Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
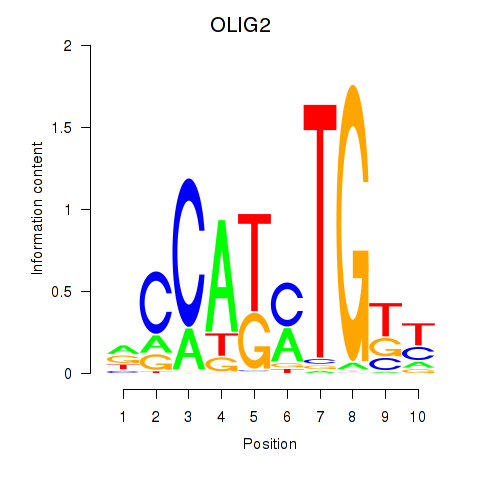
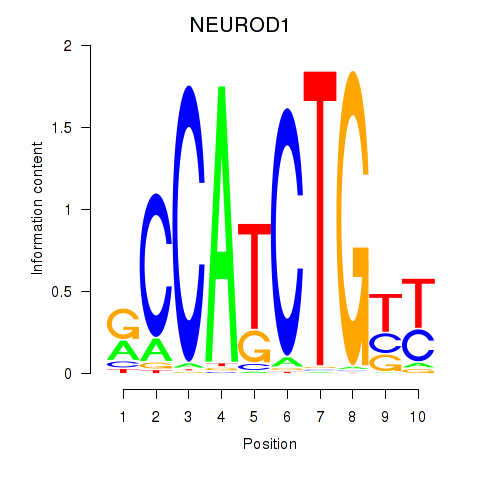
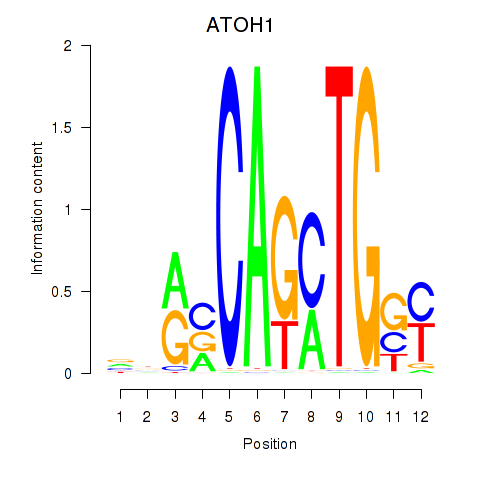
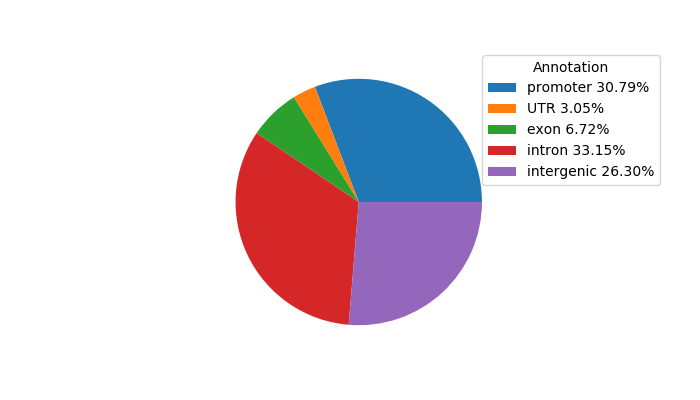
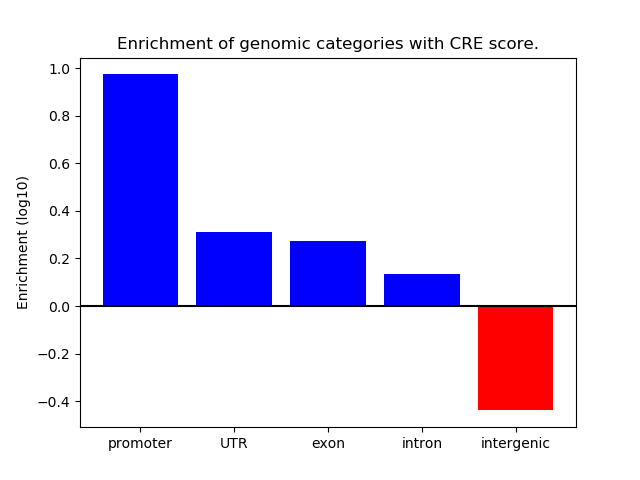
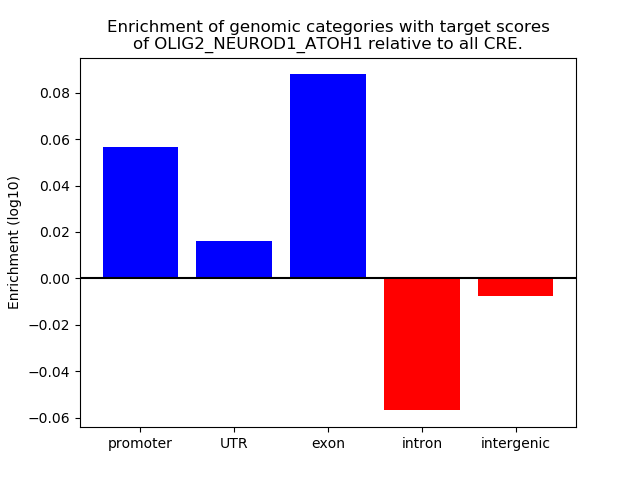
Results for OLIG2_NEUROD1_ATOH1
Z-value: 1.26
Transcription factors associated with OLIG2_NEUROD1_ATOH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG2
|
ENSG00000205927.4 | oligodendrocyte transcription factor 2 |
|
NEUROD1
|
ENSG00000162992.3 | neuronal differentiation 1 |
|
ATOH1
|
ENSG00000172238.3 | atonal bHLH transcription factor 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_94749488_94749739 | ATOH1 | 429 | 0.879995 | 0.64 | 6.3e-02 | Click! |
| chr4_94750731_94750882 | ATOH1 | 764 | 0.732053 | 0.59 | 9.5e-02 | Click! |
| chr4_94749767_94750023 | ATOH1 | 147 | 0.970769 | 0.59 | 9.6e-02 | Click! |
| chr4_94750171_94750322 | ATOH1 | 204 | 0.958334 | 0.34 | 3.7e-01 | Click! |
| chr4_94750547_94750698 | ATOH1 | 580 | 0.814317 | 0.23 | 5.6e-01 | Click! |
| chr2_182546068_182546219 | NEUROD1 | 540 | 0.716081 | -0.21 | 5.9e-01 | Click! |
| chr2_182545553_182545957 | NEUROD1 | 152 | 0.947177 | -0.07 | 8.5e-01 | Click! |
| chr21_34415498_34415672 | OLIG2 | 17342 | 0.163952 | 0.93 | 2.4e-04 | Click! |
| chr21_34415086_34415237 | OLIG2 | 16918 | 0.165161 | 0.93 | 3.3e-04 | Click! |
| chr21_34398837_34398988 | OLIG2 | 669 | 0.716984 | 0.70 | 3.6e-02 | Click! |
| chr21_34398214_34398768 | OLIG2 | 248 | 0.929091 | 0.68 | 4.5e-02 | Click! |
| chr21_34399098_34399249 | OLIG2 | 930 | 0.592307 | 0.63 | 7.1e-02 | Click! |
Activity of the OLIG2_NEUROD1_ATOH1 motif across conditions
Conditions sorted by the z-value of the OLIG2_NEUROD1_ATOH1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
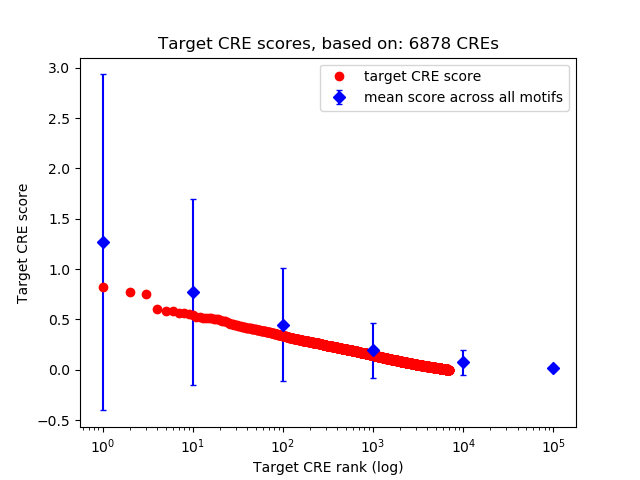
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_127822596_127822803 | 0.82 |
BIN1 |
bridging integrator 1 |
41878 |
0.16 |
| chr2_164592268_164592669 | 0.77 |
FIGN |
fidgetin |
49 |
0.99 |
| chr15_39871853_39872515 | 0.75 |
THBS1 |
thrombospondin 1 |
1096 |
0.52 |
| chr1_230223080_230223231 | 0.60 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
20137 |
0.23 |
| chr1_8932959_8933177 | 0.59 |
ENO1-IT1 |
ENO1 intronic transcript 1 (non-protein coding) |
4998 |
0.14 |
| chr19_41104181_41104507 | 0.58 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
1203 |
0.38 |
| chr9_109459969_109460612 | 0.57 |
ENSG00000200131 |
. |
18032 |
0.28 |
| chr14_95235148_95235528 | 0.56 |
GSC |
goosecoid homeobox |
1224 |
0.6 |
| chr2_239757510_239758069 | 0.55 |
TWIST2 |
twist family bHLH transcription factor 2 |
1116 |
0.62 |
| chr10_6779919_6780270 | 0.55 |
PRKCQ |
protein kinase C, theta |
157831 |
0.04 |
| chr6_48037466_48037693 | 0.53 |
PTCHD4 |
patched domain containing 4 |
1154 |
0.67 |
| chr17_72426935_72427098 | 0.53 |
GPRC5C |
G protein-coupled receptor, family C, group 5, member C |
36 |
0.97 |
| chr3_99349211_99349490 | 0.52 |
COL8A1 |
collagen, type VIII, alpha 1 |
7969 |
0.28 |
| chr1_32042837_32043215 | 0.52 |
TINAGL1 |
tubulointerstitial nephritis antigen-like 1 |
887 |
0.47 |
| chr20_52789675_52790234 | 0.51 |
CYP24A1 |
cytochrome P450, family 24, subfamily A, polypeptide 1 |
234 |
0.95 |
| chr1_17560057_17560274 | 0.51 |
PADI1 |
peptidyl arginine deiminase, type I |
315 |
0.88 |
| chr11_460592_460743 | 0.51 |
RP13-317D12.3 |
|
3232 |
0.12 |
| chr17_55945610_55945963 | 0.51 |
CUEDC1 |
CUE domain containing 1 |
1010 |
0.52 |
| chr17_7492744_7493045 | 0.50 |
SOX15 |
SRY (sex determining region Y)-box 15 |
496 |
0.46 |
| chr11_13119960_13120111 | 0.49 |
ENSG00000266625 |
. |
134848 |
0.05 |
| chr18_8615592_8616115 | 0.49 |
RAB12 |
RAB12, member RAS oncogene family |
6410 |
0.21 |
| chr20_60892287_60892438 | 0.48 |
RP11-157P1.4 |
|
10981 |
0.12 |
| chr4_115520348_115520872 | 0.48 |
UGT8 |
UDP glycosyltransferase 8 |
684 |
0.8 |
| chr12_50482265_50482493 | 0.47 |
SMARCD1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 1 |
361 |
0.78 |
| chr7_150784606_150785150 | 0.47 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
724 |
0.46 |
| chr20_6032553_6033205 | 0.46 |
LRRN4 |
leucine rich repeat neuronal 4 |
1816 |
0.35 |
| chr3_127174283_127174496 | 0.45 |
TPRA1 |
transmembrane protein, adipocyte asscociated 1 |
124891 |
0.05 |
| chr6_27584810_27585278 | 0.45 |
ENSG00000238648 |
. |
14144 |
0.19 |
| chr10_99648580_99648731 | 0.45 |
CRTAC1 |
cartilage acidic protein 1 |
4656 |
0.2 |
| chr8_67434316_67434584 | 0.44 |
ENSG00000206949 |
. |
20904 |
0.17 |
| chr2_75426626_75426804 | 0.44 |
TACR1 |
tachykinin receptor 1 |
111 |
0.98 |
| chr22_51135201_51135410 | 0.44 |
ENSG00000206841 |
. |
5514 |
0.12 |
| chr13_31439124_31439427 | 0.44 |
MEDAG |
mesenteric estrogen-dependent adipogenesis |
41053 |
0.16 |
| chr1_32135114_32135265 | 0.43 |
COL16A1 |
collagen, type XVI, alpha 1 |
3507 |
0.16 |
| chr6_169278047_169278198 | 0.43 |
SMOC2 |
SPARC related modular calcium binding 2 |
224358 |
0.02 |
| chr2_231191950_231192650 | 0.42 |
SP140L |
SP140 nuclear body protein-like |
315 |
0.92 |
| chr6_152701966_152702349 | 0.42 |
SYNE1-AS1 |
SYNE1 antisense RNA 1 |
476 |
0.79 |
| chr14_106369996_106370147 | 0.42 |
ENSG00000211923 |
. |
314 |
0.34 |
| chr7_29324592_29324856 | 0.42 |
AC004593.3 |
|
76138 |
0.1 |
| chr13_76259054_76259476 | 0.42 |
LMO7 |
LIM domain 7 |
48806 |
0.13 |
| chr20_62328885_62329036 | 0.42 |
TNFRSF6B |
tumor necrosis factor receptor superfamily, member 6b, decoy |
939 |
0.35 |
| chr1_207202821_207202972 | 0.41 |
C1orf116 |
chromosome 1 open reading frame 116 |
3196 |
0.17 |
| chr6_90928649_90929126 | 0.41 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
77574 |
0.1 |
| chr8_90323500_90323651 | 0.41 |
RIPK2 |
receptor-interacting serine-threonine kinase 2 |
446400 |
0.01 |
| chr7_42275396_42275690 | 0.41 |
GLI3 |
GLI family zinc finger 3 |
1069 |
0.7 |
| chr5_2750019_2750705 | 0.41 |
IRX2 |
iroquois homeobox 2 |
1407 |
0.43 |
| chr16_121276_121527 | 0.40 |
RHBDF1 |
rhomboid 5 homolog 1 (Drosophila) |
1228 |
0.28 |
| chr1_230223344_230223516 | 0.40 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
20412 |
0.23 |
| chr8_99836773_99837010 | 0.40 |
STK3 |
serine/threonine kinase 3 |
1008 |
0.66 |
| chr17_62051929_62052085 | 0.40 |
SCN4A |
sodium channel, voltage-gated, type IV, alpha subunit |
1729 |
0.26 |
| chr3_98609388_98609782 | 0.40 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
10430 |
0.18 |
| chr10_13748985_13749251 | 0.40 |
ENSG00000222235 |
. |
2140 |
0.21 |
| chr1_49259010_49259161 | 0.40 |
BEND5 |
BEN domain containing 5 |
16495 |
0.28 |
| chrX_19716745_19716896 | 0.40 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
27704 |
0.25 |
| chr5_66334972_66335234 | 0.39 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
34620 |
0.19 |
| chr10_105599590_105599741 | 0.39 |
SH3PXD2A |
SH3 and PX domains 2A |
15499 |
0.19 |
| chr19_46270378_46270670 | 0.39 |
AC074212.6 |
|
530 |
0.46 |
| chr17_39779390_39779541 | 0.39 |
KRT17 |
keratin 17 |
55 |
0.95 |
| chr1_116380584_116380832 | 0.39 |
NHLH2 |
nescient helix loop helix 2 |
2654 |
0.36 |
| chr10_88728248_88728709 | 0.38 |
ADIRF |
adipogenesis regulatory factor |
289 |
0.5 |
| chr7_45002030_45002849 | 0.38 |
RP4-647J21.1 |
|
1931 |
0.25 |
| chr15_90380123_90380631 | 0.38 |
ENSG00000264966 |
. |
13576 |
0.11 |
| chr1_152082781_152083810 | 0.38 |
TCHH |
trichohyalin |
3261 |
0.18 |
| chr21_18214928_18215293 | 0.38 |
ENSG00000239023 |
. |
123793 |
0.06 |
| chr2_131594749_131594981 | 0.38 |
ARHGEF4 |
Rho guanine nucleotide exchange factor (GEF) 4 |
163 |
0.67 |
| chr5_172659939_172660230 | 0.38 |
NKX2-5 |
NK2 homeobox 5 |
2125 |
0.3 |
| chr9_89952337_89952906 | 0.38 |
ENSG00000212421 |
. |
77256 |
0.11 |
| chr2_208105325_208105697 | 0.38 |
AC007879.5 |
|
13465 |
0.21 |
| chr10_95143500_95143651 | 0.38 |
MYOF |
myoferlin |
98376 |
0.07 |
| chr2_162275011_162275548 | 0.37 |
TBR1 |
T-box, brain, 1 |
873 |
0.56 |
| chr6_111947186_111947610 | 0.37 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
19917 |
0.16 |
| chr1_24469240_24469593 | 0.37 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
195 |
0.94 |
| chr2_129063011_129063600 | 0.37 |
HS6ST1 |
heparan sulfate 6-O-sulfotransferase 1 |
12846 |
0.24 |
| chr19_17940819_17940970 | 0.37 |
INSL3 |
insulin-like 3 (Leydig cell) |
8511 |
0.11 |
| chr22_42710235_42710386 | 0.37 |
TCF20 |
transcription factor 20 (AR1) |
29312 |
0.18 |
| chr12_112196119_112196270 | 0.37 |
RP11-162P23.2 |
|
4500 |
0.2 |
| chr4_85886471_85886934 | 0.36 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
801 |
0.75 |
| chr4_125065453_125065672 | 0.36 |
ANKRD50 |
ankyrin repeat domain 50 |
568325 |
0.0 |
| chr17_74525686_74525960 | 0.36 |
CYGB |
cytoglobin |
2324 |
0.14 |
| chr4_10107960_10108187 | 0.36 |
ENSG00000223086 |
. |
9435 |
0.16 |
| chr12_54393540_54393854 | 0.36 |
HOXC-AS1 |
HOXC cluster antisense RNA 1 |
97 |
0.62 |
| chr6_34191164_34191315 | 0.36 |
HMGA1 |
high mobility group AT-hook 1 |
13411 |
0.2 |
| chr20_9389767_9389966 | 0.36 |
PLCB4 |
phospholipase C, beta 4 |
101419 |
0.07 |
| chr8_142083367_142083518 | 0.36 |
DENND3 |
DENN/MADD domain containing 3 |
43935 |
0.14 |
| chr1_956379_957022 | 0.36 |
AGRN |
agrin |
1197 |
0.27 |
| chr8_37372289_37372565 | 0.36 |
RP11-150O12.6 |
|
2112 |
0.43 |
| chr4_124426780_124427488 | 0.35 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
106011 |
0.08 |
| chr19_34972293_34972513 | 0.35 |
WTIP |
Wilms tumor 1 interacting protein |
140 |
0.95 |
| chr19_39142719_39142948 | 0.35 |
ACTN4 |
actinin, alpha 4 |
4455 |
0.14 |
| chrX_153029718_153029902 | 0.35 |
PLXNB3 |
plexin B3 |
84 |
0.93 |
| chr4_88029433_88029643 | 0.35 |
AFF1 |
AF4/FMR2 family, member 1 |
58734 |
0.14 |
| chr9_688099_688250 | 0.35 |
RP11-130C19.3 |
|
2619 |
0.32 |
| chr19_11372274_11372425 | 0.34 |
DOCK6 |
dedicator of cytokinesis 6 |
779 |
0.49 |
| chr11_57406082_57406969 | 0.34 |
AP000662.4 |
|
676 |
0.5 |
| chr4_80572253_80572434 | 0.34 |
OR7E94P |
olfactory receptor, family 7, subfamily E, member 94 pseudogene |
63059 |
0.15 |
| chr4_53507302_53507475 | 0.34 |
USP46 |
ubiquitin specific peptidase 46 |
15369 |
0.2 |
| chr3_150089259_150089633 | 0.34 |
TSC22D2 |
TSC22 domain family, member 2 |
36676 |
0.21 |
| chr15_86329767_86329918 | 0.34 |
KLHL25 |
kelch-like family member 25 |
8258 |
0.12 |
| chr11_2738826_2738977 | 0.34 |
KCNQ1OT1 |
KCNQ1 opposite strand/antisense transcript 1 (non-protein coding) |
17677 |
0.22 |
| chr1_170630467_170630618 | 0.34 |
PRRX1 |
paired related homeobox 1 |
1733 |
0.5 |
| chr12_71833793_71834525 | 0.34 |
LGR5 |
leucine-rich repeat containing G protein-coupled receptor 5 |
344 |
0.86 |
| chr5_10077992_10078143 | 0.34 |
CTD-2256P15.1 |
|
170370 |
0.03 |
| chr10_73019820_73020069 | 0.34 |
UNC5B-AS1 |
UNC5B antisense RNA 1 |
41959 |
0.12 |
| chr14_78447028_78447687 | 0.34 |
ENSG00000199440 |
. |
159872 |
0.03 |
| chr11_119303601_119304219 | 0.34 |
THY1 |
Thy-1 cell surface antigen |
8215 |
0.14 |
| chr12_6420537_6420688 | 0.33 |
PLEKHG6 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 6 |
513 |
0.73 |
| chr6_12573623_12573848 | 0.33 |
PHACTR1 |
phosphatase and actin regulator 1 |
144158 |
0.05 |
| chr2_241374262_241374834 | 0.33 |
GPC1 |
glypican 1 |
540 |
0.75 |
| chr19_42710829_42711018 | 0.33 |
DEDD2 |
death effector domain containing 2 |
10898 |
0.09 |
| chr7_44298624_44298775 | 0.33 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
16794 |
0.18 |
| chr10_21625398_21625641 | 0.33 |
ENSG00000207264 |
. |
14629 |
0.25 |
| chr10_126389149_126389300 | 0.33 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
2970 |
0.28 |
| chrX_19906774_19906925 | 0.32 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
1130 |
0.6 |
| chr11_1897109_1897260 | 0.32 |
LSP1 |
lymphocyte-specific protein 1 |
523 |
0.63 |
| chr10_43248840_43249724 | 0.32 |
BMS1 |
BMS1 ribosome biogenesis factor |
28967 |
0.19 |
| chr11_57087212_57087483 | 0.32 |
TNKS1BP1 |
tankyrase 1 binding protein 1, 182kDa |
2324 |
0.17 |
| chr5_106741120_106741311 | 0.32 |
EFNA5 |
ephrin-A5 |
265113 |
0.02 |
| chr14_91735710_91735943 | 0.32 |
GPR68 |
G protein-coupled receptor 68 |
15557 |
0.16 |
| chr15_59560035_59560196 | 0.32 |
RP11-429D19.1 |
|
3246 |
0.19 |
| chr2_174983381_174983564 | 0.32 |
OLA1 |
Obg-like ATPase 1 |
99202 |
0.07 |
| chr3_73673648_73674178 | 0.32 |
PDZRN3 |
PDZ domain containing ring finger 3 |
78 |
0.96 |
| chr6_101841770_101842075 | 0.32 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
4742 |
0.37 |
| chr22_50744615_50744936 | 0.32 |
PLXNB2 |
plexin B2 |
1242 |
0.27 |
| chr9_140352440_140352691 | 0.32 |
NSMF |
NMDA receptor synaptonuclear signaling and neuronal migration factor |
576 |
0.63 |
| chr5_135364769_135365006 | 0.31 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
196 |
0.95 |
| chr22_30087528_30087679 | 0.31 |
RP1-76B20.12 |
|
28100 |
0.11 |
| chr13_86372735_86372886 | 0.31 |
SLITRK6 |
SLIT and NTRK-like family, member 6 |
813 |
0.79 |
| chr7_102632036_102632223 | 0.31 |
NFE4 |
nuclear factor, erythroid 4 |
18160 |
0.14 |
| chr20_62085538_62085770 | 0.31 |
RP11-358D14.2 |
|
6250 |
0.13 |
| chr21_30517898_30518405 | 0.31 |
ENSG00000201984 |
. |
6446 |
0.14 |
| chr8_32077875_32078317 | 0.31 |
NRG1-IT2 |
NRG1 intronic transcript 2 (non-protein coding) |
348 |
0.9 |
| chr17_27506561_27506872 | 0.31 |
MYO18A |
myosin XVIIIA |
666 |
0.68 |
| chr6_158181766_158181950 | 0.31 |
SNX9 |
sorting nexin 9 |
62438 |
0.11 |
| chr11_126870507_126870686 | 0.31 |
KIRREL3 |
kin of IRRE like 3 (Drosophila) |
59 |
0.96 |
| chr19_41729247_41729598 | 0.31 |
CTD-2195B23.3 |
|
1466 |
0.28 |
| chr3_58012091_58012242 | 0.31 |
FLNB |
filamin B, beta |
18039 |
0.25 |
| chr20_61297836_61297987 | 0.31 |
RP11-93B14.5 |
|
63 |
0.94 |
| chr20_47299051_47299422 | 0.31 |
ENSG00000251876 |
. |
56749 |
0.16 |
| chr8_86937269_86937426 | 0.31 |
ATP6V0D2 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
62205 |
0.14 |
| chr3_51425020_51425499 | 0.31 |
MANF |
mesencephalic astrocyte-derived neurotrophic factor |
2781 |
0.26 |
| chr11_61047532_61047683 | 0.30 |
VWCE |
von Willebrand factor C and EGF domains |
6329 |
0.14 |
| chr17_6797470_6797621 | 0.30 |
ALOX12P2 |
arachidonate 12-lipoxygenase pseudogene 2 |
444 |
0.79 |
| chr20_61463695_61464080 | 0.30 |
COL9A3 |
collagen, type IX, alpha 3 |
14724 |
0.11 |
| chr20_43921897_43922225 | 0.30 |
MATN4 |
matrilin 4 |
12170 |
0.12 |
| chr3_64801656_64801807 | 0.30 |
ADAMTS9 |
ADAM metallopeptidase with thrombospondin type 1 motif, 9 |
128055 |
0.05 |
| chr17_17726041_17726867 | 0.30 |
SREBF1 |
sterol regulatory element binding transcription factor 1 |
478 |
0.73 |
| chr8_22023419_22024083 | 0.30 |
BMP1 |
bone morphogenetic protein 1 |
951 |
0.4 |
| chr20_18039503_18040184 | 0.30 |
RP4-726N1.2 |
|
294 |
0.83 |
| chr17_38614815_38615028 | 0.30 |
IGFBP4 |
insulin-like growth factor binding protein 4 |
15208 |
0.13 |
| chr9_71177567_71177718 | 0.30 |
TMEM252 |
transmembrane protein 252 |
21859 |
0.24 |
| chr12_54402958_54403134 | 0.30 |
HOXC8 |
homeobox C8 |
214 |
0.8 |
| chr1_235133406_235133693 | 0.30 |
ENSG00000239690 |
. |
93616 |
0.08 |
| chr17_74666825_74666976 | 0.30 |
RP11-318A15.2 |
|
1124 |
0.31 |
| chr2_163174641_163175537 | 0.30 |
IFIH1 |
interferon induced with helicase C domain 1 |
105 |
0.76 |
| chr12_109729676_109729841 | 0.30 |
FOXN4 |
forkhead box N4 |
5230 |
0.23 |
| chr13_30168298_30169638 | 0.29 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
857 |
0.74 |
| chr19_49654515_49654666 | 0.29 |
HRC |
histidine rich calcium binding protein |
2102 |
0.15 |
| chr12_54390992_54391173 | 0.29 |
HOXC-AS2 |
HOXC cluster antisense RNA 2 |
513 |
0.46 |
| chr10_112113893_112114201 | 0.29 |
SMNDC1 |
survival motor neuron domain containing 1 |
49338 |
0.14 |
| chr9_80499174_80499388 | 0.29 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
61366 |
0.15 |
| chr19_32477992_32478143 | 0.29 |
ENSG00000221504 |
. |
60588 |
0.15 |
| chr2_115384761_115384912 | 0.29 |
DPP10 |
dipeptidyl-peptidase 10 (non-functional) |
165656 |
0.04 |
| chr4_75230404_75230837 | 0.29 |
EREG |
epiregulin |
240 |
0.93 |
| chr17_1618285_1618721 | 0.29 |
ENSG00000186594 |
. |
1218 |
0.24 |
| chr13_110521954_110522217 | 0.29 |
ENSG00000201161 |
. |
45358 |
0.18 |
| chr1_224804905_224805156 | 0.29 |
CNIH3 |
cornichon family AMPA receptor auxiliary protein 3 |
1035 |
0.41 |
| chr7_6437932_6438083 | 0.29 |
RAC1 |
ras-related C3 botulinum toxin substrate 1 (rho family, small GTP binding protein Rac1) |
23837 |
0.15 |
| chr3_28617795_28617996 | 0.29 |
LINC00693 |
long intergenic non-protein coding RNA 693 |
7 |
0.99 |
| chr12_78334342_78334902 | 0.29 |
NAV3 |
neuron navigator 3 |
25434 |
0.27 |
| chr3_42113480_42113939 | 0.29 |
TRAK1 |
trafficking protein, kinesin binding 1 |
18853 |
0.25 |
| chr16_11295551_11295900 | 0.29 |
RMI2 |
RecQ mediated genome instability 2 |
47781 |
0.08 |
| chr12_10874089_10875353 | 0.29 |
YBX3 |
Y box binding protein 3 |
1185 |
0.46 |
| chr17_73718603_73718754 | 0.29 |
ITGB4 |
integrin, beta 4 |
1101 |
0.34 |
| chr1_197880677_197880955 | 0.29 |
LHX9 |
LIM homeobox 9 |
805 |
0.69 |
| chr1_116381723_116382451 | 0.29 |
NHLH2 |
nescient helix loop helix 2 |
1275 |
0.57 |
| chr3_33319990_33320141 | 0.29 |
FBXL2 |
F-box and leucine-rich repeat protein 2 |
1097 |
0.59 |
| chr10_74436092_74436294 | 0.29 |
MCU |
mitochondrial calcium uniporter |
15696 |
0.15 |
| chr7_96653818_96654015 | 0.29 |
DLX5 |
distal-less homeobox 5 |
346 |
0.86 |
| chr9_113800702_113801367 | 0.29 |
LPAR1 |
lysophosphatidic acid receptor 1 |
53 |
0.98 |
| chr1_153587674_153587825 | 0.29 |
S100A14 |
S100 calcium binding protein A14 |
665 |
0.44 |
| chr19_3346586_3346737 | 0.29 |
NFIC |
nuclear factor I/C (CCAAT-binding transcription factor) |
12900 |
0.17 |
| chr17_694312_694463 | 0.29 |
RP11-676J12.8 |
|
622 |
0.63 |
| chr14_37050333_37050591 | 0.29 |
NKX2-8 |
NK2 homeobox 8 |
1350 |
0.4 |
| chr12_122235813_122236047 | 0.29 |
RHOF |
ras homolog family member F (in filopodia) |
2653 |
0.22 |
| chr9_139240222_139240529 | 0.28 |
GPSM1 |
G-protein signaling modulator 1 |
7134 |
0.12 |
| chr1_26127054_26127674 | 0.28 |
SEPN1 |
selenoprotein N, 1 |
680 |
0.54 |
| chr2_160918347_160919586 | 0.28 |
PLA2R1 |
phospholipase A2 receptor 1, 180kDa |
155 |
0.98 |
| chr6_33636840_33636991 | 0.28 |
SBP1 |
SBP1; Uncharacterized protein |
26559 |
0.11 |
| chr18_9825005_9825225 | 0.28 |
ENSG00000242651 |
. |
5509 |
0.21 |
| chr17_34611752_34612896 | 0.28 |
CCL3L1 |
chemokine (C-C motif) ligand 3-like 1 |
13395 |
0.14 |
| chr4_85873557_85873708 | 0.28 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
13871 |
0.26 |
| chr2_227623803_227624269 | 0.28 |
IRS1 |
insulin receptor substrate 1 |
40439 |
0.16 |
| chr16_14597742_14597893 | 0.28 |
AC092291.2 |
|
32975 |
0.17 |
| chr17_65361877_65362420 | 0.28 |
PSMD12 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 |
573 |
0.7 |
| chr19_4061458_4061934 | 0.28 |
CTD-2622I13.3 |
|
1051 |
0.37 |
| chr16_62068318_62068835 | 0.28 |
CDH8 |
cadherin 8, type 2 |
462 |
0.9 |
| chr2_106012197_106012348 | 0.28 |
FHL2 |
four and a half LIM domains 2 |
970 |
0.58 |
| chr14_52116783_52117087 | 0.28 |
FRMD6 |
FERM domain containing 6 |
1641 |
0.39 |
| chr5_160265170_160265321 | 0.28 |
ATP10B |
ATPase, class V, type 10B |
13974 |
0.28 |
| chr1_205418778_205418941 | 0.28 |
LEMD1 |
LEM domain containing 1 |
200 |
0.91 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0003079 | obsolete positive regulation of natriuresis(GO:0003079) |
| 0.1 | 0.4 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.4 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.1 | 0.4 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.1 | 0.5 | GO:0070561 | vitamin D receptor signaling pathway(GO:0070561) |
| 0.1 | 0.5 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.1 | 0.4 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.1 | 0.3 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.3 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.3 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.1 | 0.4 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.1 | 0.3 | GO:0032347 | regulation of ketone biosynthetic process(GO:0010566) regulation of aldosterone metabolic process(GO:0032344) regulation of aldosterone biosynthetic process(GO:0032347) |
| 0.1 | 0.4 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.1 | 0.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.2 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.1 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.1 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.5 | GO:0006554 | lysine metabolic process(GO:0006553) lysine catabolic process(GO:0006554) |
| 0.1 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.1 | 0.2 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.2 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.1 | 0.2 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.1 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.1 | 0.2 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.2 | GO:0046628 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.1 | 0.2 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.2 | GO:0018119 | protein nitrosylation(GO:0017014) peptidyl-cysteine S-nitrosylation(GO:0018119) |
| 0.1 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.1 | 0.2 | GO:0003283 | atrial septum development(GO:0003283) atrial septum morphogenesis(GO:0060413) |
| 0.1 | 0.2 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.1 | 0.3 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.1 | 0.2 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.1 | 0.3 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.1 | GO:0006296 | nucleotide-excision repair, DNA incision, 5'-to lesion(GO:0006296) |
| 0.0 | 0.0 | GO:0031392 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.2 | GO:0007028 | cytoplasm organization(GO:0007028) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0044266 | multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) multicellular organismal protein metabolic process(GO:0044268) |
| 0.0 | 0.7 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.2 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.1 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.2 | GO:0090026 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.7 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.2 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.2 | GO:0045815 | positive regulation of gene expression, epigenetic(GO:0045815) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.2 | GO:0032331 | negative regulation of chondrocyte differentiation(GO:0032331) |
| 0.0 | 0.1 | GO:0070977 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.2 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.3 | GO:0045843 | negative regulation of striated muscle tissue development(GO:0045843) |
| 0.0 | 0.1 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.0 | GO:0070099 | regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.0 | 0.1 | GO:0033567 | DNA replication, Okazaki fragment processing(GO:0033567) DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0072141 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.1 | GO:0032415 | regulation of sodium:proton antiporter activity(GO:0032415) regulation of sodium ion transmembrane transport(GO:1902305) regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0010269 | response to selenium ion(GO:0010269) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0014848 | urinary tract smooth muscle contraction(GO:0014848) |
| 0.0 | 0.1 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.2 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.1 | GO:0010803 | regulation of tumor necrosis factor-mediated signaling pathway(GO:0010803) |
| 0.0 | 0.2 | GO:0042487 | regulation of odontogenesis of dentin-containing tooth(GO:0042487) |
| 0.0 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.2 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.4 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.0 | GO:0002407 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0010719 | negative regulation of epithelial to mesenchymal transition(GO:0010719) |
| 0.0 | 0.2 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.2 | GO:0055003 | cardiac myofibril assembly(GO:0055003) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.2 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.1 | GO:0072367 | regulation of lipid transport by regulation of transcription from RNA polymerase II promoter(GO:0072367) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0046322 | negative regulation of fatty acid oxidation(GO:0046322) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0060291 | long-term synaptic potentiation(GO:0060291) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0048087 | positive regulation of developmental pigmentation(GO:0048087) |
| 0.0 | 0.1 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.2 | GO:0051597 | response to methylmercury(GO:0051597) |
| 0.0 | 0.2 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.1 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.2 | GO:0055093 | response to increased oxygen levels(GO:0036296) response to hyperoxia(GO:0055093) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.0 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.0 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0032682 | negative regulation of chemokine production(GO:0032682) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.1 | GO:0043032 | positive regulation of macrophage activation(GO:0043032) |
| 0.0 | 0.0 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 1.1 | GO:0034332 | adherens junction organization(GO:0034332) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0045071 | negative regulation of viral genome replication(GO:0045071) |
| 0.0 | 0.0 | GO:0060068 | vagina development(GO:0060068) |
| 0.0 | 0.0 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0009188 | ADP biosynthetic process(GO:0006172) purine nucleoside diphosphate biosynthetic process(GO:0009136) purine ribonucleoside diphosphate biosynthetic process(GO:0009180) ribonucleoside diphosphate biosynthetic process(GO:0009188) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.3 | GO:0090263 | positive regulation of canonical Wnt signaling pathway(GO:0090263) |
| 0.0 | 0.1 | GO:0022011 | Schwann cell development(GO:0014044) myelination in peripheral nervous system(GO:0022011) peripheral nervous system axon ensheathment(GO:0032292) |
| 0.0 | 0.2 | GO:1902667 | regulation of axon guidance(GO:1902667) |
| 0.0 | 0.1 | GO:0051797 | positive regulation of hair cycle(GO:0042635) regulation of hair follicle development(GO:0051797) positive regulation of hair follicle development(GO:0051798) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.0 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.2 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.3 | GO:0035195 | gene silencing by miRNA(GO:0035195) |
| 0.0 | 0.2 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0060287 | epithelial cilium movement(GO:0003351) epithelial cilium movement involved in determination of left/right asymmetry(GO:0060287) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.0 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0045916 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.2 | GO:0006558 | L-phenylalanine metabolic process(GO:0006558) L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid metabolic process(GO:1902221) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.2 | GO:0032228 | regulation of synaptic transmission, GABAergic(GO:0032228) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0003309 | type B pancreatic cell differentiation(GO:0003309) |
| 0.0 | 0.1 | GO:0085029 | extracellular matrix assembly(GO:0085029) |
| 0.0 | 0.0 | GO:0070572 | positive regulation of axon regeneration(GO:0048680) positive regulation of neuron projection regeneration(GO:0070572) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.0 | GO:0030423 | targeting of mRNA for destruction involved in RNA interference(GO:0030423) |
| 0.0 | 0.0 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.0 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.1 | GO:0035025 | positive regulation of Rho protein signal transduction(GO:0035025) |
| 0.0 | 0.1 | GO:0051446 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0032202 | telomere assembly(GO:0032202) |
| 0.0 | 0.1 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.3 | GO:0032006 | regulation of TOR signaling(GO:0032006) |
| 0.0 | 0.0 | GO:0021892 | cerebral cortex GABAergic interneuron differentiation(GO:0021892) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.0 | GO:0048702 | embryonic neurocranium morphogenesis(GO:0048702) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0014911 | positive regulation of smooth muscle cell migration(GO:0014911) |
| 0.0 | 0.0 | GO:0021826 | substrate-independent telencephalic tangential migration(GO:0021826) substrate-independent telencephalic tangential interneuron migration(GO:0021843) |
| 0.0 | 0.3 | GO:0006662 | glycerol ether metabolic process(GO:0006662) ether metabolic process(GO:0018904) |
| 0.0 | 0.1 | GO:0031272 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.0 | GO:0016999 | antibiotic metabolic process(GO:0016999) antibiotic biosynthetic process(GO:0017000) |
| 0.0 | 0.0 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0006726 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) eye pigmentation(GO:0048069) |
| 0.0 | 0.0 | GO:0045617 | negative regulation of keratinocyte differentiation(GO:0045617) |
| 0.0 | 0.0 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.0 | GO:0046639 | negative regulation of alpha-beta T cell differentiation(GO:0046639) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) negative regulation of kidney development(GO:0090185) |
| 0.0 | 0.0 | GO:0060872 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0042756 | drinking behavior(GO:0042756) |
| 0.0 | 0.0 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.0 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.2 | GO:0001502 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.2 | GO:1901661 | quinone metabolic process(GO:1901661) |
| 0.0 | 0.2 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.1 | GO:1904729 | regulation of intestinal cholesterol absorption(GO:0030300) regulation of intestinal absorption(GO:1904478) regulation of intestinal lipid absorption(GO:1904729) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.0 | GO:0033197 | response to vitamin E(GO:0033197) |
| 0.0 | 0.1 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.0 | GO:0021978 | telencephalon regionalization(GO:0021978) |
| 0.0 | 0.0 | GO:0010916 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.2 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.0 | GO:0046958 | nonassociative learning(GO:0046958) |
| 0.0 | 0.1 | GO:0051294 | establishment of mitotic spindle orientation(GO:0000132) establishment of spindle orientation(GO:0051294) |
| 0.0 | 0.2 | GO:0007405 | neuroblast proliferation(GO:0007405) |
| 0.0 | 0.0 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0014819 | regulation of skeletal muscle contraction(GO:0014819) |
| 0.0 | 0.2 | GO:0045103 | intermediate filament-based process(GO:0045103) |
| 0.0 | 0.1 | GO:0010894 | negative regulation of steroid biosynthetic process(GO:0010894) |
| 0.0 | 0.1 | GO:0006067 | ethanol metabolic process(GO:0006067) ethanol oxidation(GO:0006069) |
| 0.0 | 0.0 | GO:0032506 | cytokinetic process(GO:0032506) |
| 0.0 | 0.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0006582 | melanin metabolic process(GO:0006582) melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.0 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.2 | GO:0006085 | acetyl-CoA biosynthetic process(GO:0006085) |
| 0.0 | 0.0 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.0 | GO:0051447 | negative regulation of meiotic cell cycle(GO:0051447) |
| 0.0 | 0.1 | GO:0045773 | positive regulation of axon extension(GO:0045773) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.0 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0072243 | metanephric nephron tubule development(GO:0072234) metanephric nephron epithelium development(GO:0072243) |
| 0.0 | 0.0 | GO:0060340 | positive regulation of type I interferon-mediated signaling pathway(GO:0060340) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.2 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.1 | GO:0002360 | T cell lineage commitment(GO:0002360) |
| 0.0 | 0.0 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.0 | GO:0042518 | negative regulation of tyrosine phosphorylation of Stat3 protein(GO:0042518) negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.0 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.0 | GO:0010535 | regulation of activation of JAK2 kinase activity(GO:0010534) positive regulation of activation of JAK2 kinase activity(GO:0010535) |
| 0.0 | 0.1 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.0 | GO:0048703 | embryonic viscerocranium morphogenesis(GO:0048703) |
| 0.0 | 0.0 | GO:0065005 | protein-lipid complex assembly(GO:0065005) |
| 0.0 | 0.1 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.1 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.0 | GO:0010224 | response to UV-B(GO:0010224) |
| 0.0 | 0.0 | GO:0030970 | retrograde protein transport, ER to cytosol(GO:0030970) |
| 0.0 | 0.5 | GO:0008584 | male gonad development(GO:0008584) development of primary male sexual characteristics(GO:0046546) |
| 0.0 | 0.0 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 0.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.2 | GO:1903561 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.1 | 0.2 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.3 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.1 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.3 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.4 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0005832 | chaperonin-containing T-complex(GO:0005832) |
| 0.0 | 0.2 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0042827 | platelet dense granule(GO:0042827) |
| 0.0 | 0.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.4 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0031205 | endoplasmic reticulum Sec complex(GO:0031205) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0005838 | proteasome regulatory particle(GO:0005838) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.3 | GO:0005801 | cis-Golgi network(GO:0005801) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.9 | GO:0016459 | myosin complex(GO:0016459) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0031527 | filopodium membrane(GO:0031527) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.1 | GO:0033162 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0030132 | clathrin coat of coated pit(GO:0030132) |
| 0.0 | 0.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.0 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.4 | GO:0031674 | I band(GO:0031674) |
| 0.0 | 0.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.4 | GO:0016592 | mediator complex(GO:0016592) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.9 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.3 | GO:0008305 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.1 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.0 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.2 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.0 | GO:0033270 | paranode region of axon(GO:0033270) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.4 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.1 | 0.5 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.1 | 0.3 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.1 | 0.3 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.1 | 0.7 | GO:0004029 | aldehyde dehydrogenase (NAD) activity(GO:0004029) |
| 0.1 | 0.4 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.1 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.3 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.1 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.1 | 0.2 | GO:0000987 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.2 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.1 | 0.4 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.1 | 0.2 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.1 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.1 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.1 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.2 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.3 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.3 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.2 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.5 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.5 | GO:0043394 | proteoglycan binding(GO:0043394) |
| 0.0 | 0.2 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.3 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.3 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.5 | GO:0035250 | UDP-galactosyltransferase activity(GO:0035250) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.3 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0016972 | thiol oxidase activity(GO:0016972) |
| 0.0 | 0.5 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.1 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0008107 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.1 | GO:0008296 | 3'-5'-exodeoxyribonuclease activity(GO:0008296) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.4 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0001619 | obsolete lysosphingolipid and lysophosphatidic acid receptor activity(GO:0001619) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.0 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.2 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
| 0.0 | 0.6 | GO:0032947 | protein complex scaffold(GO:0032947) |
| 0.0 | 0.1 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.1 | GO:0031435 | mitogen-activated protein kinase kinase kinase binding(GO:0031435) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.1 | GO:0003941 | L-serine ammonia-lyase activity(GO:0003941) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.2 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0031628 | opioid receptor binding(GO:0031628) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.1 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.2 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.0 | GO:0071208 | histone pre-mRNA DCP binding(GO:0071208) |
| 0.0 | 0.0 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.0 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.0 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.0 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors(GO:0016679) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.0 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.1 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.0 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.0 | GO:0004516 | nicotinate phosphoribosyltransferase activity(GO:0004516) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0016620 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, NAD or NADP as acceptor(GO:0016620) |
| 0.0 | 0.0 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.1 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.3 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.0 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.0 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.0 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.0 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.1 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0031543 | peptidyl-proline dioxygenase activity(GO:0031543) |
| 0.0 | 0.1 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.0 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.1 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.2 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.5 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.6 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.3 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.5 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.3 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.4 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.1 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.2 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.3 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.1 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.1 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.1 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.1 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.3 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 0.3 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.3 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.0 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.1 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.3 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.1 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.1 | 0.1 | REACTOME PLATELET AGGREGATION PLUG FORMATION | Genes involved in Platelet Aggregation (Plug Formation) |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 1.0 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 0.5 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 MUTANTS | Genes involved in Signaling by FGFR1 mutants |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.5 | REACTOME STEROID HORMONES | Genes involved in Steroid hormones |
| 0.0 | 0.1 | REACTOME SEMA3A PAK DEPENDENT AXON REPULSION | Genes involved in Sema3A PAK dependent Axon repulsion |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.4 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.0 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.4 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.0 | REACTOME UNWINDING OF DNA | Genes involved in Unwinding of DNA |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.1 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.0 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.2 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.2 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.1 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME FRS2 MEDIATED CASCADE | Genes involved in FRS2-mediated cascade |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |