Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
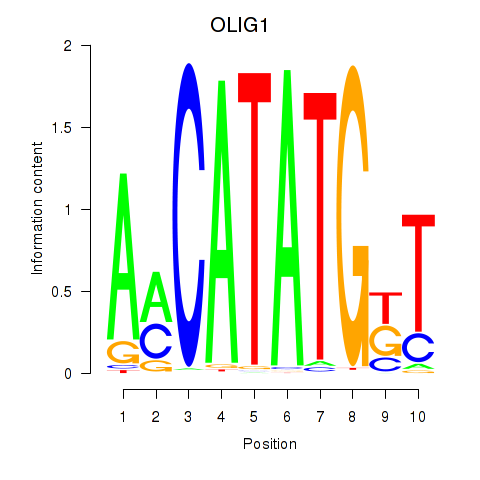
Results for OLIG1
Z-value: 0.60
Transcription factors associated with OLIG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG1
|
ENSG00000184221.8 | oligodendrocyte transcription factor 1 |
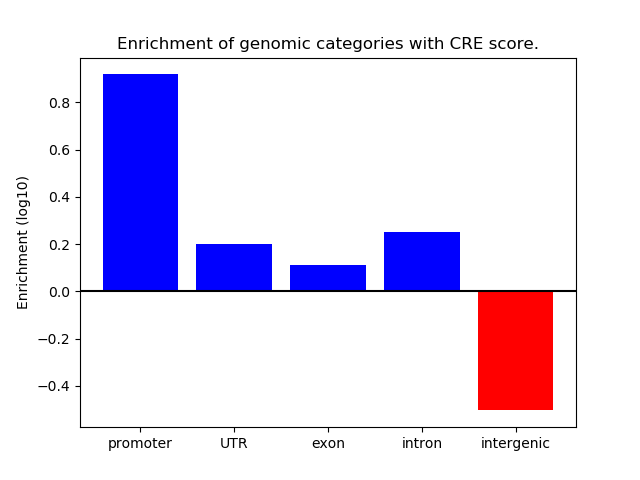
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr21_34437191_34437342 | OLIG1 | 5184 | 0.189671 | -0.44 | 2.4e-01 | Click! |
| chr21_34439565_34439843 | OLIG1 | 2746 | 0.235935 | -0.42 | 2.7e-01 | Click! |
| chr21_34431903_34432054 | OLIG1 | 10472 | 0.170916 | -0.37 | 3.3e-01 | Click! |
| chr21_34431477_34431628 | OLIG1 | 10898 | 0.170100 | 0.33 | 3.8e-01 | Click! |
| chr21_34428465_34428616 | OLIG1 | 13910 | 0.164800 | -0.33 | 3.9e-01 | Click! |
Activity of the OLIG1 motif across conditions
Conditions sorted by the z-value of the OLIG1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
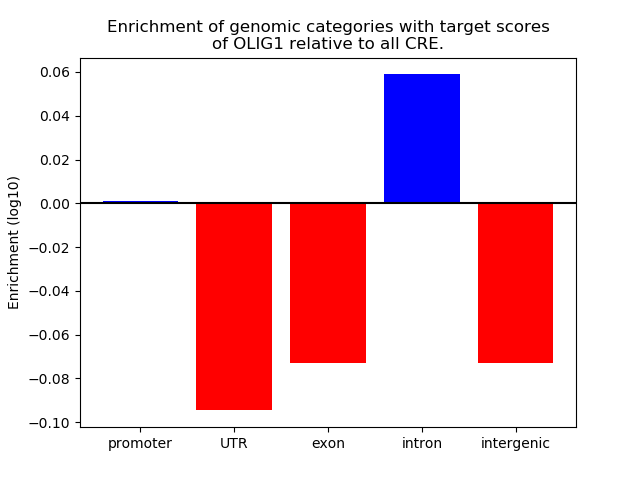
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_128913998_128914362 | 0.39 |
SASH3 |
SAM and SH3 domain containing 3 |
220 |
0.94 |
| chr1_25254983_25255134 | 0.36 |
RUNX3 |
runt-related transcription factor 3 |
554 |
0.81 |
| chr19_42056193_42056486 | 0.35 |
CEACAM21 |
carcinoembryonic antigen-related cell adhesion molecule 21 |
453 |
0.8 |
| chr1_185283352_185283831 | 0.33 |
IVNS1ABP |
influenza virus NS1A binding protein |
2870 |
0.28 |
| chr12_62657019_62657170 | 0.31 |
USP15 |
ubiquitin specific peptidase 15 |
2885 |
0.26 |
| chr1_248729435_248729591 | 0.30 |
RP11-438F14.3 |
|
3612 |
0.13 |
| chr1_84610455_84610717 | 0.30 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
632 |
0.82 |
| chr5_162930555_162931177 | 0.30 |
MAT2B |
methionine adenosyltransferase II, beta |
746 |
0.62 |
| chr2_127415089_127415240 | 0.30 |
GYPC |
glycophorin C (Gerbich blood group) |
1404 |
0.55 |
| chr12_57077777_57078354 | 0.29 |
PTGES3 |
prostaglandin E synthase 3 (cytosolic) |
3881 |
0.14 |
| chr14_53593743_53594005 | 0.29 |
DDHD1 |
DDHD domain containing 1 |
22930 |
0.17 |
| chr13_43564402_43564714 | 0.29 |
EPSTI1 |
epithelial stromal interaction 1 (breast) |
809 |
0.73 |
| chr8_66863582_66864279 | 0.29 |
DNAJC5B |
DnaJ (Hsp40) homolog, subfamily C, member 5 beta |
69865 |
0.12 |
| chr2_241630055_241630353 | 0.29 |
AQP12A |
aquaporin 12A |
1058 |
0.43 |
| chr13_46754554_46754849 | 0.28 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
1758 |
0.31 |
| chr1_248644857_248645289 | 0.28 |
RP11-407H12.8 |
|
3712 |
0.13 |
| chr10_28656896_28657047 | 0.28 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
33556 |
0.16 |
| chr5_171612568_171612893 | 0.28 |
STK10 |
serine/threonine kinase 10 |
2660 |
0.27 |
| chr3_67735991_67736422 | 0.27 |
SUCLG2 |
succinate-CoA ligase, GDP-forming, beta subunit |
31168 |
0.25 |
| chr1_25875695_25876149 | 0.27 |
LDLRAP1 |
low density lipoprotein receptor adaptor protein 1 |
5851 |
0.23 |
| chr11_118087619_118088223 | 0.27 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
2315 |
0.23 |
| chr7_2739070_2739221 | 0.27 |
AMZ1 |
archaelysin family metallopeptidase 1 |
11309 |
0.21 |
| chr17_263982_264133 | 0.27 |
AC108004.3 |
|
243 |
0.89 |
| chr18_9138515_9138757 | 0.26 |
ANKRD12 |
ankyrin repeat domain 12 |
1075 |
0.42 |
| chr13_52396131_52396530 | 0.26 |
RP11-327P2.5 |
|
17897 |
0.18 |
| chr22_24824729_24824978 | 0.26 |
ADORA2A |
adenosine A2a receptor |
1323 |
0.44 |
| chr4_89534455_89534711 | 0.26 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
7638 |
0.19 |
| chr12_9806335_9806486 | 0.26 |
RP11-705C15.2 |
|
131 |
0.93 |
| chr2_182325663_182326116 | 0.26 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
3734 |
0.36 |
| chr8_66751756_66751954 | 0.26 |
PDE7A |
phosphodiesterase 7A |
872 |
0.73 |
| chr5_148189087_148189390 | 0.26 |
ADRB2 |
adrenoceptor beta 2, surface |
16918 |
0.25 |
| chr16_29674660_29674967 | 0.25 |
QPRT |
quinolinate phosphoribosyltransferase |
213 |
0.51 |
| chr16_10971555_10971960 | 0.25 |
CIITA |
class II, major histocompatibility complex, transactivator |
690 |
0.64 |
| chr17_38719353_38719597 | 0.25 |
CCR7 |
chemokine (C-C motif) receptor 7 |
2210 |
0.28 |
| chr1_26872331_26872899 | 0.25 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
272 |
0.88 |
| chr11_14668160_14668311 | 0.25 |
PDE3B |
phosphodiesterase 3B, cGMP-inhibited |
2858 |
0.29 |
| chr20_24046827_24047099 | 0.25 |
GGTLC1 |
gamma-glutamyltransferase light chain 1 |
77547 |
0.11 |
| chr6_135408926_135409077 | 0.24 |
HBS1L |
HBS1-like (S. cerevisiae) |
15193 |
0.21 |
| chr17_35860978_35861129 | 0.24 |
DUSP14 |
dual specificity phosphatase 14 |
8928 |
0.19 |
| chr7_26895188_26895339 | 0.24 |
SKAP2 |
src kinase associated phosphoprotein 2 |
1976 |
0.43 |
| chr5_43310867_43311162 | 0.24 |
HMGCS1 |
3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) |
2312 |
0.32 |
| chr14_61814847_61814998 | 0.23 |
PRKCH |
protein kinase C, eta |
313 |
0.91 |
| chr6_159465378_159465638 | 0.23 |
TAGAP |
T-cell activation RhoGTPase activating protein |
542 |
0.78 |
| chr12_53616534_53616822 | 0.23 |
RARG |
retinoic acid receptor, gamma |
2481 |
0.17 |
| chr14_90086040_90086276 | 0.23 |
FOXN3 |
forkhead box N3 |
665 |
0.59 |
| chr14_22978649_22978851 | 0.23 |
TRAJ15 |
T cell receptor alpha joining 15 |
19830 |
0.09 |
| chr13_50114703_50115207 | 0.23 |
RCBTB1 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
26504 |
0.17 |
| chr17_10603321_10603720 | 0.23 |
ADPRM |
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
2609 |
0.18 |
| chr4_6677782_6677978 | 0.23 |
AC093323.1 |
Uncharacterized protein |
16309 |
0.14 |
| chr3_16330847_16330998 | 0.23 |
OXNAD1 |
oxidoreductase NAD-binding domain containing 1 |
20174 |
0.15 |
| chr6_153321108_153321259 | 0.23 |
MTRF1L |
mitochondrial translational release factor 1-like |
2637 |
0.24 |
| chr2_158291882_158292250 | 0.23 |
CYTIP |
cytohesin 1 interacting protein |
3860 |
0.26 |
| chr17_74188552_74188703 | 0.23 |
RNF157 |
ring finger protein 157 |
25464 |
0.11 |
| chr1_169672728_169672969 | 0.22 |
SELL |
selectin L |
7991 |
0.2 |
| chr4_6753172_6753335 | 0.22 |
KIAA0232 |
KIAA0232 |
31116 |
0.13 |
| chr20_35575234_35575857 | 0.22 |
SAMHD1 |
SAM domain and HD domain 1 |
4566 |
0.25 |
| chr13_97879341_97880219 | 0.22 |
MBNL2 |
muscleblind-like splicing regulator 2 |
5171 |
0.33 |
| chrX_118811810_118811961 | 0.22 |
SEPT6 |
septin 6 |
14907 |
0.16 |
| chr5_134712559_134712710 | 0.22 |
H2AFY |
H2A histone family, member Y |
22267 |
0.15 |
| chr4_143325352_143326244 | 0.22 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
26614 |
0.28 |
| chr4_40199107_40199293 | 0.22 |
RHOH |
ras homolog family member H |
673 |
0.73 |
| chrX_53741093_53741244 | 0.22 |
HUWE1 |
HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase |
27495 |
0.23 |
| chr14_98443744_98443895 | 0.21 |
C14orf64 |
chromosome 14 open reading frame 64 |
564 |
0.87 |
| chr1_23668742_23669298 | 0.21 |
HNRNPR |
heterogeneous nuclear ribonucleoprotein R |
1735 |
0.29 |
| chr10_121352939_121353112 | 0.21 |
TIAL1 |
TIA1 cytotoxic granule-associated RNA binding protein-like 1 |
2982 |
0.3 |
| chr7_150414088_150414577 | 0.21 |
GIMAP1 |
GTPase, IMAP family member 1 |
687 |
0.65 |
| chr10_13828862_13829013 | 0.21 |
RP11-353M9.1 |
|
57554 |
0.1 |
| chr1_229642144_229642295 | 0.21 |
NUP133 |
nucleoporin 133kDa |
1854 |
0.24 |
| chr14_64974613_64974785 | 0.21 |
ZBTB25 |
zinc finger and BTB domain containing 25 |
2768 |
0.16 |
| chr9_80524074_80524374 | 0.21 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
86309 |
0.1 |
| chr14_102290786_102291012 | 0.21 |
CTD-2017C7.2 |
|
14241 |
0.14 |
| chr14_64204420_64204571 | 0.21 |
ENSG00000252749 |
. |
1333 |
0.43 |
| chr12_46659525_46659676 | 0.21 |
SLC38A1 |
solute carrier family 38, member 1 |
1884 |
0.48 |
| chr5_44713608_44713759 | 0.21 |
ENSG00000263556 |
. |
2609 |
0.4 |
| chr2_202050412_202050706 | 0.21 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
2641 |
0.22 |
| chr22_40858602_40858925 | 0.20 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
659 |
0.7 |
| chr14_91879481_91879632 | 0.20 |
CCDC88C |
coiled-coil domain containing 88C |
4134 |
0.28 |
| chr18_53002274_53002527 | 0.20 |
TCF4 |
transcription factor 4 |
12875 |
0.28 |
| chr13_48984563_48985023 | 0.20 |
LPAR6 |
lysophosphatidic acid receptor 6 |
16250 |
0.26 |
| chr11_67176258_67176520 | 0.20 |
TBC1D10C |
TBC1 domain family, member 10C |
4729 |
0.07 |
| chr5_156731259_156731410 | 0.20 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
6711 |
0.16 |
| chr21_43516995_43517146 | 0.20 |
C21orf128 |
chromosome 21 open reading frame 128 |
11574 |
0.19 |
| chr1_150847141_150847524 | 0.20 |
ARNT |
aryl hydrocarbon receptor nuclear translocator |
1723 |
0.26 |
| chr7_5518487_5518712 | 0.20 |
ENSG00000238394 |
. |
6427 |
0.13 |
| chr18_8610996_8611147 | 0.20 |
RAB12 |
RAB12, member RAS oncogene family |
1628 |
0.4 |
| chr1_84611671_84611822 | 0.20 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
1792 |
0.48 |
| chr5_58880144_58880439 | 0.20 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
1928 |
0.5 |
| chr8_29948388_29948539 | 0.19 |
LEPROTL1 |
leptin receptor overlapping transcript-like 1 |
4451 |
0.17 |
| chr1_89737127_89737489 | 0.19 |
GBP5 |
guanylate binding protein 5 |
762 |
0.66 |
| chr18_60824857_60825062 | 0.19 |
RP11-299P2.1 |
|
6406 |
0.25 |
| chr22_40858328_40858544 | 0.19 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
986 |
0.54 |
| chr10_112624321_112624902 | 0.19 |
PDCD4 |
programmed cell death 4 (neoplastic transformation inhibitor) |
6954 |
0.15 |
| chr6_26442034_26442281 | 0.19 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
1371 |
0.29 |
| chr3_45706571_45706792 | 0.19 |
LIMD1-AS1 |
LIMD1 antisense RNA 1 |
23693 |
0.16 |
| chr1_167891180_167891331 | 0.19 |
ADCY10 |
adenylate cyclase 10 (soluble) |
7802 |
0.18 |
| chr1_211502580_211502753 | 0.19 |
TRAF5 |
TNF receptor-associated factor 5 |
2487 |
0.35 |
| chr20_24090206_24090357 | 0.18 |
GGTLC1 |
gamma-glutamyltransferase light chain 1 |
120865 |
0.06 |
| chr9_98782803_98783372 | 0.18 |
ERCC6L2 |
excision repair cross-complementing rodent repair deficiency, complementation group 6-like 2 |
47968 |
0.15 |
| chr1_150737104_150737638 | 0.18 |
CTSS |
cathepsin S |
897 |
0.5 |
| chr6_143249356_143249507 | 0.18 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
16907 |
0.25 |
| chr18_13277168_13277319 | 0.18 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
859 |
0.63 |
| chr15_67374785_67374936 | 0.18 |
SMAD3 |
SMAD family member 3 |
16060 |
0.25 |
| chr11_58342434_58343027 | 0.18 |
LPXN |
leupaxin |
604 |
0.68 |
| chr5_114193091_114193242 | 0.18 |
RP11-492A10.1 |
|
189059 |
0.03 |
| chr3_63923691_63923842 | 0.18 |
ATXN7 |
ataxin 7 |
25491 |
0.15 |
| chr7_142353602_142353808 | 0.18 |
MTRNR2L6 |
MT-RNR2-like 6 |
20399 |
0.23 |
| chr14_99726709_99727240 | 0.18 |
AL109767.1 |
|
2311 |
0.33 |
| chr12_66712096_66712247 | 0.18 |
HELB |
helicase (DNA) B |
15846 |
0.16 |
| chr3_141218856_141219007 | 0.18 |
RASA2 |
RAS p21 protein activator 2 |
13040 |
0.21 |
| chr9_3528863_3529151 | 0.18 |
RFX3 |
regulatory factor X, 3 (influences HLA class II expression) |
3003 |
0.37 |
| chr4_129026129_129026280 | 0.18 |
LARP1B |
La ribonucleoprotein domain family, member 1B |
27208 |
0.24 |
| chr9_117150863_117151014 | 0.18 |
AKNA |
AT-hook transcription factor |
577 |
0.77 |
| chr5_148726675_148726837 | 0.18 |
GRPEL2 |
GrpE-like 2, mitochondrial (E. coli) |
1691 |
0.25 |
| chr3_176915816_176916502 | 0.18 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
898 |
0.7 |
| chr15_90119818_90119983 | 0.18 |
TICRR |
TOPBP1-interacting checkpoint and replication regulator |
1082 |
0.46 |
| chr6_99380366_99380517 | 0.18 |
FBXL4 |
F-box and leucine-rich repeat protein 4 |
15361 |
0.28 |
| chr1_169662937_169663139 | 0.18 |
SELL |
selectin L |
17801 |
0.18 |
| chr5_35857770_35858187 | 0.18 |
IL7R |
interleukin 7 receptor |
984 |
0.58 |
| chr15_77287169_77287320 | 0.18 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
182 |
0.95 |
| chr1_19717605_19717920 | 0.17 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
5471 |
0.18 |
| chr1_32716475_32716711 | 0.17 |
LCK |
lymphocyte-specific protein tyrosine kinase |
247 |
0.82 |
| chr2_182329799_182329950 | 0.17 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
7719 |
0.31 |
| chr2_237446083_237446280 | 0.17 |
IQCA1 |
IQ motif containing with AAA domain 1 |
29996 |
0.19 |
| chr10_126431099_126431666 | 0.17 |
FAM53B |
family with sequence similarity 53, member B |
157 |
0.95 |
| chr10_63979683_63979834 | 0.17 |
RTKN2 |
rhotekin 2 |
16264 |
0.26 |
| chr2_70321937_70322218 | 0.17 |
PCBP1-AS1 |
PCBP1 antisense RNA 1 |
6099 |
0.17 |
| chr6_74232761_74232916 | 0.17 |
RP11-505P4.7 |
|
503 |
0.51 |
| chr7_148408377_148408528 | 0.17 |
CUL1 |
cullin 1 |
12448 |
0.17 |
| chr3_27922887_27923131 | 0.17 |
EOMES |
eomesodermin |
158803 |
0.04 |
| chr1_239883177_239883989 | 0.17 |
CHRM3 |
cholinergic receptor, muscarinic 3 |
740 |
0.59 |
| chr4_109081890_109082041 | 0.17 |
LEF1 |
lymphoid enhancer-binding factor 1 |
5492 |
0.25 |
| chr13_36407967_36408118 | 0.17 |
DCLK1 |
doublecortin-like kinase 1 |
21737 |
0.28 |
| chr1_89457123_89457381 | 0.17 |
CCBL2 |
cysteine conjugate-beta lyase 2 |
1035 |
0.37 |
| chr3_149419430_149419681 | 0.17 |
WWTR1 |
WW domain containing transcription regulator 1 |
1505 |
0.39 |
| chr18_56215999_56216150 | 0.17 |
RP11-126O1.2 |
|
8228 |
0.17 |
| chr2_55236798_55237891 | 0.17 |
RTN4 |
reticulon 4 |
241 |
0.93 |
| chr3_66514964_66515132 | 0.17 |
LRIG1 |
leucine-rich repeats and immunoglobulin-like domains 1 |
36308 |
0.22 |
| chr19_42432968_42433149 | 0.17 |
ARHGEF1 |
Rho guanine nucleotide exchange factor (GEF) 1 |
5812 |
0.14 |
| chr12_68044712_68044863 | 0.17 |
DYRK2 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 2 |
462 |
0.87 |
| chr3_59996849_59997073 | 0.17 |
NPCDR1 |
nasopharyngeal carcinoma, down-regulated 1 |
39378 |
0.24 |
| chr5_110560941_110561277 | 0.17 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
1325 |
0.51 |
| chr19_1465997_1466148 | 0.17 |
ENSG00000267317 |
. |
3308 |
0.09 |
| chr3_18475636_18475787 | 0.17 |
SATB1 |
SATB homeobox 1 |
1034 |
0.57 |
| chr1_230307234_230307385 | 0.17 |
RP5-956O18.2 |
|
96920 |
0.07 |
| chr6_75924948_75925099 | 0.17 |
COL12A1 |
collagen, type XII, alpha 1 |
9256 |
0.22 |
| chr6_143264256_143264520 | 0.17 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
1950 |
0.43 |
| chr2_106372259_106372410 | 0.17 |
NCK2 |
NCK adaptor protein 2 |
10146 |
0.29 |
| chr9_4664226_4664377 | 0.17 |
PPAPDC2 |
phosphatidic acid phosphatase type 2 domain containing 2 |
2003 |
0.26 |
| chr11_62367040_62367191 | 0.17 |
MTA2 |
metastasis associated 1 family, member 2 |
326 |
0.7 |
| chr2_38825620_38825771 | 0.17 |
HNRNPLL |
heterogeneous nuclear ribonucleoprotein L-like |
1473 |
0.39 |
| chr12_133083232_133083480 | 0.16 |
FBRSL1 |
fibrosin-like 1 |
16199 |
0.2 |
| chr5_75842727_75842878 | 0.16 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
432 |
0.88 |
| chr1_198873027_198873178 | 0.16 |
ENSG00000207759 |
. |
44820 |
0.17 |
| chr11_117685491_117685642 | 0.16 |
DSCAML1 |
Down syndrome cell adhesion molecule like 1 |
2674 |
0.19 |
| chr1_158903283_158903483 | 0.16 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
2025 |
0.36 |
| chr12_10019991_10020525 | 0.16 |
RP11-290C10.1 |
|
632 |
0.61 |
| chr3_141745583_141745864 | 0.16 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
1716 |
0.46 |
| chr7_7960072_7960471 | 0.16 |
ENSG00000201747 |
. |
14121 |
0.18 |
| chr1_156722427_156722776 | 0.16 |
HDGF |
hepatoma-derived growth factor |
361 |
0.75 |
| chr3_151989454_151989605 | 0.16 |
MBNL1-AS1 |
MBNL1 antisense RNA 1 |
2185 |
0.3 |
| chr15_33493746_33494182 | 0.16 |
FMN1 |
formin 1 |
7067 |
0.29 |
| chr11_62358316_62359010 | 0.16 |
TUT1 |
terminal uridylyl transferase 1, U6 snRNA-specific |
332 |
0.71 |
| chr5_95170159_95170387 | 0.16 |
GLRX |
glutaredoxin (thioltransferase) |
11564 |
0.14 |
| chr19_16195500_16195773 | 0.16 |
TPM4 |
tropomyosin 4 |
3103 |
0.21 |
| chr1_204490030_204490190 | 0.16 |
MDM4 |
Mdm4 p53 binding protein homolog (mouse) |
4491 |
0.2 |
| chr21_16816003_16816154 | 0.16 |
ENSG00000212564 |
. |
170524 |
0.04 |
| chr1_244487830_244488005 | 0.16 |
C1orf100 |
chromosome 1 open reading frame 100 |
28020 |
0.22 |
| chr9_102972411_102972625 | 0.16 |
ENSG00000239908 |
. |
107677 |
0.06 |
| chr5_74965499_74965780 | 0.16 |
ENSG00000207333 |
. |
39785 |
0.14 |
| chr11_11865444_11865596 | 0.16 |
USP47 |
ubiquitin specific peptidase 47 |
1907 |
0.47 |
| chr7_106505707_106506635 | 0.16 |
PIK3CG |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit gamma |
247 |
0.95 |
| chr4_56773645_56773796 | 0.16 |
ENSG00000202358 |
. |
21959 |
0.16 |
| chr22_40914778_40915024 | 0.16 |
RP4-591N18.2 |
|
2903 |
0.23 |
| chrX_153023566_153023717 | 0.16 |
PLXNB3 |
plexin B3 |
6010 |
0.1 |
| chr21_37670145_37670473 | 0.16 |
MORC3 |
MORC family CW-type zinc finger 3 |
22178 |
0.13 |
| chr5_118623311_118623849 | 0.16 |
ENSG00000243333 |
. |
18746 |
0.17 |
| chr3_71503791_71503942 | 0.16 |
ENSG00000221264 |
. |
87374 |
0.09 |
| chr12_121829638_121829987 | 0.16 |
ANAPC5 |
anaphase promoting complex subunit 5 |
7887 |
0.2 |
| chr22_50631230_50631436 | 0.16 |
TRABD |
TraB domain containing |
110 |
0.82 |
| chr5_79251776_79251927 | 0.16 |
RP11-168A11.1 |
|
19449 |
0.2 |
| chr1_108328348_108328499 | 0.16 |
ENSG00000265536 |
. |
9545 |
0.26 |
| chr2_197030897_197031094 | 0.16 |
STK17B |
serine/threonine kinase 17b |
4729 |
0.22 |
| chr11_11865222_11865373 | 0.16 |
USP47 |
ubiquitin specific peptidase 47 |
1684 |
0.51 |
| chr12_55372250_55372401 | 0.16 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
3297 |
0.29 |
| chr1_225968168_225968319 | 0.15 |
SRP9 |
signal recognition particle 9kDa |
2606 |
0.22 |
| chr6_26304754_26304905 | 0.15 |
HIST1H4H |
histone cluster 1, H4h |
19067 |
0.08 |
| chr20_10481692_10481843 | 0.15 |
SLX4IP |
SLX4 interacting protein |
65816 |
0.11 |
| chr14_25134813_25134997 | 0.15 |
GZMB |
granzyme B (granzyme 2, cytotoxic T-lymphocyte-associated serine esterase 1) |
31432 |
0.15 |
| chr4_146098361_146098512 | 0.15 |
OTUD4 |
OTU domain containing 4 |
871 |
0.65 |
| chr4_153022445_153022684 | 0.15 |
ENSG00000266244 |
. |
127056 |
0.05 |
| chr1_26644827_26645344 | 0.15 |
UBXN11 |
UBX domain protein 11 |
231 |
0.76 |
| chr14_91869059_91869463 | 0.15 |
CCDC88C |
coiled-coil domain containing 88C |
14429 |
0.23 |
| chr18_56355169_56355320 | 0.15 |
RP11-126O1.4 |
|
10398 |
0.15 |
| chr2_200490274_200490425 | 0.15 |
SATB2 |
SATB homeobox 2 |
154360 |
0.04 |
| chr5_130881277_130881428 | 0.15 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
12626 |
0.29 |
| chr4_178237433_178237648 | 0.15 |
NEIL3 |
nei endonuclease VIII-like 3 (E. coli) |
6550 |
0.23 |
| chr4_146541198_146541349 | 0.15 |
MMAA |
methylmalonic aciduria (cobalamin deficiency) cblA type |
1858 |
0.37 |
| chr12_118795905_118796056 | 0.15 |
TAOK3 |
TAO kinase 3 |
930 |
0.65 |
| chr6_12065087_12065238 | 0.15 |
HIVEP1 |
human immunodeficiency virus type I enhancer binding protein 1 |
49342 |
0.17 |
| chr4_113444412_113444838 | 0.15 |
NEUROG2 |
neurogenin 2 |
7297 |
0.19 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 0.2 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.1 | 0.4 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.1 | 0.2 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.4 | GO:0002710 | negative regulation of T cell mediated immunity(GO:0002710) |
| 0.1 | 0.2 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.1 | 0.2 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.0 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0052257 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0031053 | primary miRNA processing(GO:0031053) |
| 0.0 | 0.1 | GO:0051570 | regulation of histone H3-K9 methylation(GO:0051570) |
| 0.0 | 0.1 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.3 | GO:0000380 | alternative mRNA splicing, via spliceosome(GO:0000380) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.1 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0017183 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0046398 | UDP-glucuronate metabolic process(GO:0046398) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0003351 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.0 | GO:0031063 | regulation of histone deacetylation(GO:0031063) positive regulation of histone deacetylation(GO:0031065) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) regulation of centromeric sister chromatid cohesion(GO:0070602) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.2 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.2 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.1 | GO:0045901 | translational frameshifting(GO:0006452) positive regulation of translational elongation(GO:0045901) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0035844 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.0 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.0 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0009713 | catechol-containing compound biosynthetic process(GO:0009713) catecholamine biosynthetic process(GO:0042423) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.0 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.0 | GO:0001831 | trophectodermal cellular morphogenesis(GO:0001831) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.2 | GO:0006378 | mRNA polyadenylation(GO:0006378) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.1 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.1 | GO:0018202 | peptidyl-histidine modification(GO:0018202) |
| 0.0 | 0.1 | GO:0045842 | positive regulation of mitotic metaphase/anaphase transition(GO:0045842) positive regulation of mitotic sister chromatid separation(GO:1901970) positive regulation of metaphase/anaphase transition of cell cycle(GO:1902101) |
| 0.0 | 0.1 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.0 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.0 | GO:0019367 | fatty acid elongation, saturated fatty acid(GO:0019367) |
| 0.0 | 0.1 | GO:0007549 | dosage compensation(GO:0007549) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.2 | GO:0043489 | RNA stabilization(GO:0043489) mRNA stabilization(GO:0048255) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.0 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.0 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.0 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.3 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.0 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0051322 | anaphase(GO:0051322) |
| 0.0 | 0.0 | GO:0007183 | SMAD protein complex assembly(GO:0007183) |
| 0.0 | 0.1 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.0 | GO:0035269 | protein mannosylation(GO:0035268) protein O-linked mannosylation(GO:0035269) mannosylation(GO:0097502) |
| 0.0 | 0.0 | GO:0070233 | negative regulation of T cell apoptotic process(GO:0070233) |
| 0.0 | 0.0 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.2 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0050860 | regulation of T cell receptor signaling pathway(GO:0050856) negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0046329 | negative regulation of JNK cascade(GO:0046329) |
| 0.0 | 0.1 | GO:0006744 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0010390 | histone monoubiquitination(GO:0010390) |
| 0.0 | 0.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.0 | GO:0035751 | lysosomal lumen acidification(GO:0007042) regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:0042403 | thyroid hormone metabolic process(GO:0042403) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.4 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0000800 | lateral element(GO:0000800) |
| 0.0 | 0.2 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0044298 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.1 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.0 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.3 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.2 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.0 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.0 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.3 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.0 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.0 | GO:0031501 | mannosyltransferase complex(GO:0031501) dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0016442 | RISC complex(GO:0016442) RNAi effector complex(GO:0031332) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.2 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.6 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.3 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.4 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.0 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.0 | 0.0 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.0 | GO:0022889 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.2 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.2 | GO:0046625 | sphingolipid binding(GO:0046625) |
| 0.0 | 0.0 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0008048 | calcium sensitive guanylate cyclase activator activity(GO:0008048) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.0 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.0 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.0 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.0 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0051635 | obsolete bacterial cell surface binding(GO:0051635) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0032452 | histone demethylase activity(GO:0032452) |
| 0.0 | 0.1 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.0 | 0.0 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.0 | GO:0035004 | phosphatidylinositol 3-kinase activity(GO:0035004) |
| 0.0 | 0.1 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.0 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.6 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.2 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.6 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.1 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.0 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.0 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME SCF BETA TRCP MEDIATED DEGRADATION OF EMI1 | Genes involved in SCF-beta-TrCP mediated degradation of Emi1 |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.1 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.3 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME PROCESSIVE SYNTHESIS ON THE LAGGING STRAND | Genes involved in Processive synthesis on the lagging strand |
| 0.0 | 0.2 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.2 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.4 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.2 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | Genes involved in Post NMDA receptor activation events |
| 0.0 | 0.0 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |