14426 |
0.18 |
| chr6_49159531_49159682 | 0.05 |
ENSG00000252457 |
. |
152915 |
0.04 |
| chr3_45010992_45011143 | 0.05 |
ZDHHC3 |
zinc finger, DHHC-type containing 3 |
6591 |
0.16 |
| chr20_48804878_48805029 | 0.05 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
2423 |
0.29 |
| chr9_92220501_92221395 | 0.05 |
GADD45G |
growth arrest and DNA-damage-inducible, gamma |
995 |
0.69 |
| chr1_27989094_27989245 | 0.05 |
RP11-288L9.4 |
|
6810 |
0.12 |
| chr6_34191993_34192468 | 0.05 |
HMGA1 |
high mobility group AT-hook 1 |
12420 |
0.2 |
| chrX_39473789_39473940 | 0.05 |
ENSG00000263730 |
. |
46606 |
0.2 |
| chr7_26345077_26345229 | 0.05 |
SNX10 |
sorting nexin 10 |
12479 |
0.23 |
| chr6_56536803_56536954 | 0.05 |
DST |
dystonin |
29084 |
0.25 |
| chr3_60061903_60062075 | 0.05 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
104406 |
0.08 |
| chr5_56114546_56115097 | 0.05 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
3420 |
0.24 |
| chr16_68306988_68307276 | 0.05 |
SLC7A6 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 6 |
250 |
0.81 |
| chrX_134471958_134472109 | 0.05 |
ZNF75D |
zinc finger protein 75D |
5979 |
0.22 |
| chr10_127688132_127688283 | 0.05 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
4221 |
0.25 |
| chr10_23831077_23831228 | 0.05 |
OTUD1 |
OTU domain containing 1 |
102954 |
0.07 |
| chr14_32672059_32672251 | 0.05 |
ENSG00000202337 |
. |
320 |
0.8 |
| chr6_111409779_111410127 | 0.05 |
SLC16A10 |
solute carrier family 16 (aromatic amino acid transporter), member 10 |
1172 |
0.45 |
| chr3_141516104_141516959 | 0.05 |
ENSG00000265391 |
. |
9778 |
0.19 |
| chr17_73583887_73584096 | 0.05 |
MYO15B |
myosin XVB pseudogene |
2867 |
0.17 |
| chr1_167451857_167452008 | 0.05 |
RP11-104L21.2 |
|
24034 |
0.18 |
| chr19_2428027_2428178 | 0.05 |
TIMM13 |
translocase of inner mitochondrial membrane 13 homolog (yeast) |
210 |
0.91 |
| chr2_30489387_30489538 | 0.05 |
LBH |
limb bud and heart development |
34416 |
0.19 |
| chr22_45065479_45065630 | 0.05 |
PRR5 |
proline rich 5 (renal) |
961 |
0.61 |
| chrX_46432817_46433047 | 0.05 |
CHST7 |
carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 |
287 |
0.92 |
| chr1_28196256_28196434 | 0.05 |
THEMIS2 |
thymocyte selection associated family member 2 |
2710 |
0.16 |
| chr16_71851540_71851781 | 0.05 |
AP1G1 |
adaptor-related protein complex 1, gamma 1 subunit |
8556 |
0.13 |
| chr2_109229032_109229223 | 0.05 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
5510 |
0.26 |
| chr2_170219735_170220376 | 0.05 |
LRP2 |
low density lipoprotein receptor-related protein 2 |
860 |
0.71 |
| chr6_159103866_159104017 | 0.05 |
SYTL3 |
synaptotagmin-like 3 |
19746 |
0.18 |
| chr21_44596901_44597052 | 0.05 |
CRYAA |
crystallin, alpha A |
6710 |
0.18 |
| chr12_8087301_8087631 | 0.05 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
1316 |
0.38 |
| chr14_75801482_75801633 | 0.05 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
54661 |
0.1 |
| chrX_1325322_1325473 | 0.04 |
CRLF2 |
cytokine receptor-like factor 2 |
6130 |
0.2 |
| chr14_75807662_75807918 | 0.04 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
60894 |
0.09 |
| chr6_128799339_128799870 | 0.04 |
RP1-86D1.5 |
|
22068 |
0.14 |
| chr21_16377585_16378233 | 0.04 |
NRIP1 |
nuclear receptor interacting protein 1 |
3220 |
0.28 |
| chrX_97323503_97323654 | 0.04 |
DIAPH2-AS1 |
DIAPH2 antisense RNA 1 |
425990 |
0.01 |
| chr6_166901943_166902308 | 0.04 |
ENSG00000222958 |
. |
20796 |
0.16 |
| chr12_120729183_120729407 | 0.04 |
ENSG00000202538 |
. |
411 |
0.71 |
| chrX_15874060_15874211 | 0.04 |
AP1S2 |
adaptor-related protein complex 1, sigma 2 subunit |
1081 |
0.58 |
| chr10_99080773_99080924 | 0.04 |
FRAT1 |
frequently rearranged in advanced T-cell lymphomas |
1826 |
0.23 |
| chr20_46180702_46180853 | 0.04 |
ENSG00000221294 |
. |
18902 |
0.18 |
| chr4_95698595_95698746 | 0.04 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
19551 |
0.3 |
| chrX_135961684_135962842 | 0.04 |
RBMX |
RNA binding motif protein, X-linked |
621 |
0.53 |
| chr22_50628354_50628718 | 0.04 |
TRABD |
TraB domain containing |
491 |
0.58 |
| chr16_71916638_71916789 | 0.04 |
RP11-498D10.3 |
|
537 |
0.48 |
| chr3_73455417_73455568 | 0.04 |
PDZRN3 |
PDZ domain containing ring finger 3 |
5376 |
0.32 |
| chr12_772231_772876 | 0.04 |
NINJ2 |
ninjurin 2 |
392 |
0.85 |
| chr6_27106968_27108062 | 0.04 |
HIST1H4I |
histone cluster 1, H4i |
439 |
0.71 |
| chr7_39994488_39994769 | 0.04 |
CDK13 |
cyclin-dependent kinase 13 |
4519 |
0.23 |
| chrX_12997161_12997312 | 0.04 |
TMSB4X |
thymosin beta 4, X-linked |
3459 |
0.29 |
| chr19_41309158_41309605 | 0.04 |
EGLN2 |
egl-9 family hypoxia-inducible factor 2 |
336 |
0.82 |
| chr4_89743291_89743450 | 0.04 |
FAM13A |
family with sequence similarity 13, member A |
982 |
0.66 |
| chr7_150441383_150441534 | 0.04 |
GIMAP5 |
GTPase, IMAP family member 5 |
7022 |
0.17 |
| chr4_38784189_38784442 | 0.04 |
TLR10 |
toll-like receptor 10 |
264 |
0.9 |
| chr9_70851276_70851465 | 0.04 |
CBWD3 |
COBW domain containing 3 |
5027 |
0.19 |
| chr2_143913103_143913254 | 0.04 |
RP11-190J23.1 |
|
16563 |
0.23 |
| chrX_24482692_24483178 | 0.04 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
403 |
0.88 |
| chr6_169990292_169990458 | 0.04 |
WDR27 |
WD repeat domain 27 |
70413 |
0.1 |
| chrY_1274756_1275442 | 0.04 |
NA |
NA |
> 106 |
NA |
| chr8_107404618_107404769 | 0.04 |
OXR1 |
oxidation resistance 1 |
32989 |
0.25 |
| chr9_116351386_116351537 | 0.04 |
RP11-168K11.2 |
|
838 |
0.65 |
| chr22_25960035_25960186 | 0.04 |
CTA-407F11.8 |
|
7 |
0.93 |
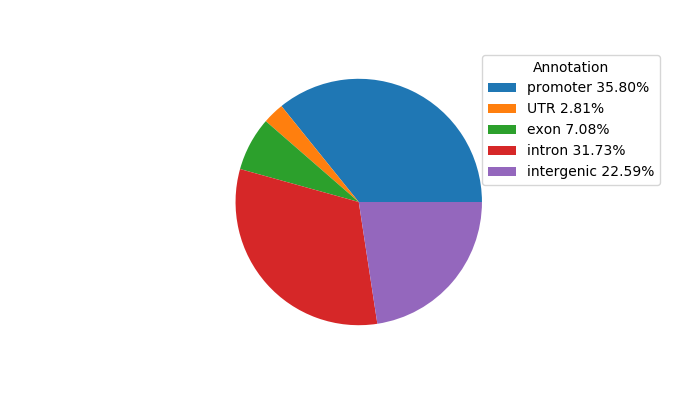
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.0 | GO:0051142 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.0 | GO:0097576 | vacuole fusion(GO:0097576) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034481 | chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.0 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |