Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
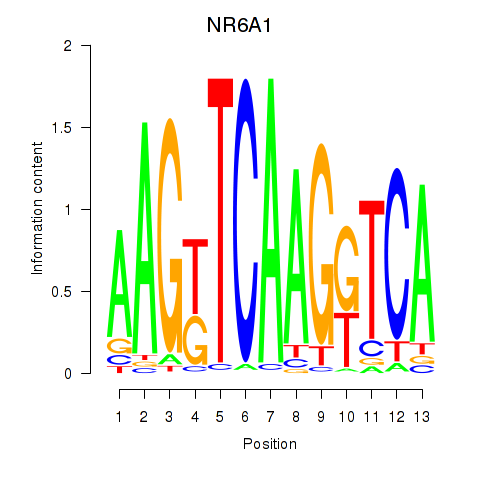
Results for NR6A1
Z-value: 1.25
Transcription factors associated with NR6A1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR6A1
|
ENSG00000148200.12 | nuclear receptor subfamily 6 group A member 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_127355583_127355734 | NR6A1 | 2429 | 0.272081 | -0.24 | 5.3e-01 | Click! |
| chr9_127533110_127533927 | NR6A1 | 38 | 0.950296 | -0.23 | 5.4e-01 | Click! |
| chr9_127355961_127356112 | NR6A1 | 2051 | 0.304428 | 0.12 | 7.6e-01 | Click! |
| chr9_127534048_127534335 | NR6A1 | 602 | 0.620366 | 0.09 | 8.1e-01 | Click! |
Activity of the NR6A1 motif across conditions
Conditions sorted by the z-value of the NR6A1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
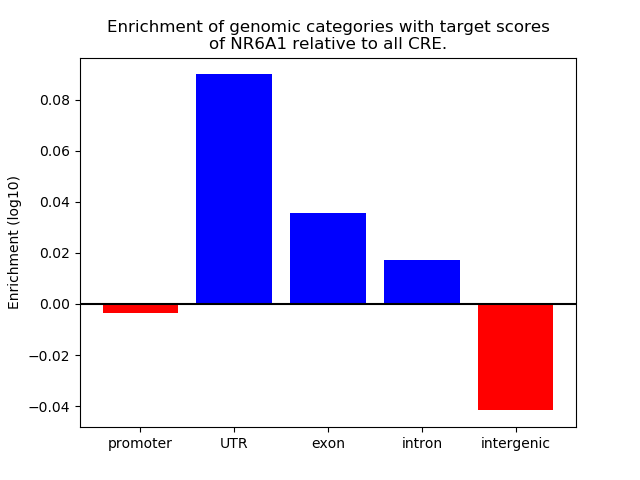
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_1874347_1875032 | 0.94 |
LSP1 |
lymphocyte-specific protein 1 |
489 |
0.65 |
| chr2_143886988_143887259 | 0.45 |
ARHGAP15 |
Rho GTPase activating protein 15 |
240 |
0.95 |
| chr5_175085682_175085863 | 0.44 |
HRH2 |
histamine receptor H2 |
739 |
0.7 |
| chr1_161038546_161039545 | 0.42 |
ARHGAP30 |
Rho GTPase activating protein 30 |
411 |
0.65 |
| chr8_134085908_134086059 | 0.41 |
SLA |
Src-like-adaptor |
13380 |
0.24 |
| chr1_207244470_207244690 | 0.41 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
6231 |
0.14 |
| chr5_70315966_70316295 | 0.40 |
NAIP |
NLR family, apoptosis inhibitory protein |
407 |
0.88 |
| chr6_159465378_159465638 | 0.38 |
TAGAP |
T-cell activation RhoGTPase activating protein |
542 |
0.78 |
| chr2_30454378_30455881 | 0.38 |
LBH |
limb bud and heart development |
83 |
0.98 |
| chrX_39968386_39969319 | 0.38 |
BCOR |
BCL6 corepressor |
12196 |
0.31 |
| chrX_111125721_111125872 | 0.37 |
TRPC5OS |
TRPC5 opposite strand |
671 |
0.81 |
| chr11_65343661_65344993 | 0.37 |
EHBP1L1 |
EH domain binding protein 1-like 1 |
810 |
0.36 |
| chr7_36653532_36653683 | 0.37 |
AOAH-IT1 |
AOAH intronic transcript 1 (non-protein coding) |
13881 |
0.19 |
| chrX_12929174_12929325 | 0.37 |
TLR8-AS1 |
TLR8 antisense RNA 1 |
2797 |
0.26 |
| chr8_61824618_61824780 | 0.35 |
RP11-33I11.2 |
|
102534 |
0.08 |
| chr16_3306416_3306567 | 0.35 |
MEFV |
Mediterranean fever |
96 |
0.94 |
| chr10_70847938_70848765 | 0.35 |
SRGN |
serglycin |
477 |
0.82 |
| chr19_2290395_2291381 | 0.34 |
LINGO3 |
leucine rich repeat and Ig domain containing 3 |
1135 |
0.28 |
| chr3_14442430_14442581 | 0.34 |
SLC6A6 |
solute carrier family 6 (neurotransmitter transporter), member 6 |
1571 |
0.43 |
| chr3_46140245_46140452 | 0.34 |
CCR3 |
chemokine (C-C motif) receptor 3 |
64748 |
0.1 |
| chr7_50246497_50246648 | 0.33 |
AC020743.2 |
|
64153 |
0.12 |
| chr19_17958305_17958511 | 0.33 |
JAK3 |
Janus kinase 3 |
418 |
0.74 |
| chr20_1926522_1926673 | 0.33 |
RP4-684O24.5 |
|
1295 |
0.5 |
| chrX_153190817_153191535 | 0.33 |
ARHGAP4 |
Rho GTPase activating protein 4 |
522 |
0.6 |
| chr19_52268680_52268969 | 0.32 |
FPR2 |
formyl peptide receptor 2 |
2807 |
0.17 |
| chr6_126069184_126070625 | 0.32 |
RP11-624M8.1 |
|
428 |
0.68 |
| chr15_81594027_81594306 | 0.32 |
IL16 |
interleukin 16 |
2409 |
0.29 |
| chr4_70626143_70626357 | 0.32 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
66 |
0.98 |
| chr1_159823893_159825316 | 0.32 |
C1orf204 |
chromosome 1 open reading frame 204 |
533 |
0.6 |
| chr2_54808515_54808696 | 0.32 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
23074 |
0.17 |
| chr1_61332602_61332753 | 0.31 |
NFIA |
nuclear factor I/A |
1746 |
0.51 |
| chr17_47467788_47467939 | 0.31 |
RP11-1079K10.4 |
|
13580 |
0.11 |
| chr6_138002128_138002279 | 0.31 |
ENSG00000216097 |
. |
35882 |
0.2 |
| chr1_161641257_161641408 | 0.31 |
RP11-25K21.1 |
|
3282 |
0.14 |
| chr21_44819752_44820164 | 0.31 |
SIK1 |
salt-inducible kinase 1 |
27050 |
0.24 |
| chr13_114271369_114271520 | 0.31 |
TFDP1 |
transcription factor Dp-1 |
31703 |
0.15 |
| chr6_144471049_144472508 | 0.30 |
STX11 |
syntaxin 11 |
115 |
0.98 |
| chr1_192780744_192780895 | 0.30 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
2648 |
0.37 |
| chr12_109025614_109025806 | 0.30 |
SELPLG |
selectin P ligand |
144 |
0.94 |
| chr17_29646389_29646953 | 0.30 |
CTD-2370N5.3 |
|
822 |
0.44 |
| chr1_1830918_1831069 | 0.30 |
RP1-140A9.1 |
|
8083 |
0.13 |
| chr2_47406474_47406729 | 0.30 |
CALM2 |
calmodulin 2 (phosphorylase kinase, delta) |
2861 |
0.29 |
| chr12_123367208_123367359 | 0.30 |
VPS37B |
vacuolar protein sorting 37 homolog B (S. cerevisiae) |
7423 |
0.17 |
| chr1_161559432_161559583 | 0.30 |
FCGR2C |
Fc fragment of IgG, low affinity IIc, receptor for (CD32) (gene/pseudogene) |
5836 |
0.13 |
| chr4_88786510_88786661 | 0.29 |
MEPE |
matrix extracellular phosphoglycoprotein |
32446 |
0.16 |
| chr2_58694154_58694305 | 0.29 |
FANCL |
Fanconi anemia, complementation group L |
225722 |
0.02 |
| chr12_8692426_8692577 | 0.29 |
CLEC4E |
C-type lectin domain family 4, member E |
650 |
0.65 |
| chr20_60760834_60760985 | 0.29 |
MTG2 |
mitochondrial ribosome-associated GTPase 2 |
2802 |
0.2 |
| chr7_130567596_130567747 | 0.29 |
ENSG00000226380 |
. |
5373 |
0.29 |
| chr6_144981294_144981445 | 0.29 |
UTRN |
utrophin |
389 |
0.93 |
| chr9_75567665_75568017 | 0.29 |
ALDH1A1 |
aldehyde dehydrogenase 1 family, member A1 |
130 |
0.98 |
| chr2_152779879_152780030 | 0.29 |
CACNB4 |
calcium channel, voltage-dependent, beta 4 subunit |
48548 |
0.17 |
| chr7_50257083_50257268 | 0.28 |
AC020743.2 |
|
74756 |
0.1 |
| chr19_42301375_42301526 | 0.28 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
365 |
0.8 |
| chr2_232479090_232479920 | 0.28 |
C2orf57 |
chromosome 2 open reading frame 57 |
21930 |
0.15 |
| chr14_93118654_93119048 | 0.28 |
RIN3 |
Ras and Rab interactor 3 |
5 |
0.99 |
| chr17_34435571_34435722 | 0.28 |
ENSG00000263488 |
. |
1897 |
0.18 |
| chr16_88768022_88768436 | 0.28 |
RNF166 |
ring finger protein 166 |
1218 |
0.22 |
| chr21_39645775_39646073 | 0.28 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
1495 |
0.53 |
| chr4_15081994_15082145 | 0.28 |
ENSG00000199420 |
. |
13649 |
0.24 |
| chr1_232715037_232715188 | 0.28 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
17808 |
0.28 |
| chr15_69073260_69073411 | 0.28 |
ENSG00000265195 |
. |
20929 |
0.21 |
| chr10_6243545_6244657 | 0.27 |
RP11-414H17.5 |
|
555 |
0.52 |
| chr14_56588992_56589252 | 0.27 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
3295 |
0.31 |
| chr7_140681725_140681876 | 0.27 |
MRPS33 |
mitochondrial ribosomal protein S33 |
32429 |
0.17 |
| chr6_5084934_5086227 | 0.27 |
PPP1R3G |
protein phosphatase 1, regulatory subunit 3G |
140 |
0.97 |
| chr17_72517580_72517731 | 0.27 |
CD300LB |
CD300 molecule-like family member b |
9950 |
0.14 |
| chr10_14920855_14921866 | 0.27 |
SUV39H2 |
suppressor of variegation 3-9 homolog 2 (Drosophila) |
439 |
0.82 |
| chr8_142131426_142131715 | 0.27 |
DENND3 |
DENN/MADD domain containing 3 |
4193 |
0.23 |
| chr2_233925207_233925694 | 0.27 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
261 |
0.92 |
| chr5_131798996_131799147 | 0.27 |
ENSG00000202533 |
. |
4768 |
0.15 |
| chr13_49788937_49789202 | 0.27 |
MLNR |
motilin receptor |
5405 |
0.25 |
| chr19_17517338_17517489 | 0.27 |
MVB12A |
multivesicular body subunit 12A |
459 |
0.54 |
| chr17_36861833_36863162 | 0.27 |
CTB-58E17.3 |
|
17 |
0.9 |
| chr15_80253251_80253402 | 0.27 |
BCL2A1 |
BCL2-related protein A1 |
10185 |
0.17 |
| chr2_135051969_135052120 | 0.27 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
40214 |
0.19 |
| chr16_28997019_28997515 | 0.27 |
LAT |
linker for activation of T cells |
266 |
0.8 |
| chr8_56824063_56824214 | 0.27 |
ENSG00000216204 |
. |
2618 |
0.21 |
| chr10_35424848_35424999 | 0.26 |
ENSG00000253054 |
. |
552 |
0.66 |
| chr9_101871320_101871471 | 0.26 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
3963 |
0.26 |
| chr12_66635842_66635993 | 0.26 |
ENSG00000266539 |
. |
6515 |
0.17 |
| chr1_44534212_44534363 | 0.26 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
37153 |
0.11 |
| chr11_78646693_78646844 | 0.26 |
TENM4 |
teneurin transmembrane protein 4 |
31837 |
0.2 |
| chr2_162270607_162270758 | 0.26 |
TBR1 |
T-box, brain, 1 |
1923 |
0.3 |
| chr1_234746129_234746973 | 0.26 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
1280 |
0.44 |
| chr17_75868336_75868487 | 0.26 |
FLJ45079 |
|
10248 |
0.24 |
| chr2_84678068_84678219 | 0.26 |
SUCLG1 |
succinate-CoA ligase, alpha subunit |
8461 |
0.29 |
| chr2_70269953_70270104 | 0.26 |
PCBP1-AS1 |
PCBP1 antisense RNA 1 |
10889 |
0.17 |
| chr21_15917916_15918619 | 0.26 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
395 |
0.89 |
| chr20_33871790_33873092 | 0.26 |
EIF6 |
eukaryotic translation initiation factor 6 |
77 |
0.88 |
| chr19_15760987_15761138 | 0.26 |
CYP4F3 |
cytochrome P450, family 4, subfamily F, polypeptide 3 |
8943 |
0.18 |
| chr7_31851156_31851370 | 0.26 |
ENSG00000223070 |
. |
17401 |
0.29 |
| chr2_69002223_69002932 | 0.26 |
ARHGAP25 |
Rho GTPase activating protein 25 |
505 |
0.84 |
| chr6_159074554_159074705 | 0.25 |
SYTL3 |
synaptotagmin-like 3 |
3583 |
0.22 |
| chr1_17630308_17630459 | 0.25 |
PADI4 |
peptidyl arginine deiminase, type IV |
4307 |
0.19 |
| chr16_81867856_81868007 | 0.25 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
48943 |
0.17 |
| chrX_12925475_12925626 | 0.25 |
TLR8 |
toll-like receptor 8 |
792 |
0.45 |
| chr17_46565994_46566145 | 0.25 |
ENSG00000206805 |
. |
9610 |
0.12 |
| chr6_37140965_37141167 | 0.25 |
PIM1 |
pim-1 oncogene |
3087 |
0.24 |
| chr9_127024548_127024933 | 0.25 |
RP11-121A14.3 |
|
415 |
0.67 |
| chr18_43785413_43785564 | 0.25 |
C18orf25 |
chromosome 18 open reading frame 25 |
31488 |
0.18 |
| chr22_23561758_23561909 | 0.25 |
BCR |
breakpoint cluster region |
34314 |
0.12 |
| chr11_112192338_112192489 | 0.25 |
ENSG00000206772 |
. |
30972 |
0.14 |
| chr12_15113209_15113383 | 0.25 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
904 |
0.55 |
| chr20_54989282_54989551 | 0.25 |
CASS4 |
Cas scaffolding protein family member 4 |
2099 |
0.24 |
| chr22_22216989_22217140 | 0.25 |
MAPK1 |
mitogen-activated protein kinase 1 |
4666 |
0.18 |
| chr2_43439997_43440275 | 0.25 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
13612 |
0.23 |
| chr10_129860321_129860472 | 0.25 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
14562 |
0.26 |
| chr1_9299570_9299924 | 0.24 |
H6PD |
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
156 |
0.96 |
| chr7_44079644_44080725 | 0.24 |
RASA4CP |
RAS p21 protein activator 4C, pseudogene |
1307 |
0.26 |
| chr13_40976108_40976619 | 0.24 |
ENSG00000252812 |
. |
5214 |
0.32 |
| chr7_36642261_36642412 | 0.24 |
AOAH-IT1 |
AOAH intronic transcript 1 (non-protein coding) |
2610 |
0.28 |
| chrX_13108426_13108692 | 0.24 |
FAM9C |
family with sequence similarity 9, member C |
45758 |
0.18 |
| chr16_53404692_53404843 | 0.24 |
RP11-44F14.1 |
|
2228 |
0.28 |
| chr12_92523772_92523923 | 0.24 |
C12orf79 |
chromosome 12 open reading frame 79 |
6950 |
0.19 |
| chr1_27438843_27438994 | 0.24 |
SLC9A1 |
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
42055 |
0.13 |
| chr16_75661718_75661869 | 0.24 |
ADAT1 |
adenosine deaminase, tRNA-specific 1 |
4595 |
0.16 |
| chr2_99080716_99081105 | 0.24 |
INPP4A |
inositol polyphosphate-4-phosphatase, type I, 107kDa |
19497 |
0.22 |
| chr12_116996820_116997035 | 0.24 |
MAP1LC3B2 |
microtubule-associated protein 1 light chain 3 beta 2 |
259 |
0.95 |
| chr6_11330504_11330655 | 0.24 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
51953 |
0.16 |
| chr4_185741986_185742137 | 0.24 |
ACSL1 |
acyl-CoA synthetase long-chain family member 1 |
5127 |
0.2 |
| chr1_36840428_36840700 | 0.24 |
STK40 |
serine/threonine kinase 40 |
10921 |
0.12 |
| chr7_134845732_134845901 | 0.24 |
RP11-134L10.1 |
|
7339 |
0.14 |
| chr17_56408975_56409725 | 0.24 |
MIR142 |
microRNA 142 |
519 |
0.63 |
| chr20_25554452_25554603 | 0.23 |
NINL |
ninein-like |
11626 |
0.18 |
| chr11_44590165_44590737 | 0.23 |
CD82 |
CD82 molecule |
3178 |
0.29 |
| chr22_35725770_35725921 | 0.23 |
ENSG00000266320 |
. |
5788 |
0.17 |
| chr19_54881402_54882266 | 0.23 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
224 |
0.88 |
| chr11_104914233_104914622 | 0.23 |
CARD16 |
caspase recruitment domain family, member 16 |
1607 |
0.35 |
| chr5_130588828_130589214 | 0.23 |
CDC42SE2 |
CDC42 small effector 2 |
10681 |
0.28 |
| chr2_96923789_96923942 | 0.23 |
TMEM127 |
transmembrane protein 127 |
2468 |
0.21 |
| chr5_118646029_118646180 | 0.23 |
ENSG00000243333 |
. |
3778 |
0.24 |
| chr15_85141845_85142241 | 0.23 |
ZSCAN2 |
zinc finger and SCAN domain containing 2 |
2174 |
0.19 |
| chr5_133860075_133861084 | 0.23 |
JADE2 |
jade family PHD finger 2 |
576 |
0.71 |
| chr3_48259036_48259187 | 0.23 |
CAMP |
cathelicidin antimicrobial peptide |
5726 |
0.13 |
| chr10_12307829_12307980 | 0.23 |
ENSG00000265824 |
. |
4234 |
0.18 |
| chr2_237884098_237884249 | 0.23 |
ENSG00000202341 |
. |
39775 |
0.18 |
| chr12_122230893_122231238 | 0.23 |
RHOF |
ras homolog family member F (in filopodia) |
201 |
0.93 |
| chr13_46743875_46744026 | 0.23 |
ENSG00000240767 |
. |
67 |
0.96 |
| chr12_56963795_56963946 | 0.23 |
ENSG00000201579 |
. |
18432 |
0.12 |
| chr14_105527834_105528220 | 0.23 |
GPR132 |
G protein-coupled receptor 132 |
3740 |
0.23 |
| chr19_54891617_54891768 | 0.23 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
9527 |
0.1 |
| chr2_175498427_175499207 | 0.23 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
490 |
0.83 |
| chrY_15016828_15017148 | 0.22 |
DDX3Y |
DEAD (Asp-Glu-Ala-Asp) box helicase 3, Y-linked |
246 |
0.96 |
| chr8_61821706_61823076 | 0.22 |
RP11-33I11.2 |
|
100226 |
0.08 |
| chr22_22215698_22215849 | 0.22 |
MAPK1 |
mitogen-activated protein kinase 1 |
5957 |
0.17 |
| chr11_3744283_3744434 | 0.22 |
ENSG00000251934 |
. |
8253 |
0.12 |
| chr1_198978699_198978850 | 0.22 |
ENSG00000207759 |
. |
150492 |
0.04 |
| chrX_19884431_19884582 | 0.22 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
21071 |
0.24 |
| chr2_112196931_112197265 | 0.22 |
ENSG00000266139 |
. |
118430 |
0.07 |
| chr20_3694963_3695114 | 0.22 |
SIGLEC1 |
sialic acid binding Ig-like lectin 1, sialoadhesin |
7263 |
0.14 |
| chr11_116857425_116857669 | 0.22 |
ENSG00000264344 |
. |
28698 |
0.13 |
| chr5_55711467_55711618 | 0.22 |
ENSG00000265665 |
. |
41423 |
0.16 |
| chr1_26954743_26954935 | 0.22 |
ENSG00000238316 |
. |
14041 |
0.13 |
| chr16_53552293_53552836 | 0.22 |
AKTIP |
AKT interacting protein |
14241 |
0.21 |
| chr5_55777832_55777983 | 0.22 |
CTC-236F12.4 |
Uncharacterized protein |
311 |
0.91 |
| chr12_120670116_120670490 | 0.22 |
PXN |
paxillin |
5656 |
0.13 |
| chr6_42009511_42009971 | 0.22 |
CCND3 |
cyclin D3 |
6683 |
0.16 |
| chr17_57074891_57075042 | 0.22 |
ENSG00000216168 |
. |
8651 |
0.16 |
| chr7_129666009_129666160 | 0.22 |
ENSG00000201109 |
. |
1033 |
0.43 |
| chr16_28204366_28204526 | 0.22 |
ENSG00000200138 |
. |
9807 |
0.17 |
| chrY_21906454_21906814 | 0.22 |
KDM5D |
lysine (K)-specific demethylase 5D |
13 |
0.99 |
| chr17_43301591_43301923 | 0.22 |
CTD-2020K17.1 |
|
2168 |
0.16 |
| chr13_100630550_100631126 | 0.22 |
ZIC2 |
Zic family member 2 |
3188 |
0.24 |
| chr10_81959862_81960013 | 0.22 |
ANXA11 |
annexin A11 |
4310 |
0.24 |
| chr6_90088751_90088902 | 0.22 |
UBE2J1 |
ubiquitin-conjugating enzyme E2, J1 |
26259 |
0.16 |
| chr2_68963054_68963510 | 0.21 |
ARHGAP25 |
Rho GTPase activating protein 25 |
1268 |
0.56 |
| chr1_231748956_231749347 | 0.21 |
LINC00582 |
long intergenic non-protein coding RNA 582 |
1315 |
0.46 |
| chr7_90224844_90225183 | 0.21 |
AC002456.2 |
|
49 |
0.94 |
| chr13_99227754_99228977 | 0.21 |
STK24 |
serine/threonine kinase 24 |
752 |
0.54 |
| chr19_42082862_42083083 | 0.21 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
371 |
0.84 |
| chr9_134147000_134147362 | 0.21 |
FAM78A |
family with sequence similarity 78, member A |
1271 |
0.44 |
| chr9_113838212_113838363 | 0.21 |
ENSG00000212409 |
. |
21406 |
0.22 |
| chrX_100640419_100640710 | 0.21 |
BTK |
Bruton agammaglobulinemia tyrosine kinase |
603 |
0.58 |
| chr7_142919179_142919330 | 0.21 |
AC073342.1 |
HCG2002387; Uncharacterized protein |
106 |
0.51 |
| chr19_52154438_52154589 | 0.21 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
4362 |
0.12 |
| chr19_18586594_18586745 | 0.21 |
ELL |
elongation factor RNA polymerase II |
74 |
0.95 |
| chr14_90526445_90527696 | 0.21 |
KCNK13 |
potassium channel, subfamily K, member 13 |
1039 |
0.63 |
| chr11_34378633_34379465 | 0.21 |
ABTB2 |
ankyrin repeat and BTB (POZ) domain containing 2 |
247 |
0.95 |
| chrX_39964334_39965624 | 0.21 |
BCOR |
BCL6 corepressor |
8323 |
0.32 |
| chr12_40551802_40551953 | 0.21 |
LRRK2 |
leucine-rich repeat kinase 2 |
38669 |
0.17 |
| chr2_102609202_102609353 | 0.21 |
IL1R2 |
interleukin 1 receptor, type II |
971 |
0.66 |
| chr10_11758024_11758175 | 0.21 |
ECHDC3 |
enoyl CoA hydratase domain containing 3 |
26266 |
0.21 |
| chr19_2619419_2619570 | 0.21 |
CTC-265F19.2 |
|
7634 |
0.14 |
| chr16_18816680_18816831 | 0.21 |
RP11-1035H13.2 |
|
2350 |
0.2 |
| chr8_42742851_42743002 | 0.21 |
ENSG00000242719 |
. |
6547 |
0.13 |
| chr16_68027365_68027982 | 0.20 |
DPEP2 |
dipeptidase 2 |
93 |
0.9 |
| chr1_174937099_174937250 | 0.20 |
RABGAP1L |
RAB GTPase activating protein 1-like |
3269 |
0.19 |
| chr9_100807481_100807632 | 0.20 |
NANS |
N-acetylneuraminic acid synthase |
11465 |
0.16 |
| chr1_204482801_204482952 | 0.20 |
MDM4 |
Mdm4 p53 binding protein homolog (mouse) |
2635 |
0.26 |
| chr7_50345445_50345596 | 0.20 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
1142 |
0.64 |
| chr10_70092145_70093579 | 0.20 |
PBLD |
phenazine biosynthesis-like protein domain containing |
56 |
0.95 |
| chr19_42705923_42706118 | 0.20 |
ENSG00000265122 |
. |
8790 |
0.09 |
| chr10_21502036_21502187 | 0.20 |
NEBL-AS1 |
NEBL antisense RNA 1 |
38828 |
0.16 |
| chr1_206747757_206747908 | 0.20 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
17339 |
0.15 |
| chr19_54899574_54899725 | 0.20 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
17484 |
0.09 |
| chr11_63370704_63370855 | 0.20 |
PLA2G16 |
phospholipase A2, group XVI |
5274 |
0.15 |
| chr1_54946073_54946224 | 0.20 |
ENSG00000265404 |
. |
24193 |
0.16 |
| chr3_119041182_119042341 | 0.20 |
ARHGAP31-AS1 |
ARHGAP31 antisense RNA 1 |
154 |
0.94 |
| chr2_103052502_103052757 | 0.20 |
AC007278.2 |
|
2212 |
0.21 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.1 | 0.3 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.1 | 0.3 | GO:1901339 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.1 | 0.6 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.1 | 0.2 | GO:0034135 | regulation of toll-like receptor 2 signaling pathway(GO:0034135) |
| 0.1 | 0.4 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.1 | 0.2 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.1 | 0.2 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.1 | 0.3 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.1 | 0.3 | GO:0071354 | cellular response to interleukin-6(GO:0071354) |
| 0.1 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.1 | 0.2 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.0 | GO:0014742 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.2 | GO:0061647 | histone H3-K9 methylation(GO:0051567) histone H3-K9 modification(GO:0061647) |
| 0.0 | 0.6 | GO:0002279 | mast cell activation involved in immune response(GO:0002279) mast cell degranulation(GO:0043303) |
| 0.0 | 0.2 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.3 | GO:0050710 | negative regulation of cytokine secretion(GO:0050710) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.1 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.1 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.2 | GO:0090049 | regulation of cell migration involved in sprouting angiogenesis(GO:0090049) regulation of sprouting angiogenesis(GO:1903670) |
| 0.0 | 0.1 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0045075 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0032493 | response to bacterial lipoprotein(GO:0032493) response to bacterial lipopeptide(GO:0070339) cellular response to bacterial lipoprotein(GO:0071220) cellular response to bacterial lipopeptide(GO:0071221) |
| 0.0 | 0.3 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.0 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.2 | GO:0070265 | necrotic cell death(GO:0070265) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0035067 | negative regulation of histone acetylation(GO:0035067) negative regulation of protein acetylation(GO:1901984) negative regulation of peptidyl-lysine acetylation(GO:2000757) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0032803 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.1 | GO:0033003 | regulation of mast cell activation(GO:0033003) |
| 0.0 | 0.1 | GO:0031620 | regulation of fever generation(GO:0031620) positive regulation of fever generation(GO:0031622) |
| 0.0 | 0.0 | GO:0002538 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.1 | GO:0009756 | carbohydrate mediated signaling(GO:0009756) |
| 0.0 | 1.1 | GO:0006968 | cellular defense response(GO:0006968) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.2 | GO:0050775 | positive regulation of dendrite morphogenesis(GO:0050775) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0034139 | regulation of toll-like receptor 3 signaling pathway(GO:0034139) |
| 0.0 | 0.1 | GO:0008615 | pyridoxine metabolic process(GO:0008614) pyridoxine biosynthetic process(GO:0008615) vitamin B6 metabolic process(GO:0042816) vitamin B6 biosynthetic process(GO:0042819) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.0 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:1901797 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0034314 | Arp2/3 complex-mediated actin nucleation(GO:0034314) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.1 | GO:1904019 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0002532 | production of molecular mediator involved in inflammatory response(GO:0002532) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0031659 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0007168 | receptor guanylyl cyclase signaling pathway(GO:0007168) |
| 0.0 | 0.1 | GO:0033860 | regulation of NAD(P)H oxidase activity(GO:0033860) |
| 0.0 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0032615 | interleukin-12 production(GO:0032615) |
| 0.0 | 0.1 | GO:0045039 | protein import into mitochondrial inner membrane(GO:0045039) |
| 0.0 | 0.1 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.2 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.1 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.1 | GO:0048679 | regulation of axon regeneration(GO:0048679) regulation of neuron projection regeneration(GO:0070570) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.0 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.4 | GO:0006692 | prostanoid metabolic process(GO:0006692) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.2 | GO:0009435 | NAD biosynthetic process(GO:0009435) |
| 0.0 | 0.0 | GO:0033591 | response to L-ascorbic acid(GO:0033591) |
| 0.0 | 0.1 | GO:0002921 | negative regulation of humoral immune response(GO:0002921) negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.2 | GO:0050702 | interleukin-1 beta secretion(GO:0050702) |
| 0.0 | 0.1 | GO:0006924 | activation-induced cell death of T cells(GO:0006924) |
| 0.0 | 0.0 | GO:0002448 | mast cell mediated immunity(GO:0002448) |
| 0.0 | 0.0 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.0 | GO:0034625 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) |
| 0.0 | 0.0 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.0 | 0.3 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.0 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0002544 | chronic inflammatory response(GO:0002544) |
| 0.0 | 0.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.0 | GO:0001820 | serotonin secretion(GO:0001820) |
| 0.0 | 0.0 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.0 | 0.0 | GO:0042535 | positive regulation of tumor necrosis factor biosynthetic process(GO:0042535) |
| 0.0 | 0.1 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.0 | GO:0002643 | regulation of tolerance induction(GO:0002643) positive regulation of tolerance induction(GO:0002645) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.0 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.0 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.1 | GO:0046348 | amino sugar catabolic process(GO:0046348) glucosamine-containing compound catabolic process(GO:1901072) |
| 0.0 | 0.1 | GO:0050849 | negative regulation of calcium-mediated signaling(GO:0050849) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0006837 | serotonin transport(GO:0006837) |
| 0.0 | 0.1 | GO:0030212 | hyaluronan metabolic process(GO:0030212) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.2 | GO:0006027 | glycosaminoglycan catabolic process(GO:0006027) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.2 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.1 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0002097 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.0 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.7 | GO:0006914 | autophagy(GO:0006914) |
| 0.0 | 0.0 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.0 | GO:0010988 | regulation of low-density lipoprotein particle clearance(GO:0010988) |
| 0.0 | 0.1 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.0 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.0 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:1902305 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.0 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.0 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.1 | GO:0071108 | protein K48-linked deubiquitination(GO:0071108) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) telomere maintenance in response to DNA damage(GO:0043247) |
| 0.0 | 0.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.0 | GO:1904063 | negative regulation of cation transmembrane transport(GO:1904063) |
| 0.0 | 0.0 | GO:0046149 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.0 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.1 | GO:0009133 | nucleoside diphosphate biosynthetic process(GO:0009133) |
| 0.0 | 0.0 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.0 | GO:0010832 | negative regulation of myotube differentiation(GO:0010832) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0021853 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 | 0.0 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.2 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.2 | GO:0042531 | positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.1 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.3 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.4 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.0 | 0.0 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.0 | 0.1 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0030870 | Mre11 complex(GO:0030870) |
| 0.0 | 0.0 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.0 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.1 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.2 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.3 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.1 | 0.3 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.1 | 0.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.2 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.1 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.2 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.1 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.1 | 0.4 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.2 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.1 | 0.2 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.1 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.4 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.3 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.2 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0008159 | obsolete positive transcription elongation factor activity(GO:0008159) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.1 | GO:0019959 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.6 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.3 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.2 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.4 | GO:0001637 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) tricarboxylic acid transmembrane transporter activity(GO:0015142) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.1 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.0 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.2 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.3 | GO:0043022 | ribosome binding(GO:0043022) |
| 0.0 | 0.0 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.2 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.1 | GO:0047429 | nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.0 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.3 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.0 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.2 | GO:0004383 | guanylate cyclase activity(GO:0004383) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0030955 | potassium ion binding(GO:0030955) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.0 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.0 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.0 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.2 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.3 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.0 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.0 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.0 | GO:0019956 | chemokine binding(GO:0019956) |
| 0.0 | 0.1 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0043008 | ATP-dependent protein binding(GO:0043008) |
| 0.0 | 0.1 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0033612 | receptor serine/threonine kinase binding(GO:0033612) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.0 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.3 | GO:0003725 | double-stranded RNA binding(GO:0003725) |
| 0.0 | 0.1 | GO:0015347 | sodium-independent organic anion transmembrane transporter activity(GO:0015347) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.5 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.2 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.4 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 1.3 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 0.1 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.4 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.3 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.3 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.6 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID IL1 PATHWAY | IL1-mediated signaling events |
| 0.0 | 0.1 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | PID FAS PATHWAY | FAS (CD95) signaling pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.4 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.1 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.4 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.3 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.1 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.1 | PID PI3KCI AKT PATHWAY | Class I PI3K signaling events mediated by Akt |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.0 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.6 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 1.0 | REACTOME IL RECEPTOR SHC SIGNALING | Genes involved in Interleukin receptor SHC signaling |
| 0.0 | 0.4 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.4 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.8 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.2 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.3 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.0 | REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | Genes involved in The role of Nef in HIV-1 replication and disease pathogenesis |
| 0.0 | 0.1 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.0 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.2 | REACTOME ACTIVATION OF CHAPERONES BY ATF6 ALPHA | Genes involved in Activation of Chaperones by ATF6-alpha |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.4 | REACTOME IL1 SIGNALING | Genes involved in Interleukin-1 signaling |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.1 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.0 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 1.4 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.0 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.0 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.3 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.5 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME ACTIVATED AMPK STIMULATES FATTY ACID OXIDATION IN MUSCLE | Genes involved in Activated AMPK stimulates fatty-acid oxidation in muscle |
| 0.0 | 0.0 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.0 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.1 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |