Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
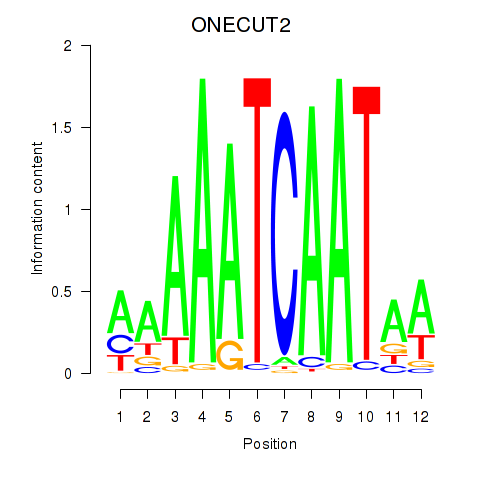
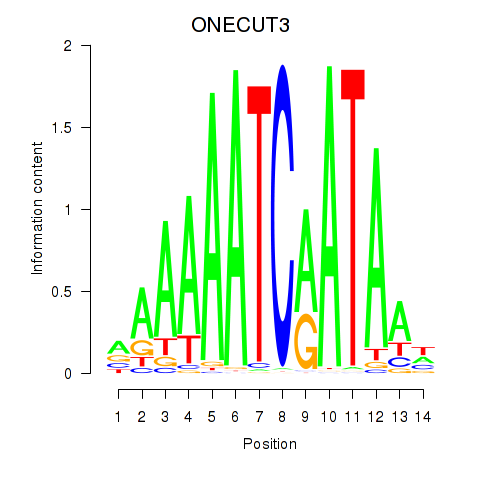
Results for ONECUT2_ONECUT3
Z-value: 1.42
Transcription factors associated with ONECUT2_ONECUT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT2
|
ENSG00000119547.5 | one cut homeobox 2 |
|
ONECUT3
|
ENSG00000205922.4 | one cut homeobox 3 |
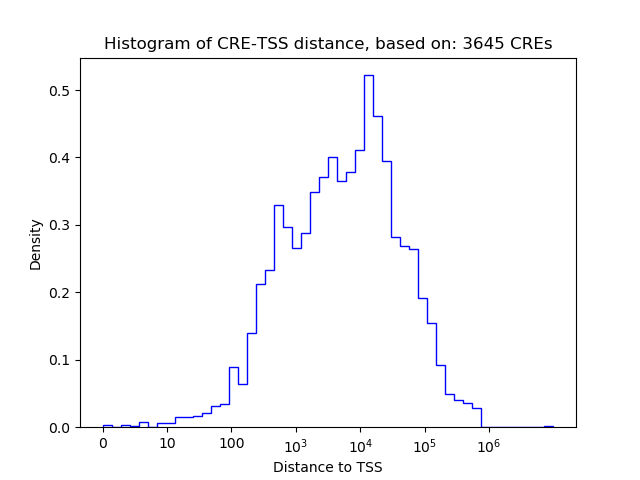
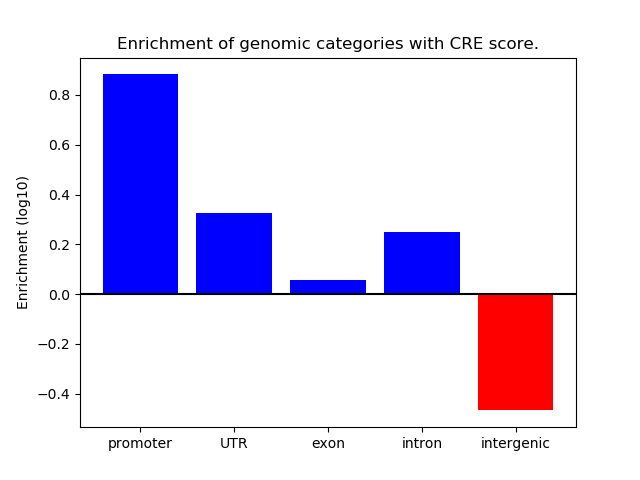
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr18_55101682_55102405 | ONECUT2 | 874 | 0.450005 | -0.65 | 5.8e-02 | Click! |
| chr18_55108207_55108358 | ONECUT2 | 3326 | 0.257933 | -0.56 | 1.2e-01 | Click! |
| chr18_55097019_55097310 | ONECUT2 | 5753 | 0.218301 | -0.45 | 2.2e-01 | Click! |
| chr18_55108476_55109011 | ONECUT2 | 2865 | 0.278155 | -0.33 | 3.8e-01 | Click! |
| chr18_55101119_55101270 | ONECUT2 | 1723 | 0.325356 | -0.28 | 4.6e-01 | Click! |
| chr19_1748094_1748245 | ONECUT3 | 4203 | 0.160848 | -0.60 | 8.9e-02 | Click! |
| chr19_1725454_1725639 | ONECUT3 | 26826 | 0.113872 | -0.59 | 9.7e-02 | Click! |
| chr19_1748614_1748765 | ONECUT3 | 3683 | 0.168872 | -0.36 | 3.4e-01 | Click! |
| chr19_1749427_1749644 | ONECUT3 | 2837 | 0.191666 | -0.35 | 3.6e-01 | Click! |
| chr19_1773658_1773809 | ONECUT3 | 21361 | 0.099840 | -0.35 | 3.6e-01 | Click! |
Activity of the ONECUT2_ONECUT3 motif across conditions
Conditions sorted by the z-value of the ONECUT2_ONECUT3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
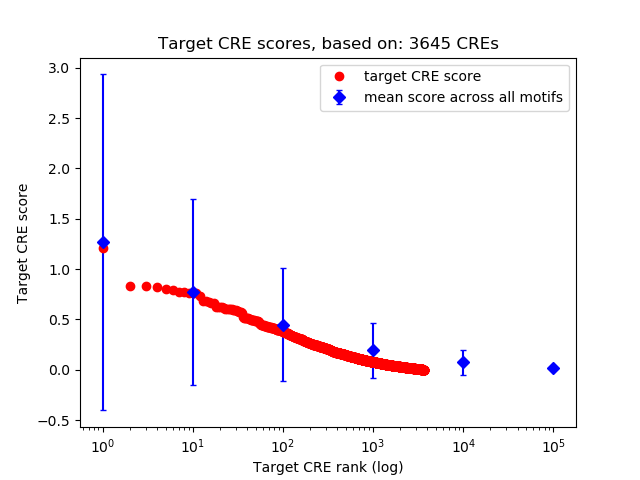
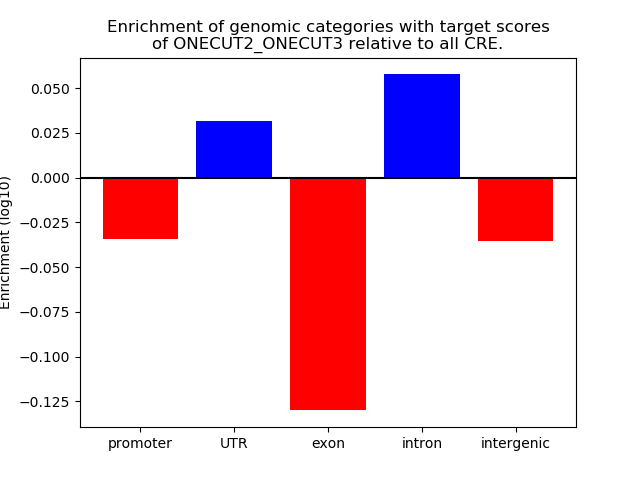
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr19_32976430_32976581 | 1.21 |
ENSG00000263780 |
. |
15342 |
0.19 |
| chr17_61727557_61727708 | 0.83 |
MAP3K3 |
mitogen-activated protein kinase kinase kinase 3 |
27562 |
0.12 |
| chr1_181474291_181474442 | 0.83 |
CACNA1E |
calcium channel, voltage-dependent, R type, alpha 1E subunit |
21485 |
0.28 |
| chr8_121761660_121761834 | 0.82 |
RP11-713M15.1 |
|
11746 |
0.25 |
| chr12_25207113_25207806 | 0.80 |
LRMP |
lymphoid-restricted membrane protein |
1785 |
0.37 |
| chr7_130610833_130610984 | 0.79 |
ENSG00000226380 |
. |
48610 |
0.15 |
| chr7_50348888_50349341 | 0.78 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
796 |
0.76 |
| chr14_89831123_89831421 | 0.78 |
RP11-356K23.2 |
|
9868 |
0.17 |
| chr11_3852411_3852562 | 0.77 |
RHOG |
ras homolog family member G |
605 |
0.57 |
| chrX_117632028_117632179 | 0.77 |
DOCK11 |
dedicator of cytokinesis 11 |
2231 |
0.39 |
| chr10_50398086_50398237 | 0.77 |
C10orf128 |
chromosome 10 open reading frame 128 |
1725 |
0.38 |
| chr6_24951057_24951285 | 0.74 |
FAM65B |
family with sequence similarity 65, member B |
14983 |
0.22 |
| chr14_24020377_24021144 | 0.68 |
ZFHX2 |
zinc finger homeobox 2 |
98 |
0.94 |
| chr6_25405502_25405752 | 0.68 |
ENSG00000253017 |
. |
2496 |
0.27 |
| chr8_18054305_18054456 | 0.67 |
NAT1 |
N-acetyltransferase 1 (arylamine N-acetyltransferase) |
13222 |
0.27 |
| chr17_441739_441890 | 0.67 |
RP5-1029F21.2 |
|
13992 |
0.17 |
| chr13_48736574_48736800 | 0.66 |
MED4 |
mediator complex subunit 4 |
67420 |
0.1 |
| chr1_114476630_114476781 | 0.63 |
HIPK1 |
homeodomain interacting protein kinase 1 |
3355 |
0.17 |
| chr2_202127394_202127951 | 0.63 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
993 |
0.56 |
| chr13_30510013_30510639 | 0.62 |
LINC00572 |
long intergenic non-protein coding RNA 572 |
9538 |
0.3 |
| chr1_66799510_66799957 | 0.62 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
1861 |
0.5 |
| chr1_206729975_206731239 | 0.62 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
114 |
0.96 |
| chr7_7298190_7298341 | 0.61 |
C1GALT1 |
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
24314 |
0.22 |
| chr2_55455458_55455609 | 0.60 |
RPS27A |
ribosomal protein S27a |
3506 |
0.17 |
| chr18_29601411_29601562 | 0.60 |
RNF125 |
ring finger protein 125, E3 ubiquitin protein ligase |
3151 |
0.21 |
| chr13_50638056_50638207 | 0.60 |
ENSG00000231607 |
. |
14794 |
0.14 |
| chr1_40849833_40850457 | 0.60 |
SMAP2 |
small ArfGAP2 |
9825 |
0.18 |
| chr2_40621643_40621794 | 0.60 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
35702 |
0.23 |
| chrX_70272475_70272626 | 0.60 |
SNX12 |
sorting nexin 12 |
15703 |
0.12 |
| chr11_104914640_104914826 | 0.59 |
CARD16 |
caspase recruitment domain family, member 16 |
1301 |
0.41 |
| chr11_85754438_85754589 | 0.59 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
25309 |
0.2 |
| chr10_60038316_60038467 | 0.58 |
CISD1 |
CDGSH iron sulfur domain 1 |
9573 |
0.19 |
| chr9_4762764_4763000 | 0.58 |
AK3 |
adenylate kinase 3 |
20839 |
0.16 |
| chr21_15917916_15918619 | 0.58 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
395 |
0.89 |
| chr2_197126307_197126458 | 0.57 |
AC020571.3 |
|
1225 |
0.49 |
| chr10_63660554_63661323 | 0.53 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
121 |
0.98 |
| chr9_132801987_132802138 | 0.52 |
FNBP1 |
formin binding protein 1 |
3379 |
0.25 |
| chr2_158299517_158299677 | 0.52 |
CYTIP |
cytohesin 1 interacting protein |
1057 |
0.48 |
| chr12_93638285_93638436 | 0.51 |
ENSG00000238361 |
. |
21148 |
0.16 |
| chrX_76924403_76924554 | 0.51 |
ATRX |
alpha thalassemia/mental retardation syndrome X-linked |
48242 |
0.18 |
| chr2_143908498_143908649 | 0.51 |
RP11-190J23.1 |
|
21168 |
0.22 |
| chr16_79197756_79198135 | 0.50 |
RP11-556H2.2 |
|
58091 |
0.12 |
| chr15_99443302_99443551 | 0.50 |
RP11-654A16.1 |
|
6662 |
0.22 |
| chr15_63777077_63777228 | 0.50 |
USP3 |
ubiquitin specific peptidase 3 |
19641 |
0.22 |
| chr2_143910568_143910719 | 0.50 |
RP11-190J23.1 |
|
19098 |
0.23 |
| chr4_36244695_36244948 | 0.50 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
740 |
0.5 |
| chr2_46688788_46688939 | 0.49 |
ENSG00000241791 |
. |
13213 |
0.16 |
| chr6_24928259_24928410 | 0.49 |
FAM65B |
family with sequence similarity 65, member B |
7854 |
0.24 |
| chr10_120965669_120965820 | 0.49 |
GRK5 |
G protein-coupled receptor kinase 5 |
1357 |
0.32 |
| chr18_10868392_10868543 | 0.49 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
13033 |
0.24 |
| chr13_97879341_97880219 | 0.49 |
MBNL2 |
muscleblind-like splicing regulator 2 |
5171 |
0.33 |
| chr12_122589685_122589889 | 0.48 |
MLXIP |
MLX interacting protein |
22640 |
0.18 |
| chr3_56627689_56627840 | 0.48 |
CCDC66 |
coiled-coil domain containing 66 |
34984 |
0.2 |
| chr17_33868045_33868636 | 0.47 |
SLFN12L |
schlafen family member 12-like |
3460 |
0.14 |
| chr5_108956915_108957066 | 0.47 |
ENSG00000266090 |
. |
64291 |
0.12 |
| chr16_71915132_71915582 | 0.45 |
RP11-498D10.3 |
|
819 |
0.43 |
| chr20_52269637_52270024 | 0.45 |
ENSG00000238468 |
. |
15467 |
0.22 |
| chr14_53208430_53208600 | 0.44 |
STYX |
serine/threonine/tyrosine interacting protein |
11615 |
0.15 |
| chr9_273424_273720 | 0.44 |
DOCK8 |
dedicator of cytokinesis 8 |
502 |
0.5 |
| chr3_152027985_152028136 | 0.44 |
MBNL1 |
muscleblind-like splicing regulator 1 |
10073 |
0.22 |
| chr4_87050749_87050921 | 0.44 |
RP11-778J15.1 |
|
9968 |
0.23 |
| chr10_105670292_105670878 | 0.44 |
OBFC1 |
oligonucleotide/oligosaccharide-binding fold containing 1 |
6842 |
0.18 |
| chr12_32114494_32114645 | 0.44 |
KIAA1551 |
KIAA1551 |
854 |
0.68 |
| chr6_130534438_130534589 | 0.43 |
SAMD3 |
sterile alpha motif domain containing 3 |
1922 |
0.46 |
| chr9_80524074_80524374 | 0.43 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
86309 |
0.1 |
| chr5_57787350_57787738 | 0.43 |
GAPT |
GRB2-binding adaptor protein, transmembrane |
280 |
0.92 |
| chr11_108443146_108443297 | 0.43 |
EXPH5 |
exophilin 5 |
4368 |
0.28 |
| chr5_98276840_98277133 | 0.43 |
ENSG00000200351 |
. |
4535 |
0.25 |
| chr18_46572008_46572159 | 0.43 |
ENSG00000263849 |
. |
4055 |
0.23 |
| chr10_7451836_7452578 | 0.43 |
SFMBT2 |
Scm-like with four mbt domains 2 |
904 |
0.71 |
| chr4_82415207_82415715 | 0.42 |
RASGEF1B |
RasGEF domain family, member 1B |
22392 |
0.28 |
| chr8_21817827_21818019 | 0.42 |
XPO7 |
exportin 7 |
5826 |
0.19 |
| chr3_71632242_71632973 | 0.42 |
FOXP1 |
forkhead box P1 |
297 |
0.91 |
| chrX_30594869_30596024 | 0.42 |
CXorf21 |
chromosome X open reading frame 21 |
515 |
0.84 |
| chr14_52328565_52328716 | 0.42 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
598 |
0.77 |
| chr2_69002223_69002932 | 0.42 |
ARHGAP25 |
Rho GTPase activating protein 25 |
505 |
0.84 |
| chr17_80084198_80084349 | 0.41 |
CCDC57 |
coiled-coil domain containing 57 |
2159 |
0.16 |
| chr1_198581577_198581728 | 0.41 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
26149 |
0.21 |
| chr8_82052502_82052653 | 0.41 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
28274 |
0.23 |
| chr10_76728189_76728340 | 0.41 |
RP11-77G23.5 |
|
55745 |
0.12 |
| chr6_128239129_128239316 | 0.41 |
THEMIS |
thymocyte selection associated |
463 |
0.89 |
| chr2_46005650_46005801 | 0.41 |
U51244.2 |
|
103110 |
0.07 |
| chr14_61793878_61794206 | 0.41 |
PRKCH |
protein kinase C, eta |
411 |
0.85 |
| chr1_32394372_32394767 | 0.40 |
PTP4A2 |
protein tyrosine phosphatase type IVA, member 2 |
8817 |
0.18 |
| chr15_56760953_56761104 | 0.40 |
MNS1 |
meiosis-specific nuclear structural 1 |
3693 |
0.33 |
| chr1_175178431_175178668 | 0.40 |
KIAA0040 |
KIAA0040 |
16470 |
0.25 |
| chr12_93529897_93530048 | 0.40 |
RP11-511B23.2 |
|
2895 |
0.3 |
| chr5_109024574_109025025 | 0.39 |
MAN2A1 |
mannosidase, alpha, class 2A, member 1 |
268 |
0.92 |
| chr1_101471229_101471380 | 0.39 |
RP11-421L21.2 |
|
10275 |
0.14 |
| chr5_177665530_177665681 | 0.39 |
PHYKPL |
5-phosphohydroxy-L-lysine phospho-lyase |
5819 |
0.22 |
| chr6_15337079_15337415 | 0.39 |
ENSG00000201519 |
. |
12544 |
0.21 |
| chr13_99857558_99857709 | 0.39 |
ENSG00000201793 |
. |
329 |
0.88 |
| chr20_32312104_32312255 | 0.39 |
PXMP4 |
peroxisomal membrane protein 4, 24kDa |
4054 |
0.13 |
| chr8_120448366_120448517 | 0.39 |
NOV |
nephroblastoma overexpressed |
19895 |
0.19 |
| chr1_144084788_144084939 | 0.38 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
61945 |
0.14 |
| chr12_49626686_49627168 | 0.38 |
TUBA1C |
tubulin, alpha 1c |
5218 |
0.14 |
| chr5_149791368_149791565 | 0.38 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
829 |
0.6 |
| chr2_28978187_28978588 | 0.38 |
PPP1CB |
protein phosphatase 1, catalytic subunit, beta isozyme |
3668 |
0.21 |
| chr16_64412662_64412813 | 0.38 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
680844 |
0.0 |
| chr7_77177930_77178081 | 0.38 |
PTPN12 |
protein tyrosine phosphatase, non-receptor type 12 |
2936 |
0.37 |
| chrX_109250264_109250544 | 0.38 |
TMEM164 |
transmembrane protein 164 |
4061 |
0.29 |
| chr15_41701772_41702069 | 0.38 |
NDUFAF1 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 1 |
7203 |
0.15 |
| chrX_38665528_38665784 | 0.38 |
MID1IP1-AS1 |
MID1IP1 antisense RNA 1 |
2520 |
0.31 |
| chr15_91955696_91955847 | 0.37 |
SV2B |
synaptic vesicle glycoprotein 2B |
186671 |
0.03 |
| chr19_2083107_2083981 | 0.37 |
MOB3A |
MOB kinase activator 3A |
1847 |
0.21 |
| chr5_39203281_39203432 | 0.37 |
FYB |
FYN binding protein |
227 |
0.96 |
| chr7_38209053_38209204 | 0.37 |
STARD3NL |
STARD3 N-terminal like |
8696 |
0.31 |
| chr1_151634486_151634721 | 0.36 |
SNX27 |
sorting nexin family member 27 |
23271 |
0.08 |
| chr2_185461919_185462338 | 0.36 |
ZNF804A |
zinc finger protein 804A |
965 |
0.73 |
| chr12_68844621_68844772 | 0.36 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
118535 |
0.06 |
| chr7_150382525_150383418 | 0.36 |
GIMAP2 |
GTPase, IMAP family member 2 |
156 |
0.95 |
| chr12_54891959_54892179 | 0.36 |
NCKAP1L |
NCK-associated protein 1-like |
499 |
0.73 |
| chr2_233948481_233948771 | 0.36 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
23437 |
0.17 |
| chr1_207496793_207497173 | 0.35 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
1847 |
0.46 |
| chr2_197026499_197026722 | 0.35 |
STK17B |
serine/threonine kinase 17b |
5239 |
0.22 |
| chr7_36192851_36193841 | 0.35 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
469 |
0.85 |
| chr5_26147309_26147460 | 0.35 |
ENSG00000222560 |
. |
134386 |
0.06 |
| chr9_34957561_34957764 | 0.35 |
KIAA1045 |
KIAA1045 |
178 |
0.94 |
| chr1_208047081_208047232 | 0.35 |
CD34 |
CD34 molecule |
16681 |
0.25 |
| chr3_108322928_108323079 | 0.34 |
DZIP3 |
DAZ interacting zinc finger protein 3 |
1305 |
0.45 |
| chr11_85758745_85758896 | 0.34 |
PICALM |
phosphatidylinositol binding clathrin assembly protein |
21002 |
0.21 |
| chr14_90108423_90108574 | 0.34 |
ENSG00000252655 |
. |
8749 |
0.14 |
| chr10_22901004_22901155 | 0.34 |
PIP4K2A |
phosphatidylinositol-5-phosphate 4-kinase, type II, alpha |
20437 |
0.27 |
| chr12_4283030_4283181 | 0.34 |
CCND2 |
cyclin D2 |
99833 |
0.07 |
| chr8_61823770_61824122 | 0.34 |
RP11-33I11.2 |
|
101781 |
0.08 |
| chr10_73974639_73974790 | 0.34 |
ASCC1 |
activating signal cointegrator 1 complex subunit 1 |
980 |
0.36 |
| chr1_183558852_183559003 | 0.33 |
NCF2 |
neutrophil cytosolic factor 2 |
789 |
0.65 |
| chr8_15861662_15861813 | 0.33 |
MSR1 |
macrophage scavenger receptor 1 |
160023 |
0.04 |
| chr1_2082433_2082584 | 0.33 |
RP5-892K4.1 |
|
5790 |
0.14 |
| chr7_79953796_79953947 | 0.33 |
ENSG00000240347 |
. |
6302 |
0.23 |
| chr7_72696264_72696415 | 0.33 |
GTF2IRD2P1 |
GTF2I repeat domain containing 2 pseudogene 1 |
2196 |
0.24 |
| chr5_148188094_148188245 | 0.33 |
ADRB2 |
adrenoceptor beta 2, surface |
17987 |
0.25 |
| chr4_103783164_103783315 | 0.33 |
ENSG00000238948 |
. |
2586 |
0.2 |
| chr2_20091613_20091764 | 0.33 |
TTC32 |
tetratricopeptide repeat domain 32 |
9714 |
0.25 |
| chr14_89882597_89883166 | 0.33 |
FOXN3 |
forkhead box N3 |
575 |
0.54 |
| chr13_107025572_107025723 | 0.33 |
EFNB2 |
ephrin-B2 |
161815 |
0.04 |
| chr2_44403792_44403943 | 0.33 |
PPM1B |
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
7818 |
0.18 |
| chr17_5484149_5484300 | 0.33 |
NLRP1 |
NLR family, pyrin domain containing 1 |
3053 |
0.28 |
| chr16_89538828_89538979 | 0.33 |
ANKRD11 |
ankyrin repeat domain 11 |
17823 |
0.11 |
| chr13_49079711_49079862 | 0.32 |
RCBTB2 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 |
27427 |
0.22 |
| chr19_52268680_52268969 | 0.32 |
FPR2 |
formyl peptide receptor 2 |
2807 |
0.17 |
| chr1_89594453_89594640 | 0.32 |
GBP2 |
guanylate binding protein 2, interferon-inducible |
2749 |
0.27 |
| chr2_185465044_185465320 | 0.32 |
ZNF804A |
zinc finger protein 804A |
2089 |
0.5 |
| chr3_18482753_18482904 | 0.32 |
SATB1 |
SATB homeobox 1 |
2563 |
0.29 |
| chr11_46136336_46136487 | 0.32 |
ENSG00000263539 |
. |
1642 |
0.4 |
| chr5_93917742_93917893 | 0.32 |
ENSG00000211498 |
. |
3343 |
0.25 |
| chr1_66798585_66798917 | 0.31 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
879 |
0.74 |
| chr7_26141262_26141558 | 0.31 |
ENSG00000266430 |
. |
41451 |
0.15 |
| chr4_71570546_71572213 | 0.31 |
RUFY3 |
RUN and FYVE domain containing 3 |
861 |
0.36 |
| chr1_207262735_207262932 | 0.31 |
C4BPB |
complement component 4 binding protein, beta |
54 |
0.97 |
| chr1_247574766_247574917 | 0.31 |
NLRP3 |
NLR family, pyrin domain containing 3 |
4617 |
0.2 |
| chr5_118355478_118355629 | 0.31 |
ENSG00000223179 |
. |
12287 |
0.16 |
| chr4_184642687_184643147 | 0.31 |
ENSG00000251739 |
. |
15833 |
0.17 |
| chr3_18478655_18478806 | 0.31 |
SATB1 |
SATB homeobox 1 |
1454 |
0.45 |
| chr2_266277_266428 | 0.31 |
ACP1 |
acid phosphatase 1, soluble |
1426 |
0.31 |
| chrX_78401406_78401924 | 0.31 |
GPR174 |
G protein-coupled receptor 174 |
24804 |
0.27 |
| chr17_30816627_30816809 | 0.31 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
1904 |
0.22 |
| chr12_4384723_4384874 | 0.31 |
CCND2 |
cyclin D2 |
432 |
0.41 |
| chr6_169612787_169612938 | 0.30 |
XXyac-YX65C7_A.2 |
|
487 |
0.87 |
| chr3_151921407_151921835 | 0.30 |
MBNL1 |
muscleblind-like splicing regulator 1 |
64208 |
0.12 |
| chr11_72647389_72647540 | 0.30 |
FCHSD2 |
FCH and double SH3 domains 2 |
52593 |
0.12 |
| chr14_34413988_34414139 | 0.30 |
EGLN3 |
egl-9 family hypoxia-inducible factor 3 |
5969 |
0.27 |
| chr7_50354880_50355316 | 0.30 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
6780 |
0.3 |
| chr10_17705012_17705163 | 0.30 |
ENSG00000251959 |
. |
16100 |
0.14 |
| chr1_117529081_117529232 | 0.30 |
CD101 |
CD101 molecule |
15226 |
0.16 |
| chr15_55582423_55582730 | 0.29 |
RAB27A |
RAB27A, member RAS oncogene family |
575 |
0.75 |
| chr7_26331698_26332123 | 0.29 |
SNX10 |
sorting nexin 10 |
225 |
0.95 |
| chr12_122124689_122124840 | 0.29 |
MORN3 |
MORN repeat containing 3 |
17204 |
0.17 |
| chrX_78619893_78620072 | 0.29 |
ITM2A |
integral membrane protein 2A |
2874 |
0.42 |
| chr8_133787215_133787383 | 0.29 |
PHF20L1 |
PHD finger protein 20-like 1 |
322 |
0.89 |
| chr9_123886056_123886207 | 0.29 |
CNTRL |
centriolin |
2052 |
0.35 |
| chrX_15934152_15934303 | 0.29 |
ENSG00000200566 |
. |
199 |
0.96 |
| chr1_198613830_198613981 | 0.28 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
5613 |
0.26 |
| chr13_46952408_46952559 | 0.28 |
KIAA0226L |
KIAA0226-like |
297 |
0.9 |
| chr20_62588051_62588276 | 0.28 |
UCKL1 |
uridine-cytidine kinase 1-like 1 |
394 |
0.66 |
| chr3_134206036_134206187 | 0.28 |
ANAPC13 |
anaphase promoting complex subunit 13 |
553 |
0.63 |
| chr4_55575983_55576134 | 0.28 |
KIT |
v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
51973 |
0.18 |
| chr8_108509086_108509237 | 0.28 |
ANGPT1 |
angiopoietin 1 |
1077 |
0.7 |
| chr2_114659464_114659615 | 0.28 |
ACTR3 |
ARP3 actin-related protein 3 homolog (yeast) |
11364 |
0.23 |
| chr4_85886471_85886934 | 0.28 |
WDFY3 |
WD repeat and FYVE domain containing 3 |
801 |
0.75 |
| chr7_96357973_96358124 | 0.28 |
SHFM1 |
split hand/foot malformation (ectrodactyly) type 1 |
18845 |
0.27 |
| chr6_6386490_6386781 | 0.28 |
F13A1 |
coagulation factor XIII, A1 polypeptide |
65389 |
0.13 |
| chr3_30699890_30700041 | 0.28 |
RP11-1024P17.1 |
|
39188 |
0.19 |
| chrX_129130468_129130619 | 0.28 |
BCORL1 |
BCL6 corepressor-like 1 |
8621 |
0.22 |
| chr4_74046909_74047092 | 0.27 |
ANKRD17 |
ankyrin repeat domain 17 |
41831 |
0.16 |
| chr14_61870519_61870670 | 0.27 |
PRKCH |
protein kinase C, eta |
13182 |
0.24 |
| chr5_88076518_88076805 | 0.27 |
MEF2C |
myocyte enhancer factor 2C |
42944 |
0.17 |
| chr8_117642188_117642339 | 0.27 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
125760 |
0.05 |
| chr3_3214293_3214763 | 0.27 |
CRBN |
cereblon |
6830 |
0.19 |
| chr12_50908341_50908492 | 0.27 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
9526 |
0.23 |
| chr17_57969956_57970188 | 0.27 |
TUBD1 |
tubulin, delta 1 |
4 |
0.87 |
| chr6_147525530_147526351 | 0.27 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
190 |
0.61 |
| chr18_18654105_18654256 | 0.27 |
ENSG00000216205 |
. |
11746 |
0.22 |
| chr5_49968694_49968985 | 0.27 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
5448 |
0.35 |
| chr16_2771529_2771859 | 0.27 |
PRSS27 |
protease, serine 27 |
1478 |
0.22 |
| chr21_15917471_15917848 | 0.27 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
1003 |
0.63 |
| chrX_70585572_70585765 | 0.27 |
TAF1 |
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
446 |
0.84 |
| chr17_74547322_74547606 | 0.27 |
RP11-666A8.12 |
|
30 |
0.69 |
| chr12_40588874_40589025 | 0.26 |
LRRK2 |
leucine-rich repeat kinase 2 |
1597 |
0.46 |
| chr15_77711149_77711991 | 0.26 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
872 |
0.51 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.1 | 0.3 | GO:0070340 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.1 | 0.3 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.4 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.3 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.1 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.5 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.3 | GO:0002495 | antigen processing and presentation of peptide antigen via MHC class II(GO:0002495) |
| 0.1 | 0.2 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.1 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.1 | 0.2 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.1 | 0.1 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.1 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.1 | 0.2 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 | 0.2 | GO:0002903 | negative regulation of B cell apoptotic process(GO:0002903) |
| 0.1 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.1 | 0.2 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.3 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.1 | 0.4 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.1 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.1 | 0.2 | GO:0010746 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) |
| 0.0 | 0.1 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.3 | GO:0070897 | RNA polymerase II transcriptional preinitiation complex assembly(GO:0051123) DNA-templated transcriptional preinitiation complex assembly(GO:0070897) |
| 0.0 | 0.4 | GO:0043383 | negative T cell selection(GO:0043383) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.4 | GO:1903078 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) positive regulation of protein localization to plasma membrane(GO:1903078) positive regulation of protein localization to cell periphery(GO:1904377) |
| 0.0 | 0.2 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.1 | GO:0002902 | regulation of B cell apoptotic process(GO:0002902) |
| 0.0 | 0.2 | GO:0045359 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.3 | GO:0051319 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.0 | GO:0015911 | plasma membrane long-chain fatty acid transport(GO:0015911) |
| 0.0 | 0.5 | GO:0006906 | vesicle fusion(GO:0006906) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.2 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.1 | GO:0045634 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.1 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.1 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0072577 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.3 | GO:0050718 | positive regulation of interleukin-1 beta secretion(GO:0050718) |
| 0.0 | 0.7 | GO:0008633 | obsolete activation of pro-apoptotic gene products(GO:0008633) |
| 0.0 | 0.2 | GO:0033005 | positive regulation of mast cell activation(GO:0033005) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:1902745 | positive regulation of lamellipodium assembly(GO:0010592) positive regulation of lamellipodium organization(GO:1902745) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0031573 | intra-S DNA damage checkpoint(GO:0031573) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.2 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0035234 | ectopic germ cell programmed cell death(GO:0035234) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.1 | GO:0035520 | monoubiquitinated protein deubiquitination(GO:0035520) |
| 0.0 | 0.1 | GO:0009186 | deoxyribonucleoside diphosphate metabolic process(GO:0009186) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.4 | GO:0007032 | endosome organization(GO:0007032) |
| 0.0 | 0.1 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:0040001 | establishment of mitotic spindle localization(GO:0040001) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0043162 | ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043162) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.1 | GO:0003084 | positive regulation of systemic arterial blood pressure(GO:0003084) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.0 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0016233 | telomere capping(GO:0016233) |
| 0.0 | 0.3 | GO:0042771 | intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:0042771) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.1 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.5 | GO:0006958 | complement activation, classical pathway(GO:0006958) |
| 0.0 | 0.1 | GO:0050860 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0060872 | semicircular canal morphogenesis(GO:0048752) semicircular canal development(GO:0060872) |
| 0.0 | 0.1 | GO:0006983 | ER overload response(GO:0006983) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.0 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.2 | GO:0051568 | histone H3-K4 methylation(GO:0051568) |
| 0.0 | 0.0 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.2 | GO:0045737 | positive regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0045737) positive regulation of cyclin-dependent protein kinase activity(GO:1904031) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.0 | GO:0051136 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.0 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.1 | GO:0043550 | regulation of lipid kinase activity(GO:0043550) |
| 0.0 | 0.1 | GO:0060123 | regulation of growth hormone secretion(GO:0060123) |
| 0.0 | 0.1 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:0097501 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0071027 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.0 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0008611 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.0 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.0 | GO:0051573 | negative regulation of histone H3-K9 methylation(GO:0051573) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.4 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.0 | GO:0009191 | nucleoside diphosphate catabolic process(GO:0009134) ribonucleoside diphosphate catabolic process(GO:0009191) |
| 0.0 | 0.1 | GO:0051489 | regulation of filopodium assembly(GO:0051489) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0048745 | smooth muscle tissue development(GO:0048745) |
| 0.0 | 0.1 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0045723 | positive regulation of fatty acid biosynthetic process(GO:0045723) |
| 0.0 | 0.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.0 | GO:0060397 | JAK-STAT cascade involved in growth hormone signaling pathway(GO:0060397) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.0 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.0 | GO:0061726 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.3 | GO:0070979 | protein K11-linked ubiquitination(GO:0070979) |
| 0.0 | 0.2 | GO:0042744 | hydrogen peroxide catabolic process(GO:0042744) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0045657 | positive regulation of monocyte differentiation(GO:0045657) |
| 0.0 | 0.1 | GO:0043568 | positive regulation of insulin-like growth factor receptor signaling pathway(GO:0043568) |
| 0.0 | 0.0 | GO:0045591 | positive regulation of regulatory T cell differentiation(GO:0045591) |
| 0.0 | 0.2 | GO:0043486 | histone exchange(GO:0043486) |
| 0.0 | 0.2 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.1 | GO:0032364 | oxygen homeostasis(GO:0032364) |
| 0.0 | 0.5 | GO:0034976 | response to endoplasmic reticulum stress(GO:0034976) |
| 0.0 | 0.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.0 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.0 | GO:0010586 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0031313 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.6 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.1 | 0.2 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.2 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.3 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.4 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
| 0.0 | 0.1 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.1 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.2 | GO:0043073 | germ cell nucleus(GO:0043073) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
| 0.0 | 0.2 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.3 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0030667 | secretory granule membrane(GO:0030667) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.4 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.0 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.1 | 0.2 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.1 | 0.2 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.1 | 0.2 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.2 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.7 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.4 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.8 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.3 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0043024 | ribosomal small subunit binding(GO:0043024) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.1 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.2 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.3 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.2 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.2 | GO:0004467 | long-chain fatty acid-CoA ligase activity(GO:0004467) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.3 | GO:0004708 | MAP kinase kinase activity(GO:0004708) |
| 0.0 | 0.1 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
| 0.0 | 0.3 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.5 | GO:0030295 | protein kinase activator activity(GO:0030295) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.3 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.0 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.8 | GO:0008301 | DNA binding, bending(GO:0008301) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.0 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.3 | GO:0051219 | phosphoprotein binding(GO:0051219) |
| 0.0 | 0.4 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.2 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.2 | GO:0031593 | polyubiquitin binding(GO:0031593) |
| 0.0 | 0.8 | GO:0004197 | cysteine-type endopeptidase activity(GO:0004197) |
| 0.0 | 0.3 | GO:0036459 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) thiol-dependent ubiquitinyl hydrolase activity(GO:0036459) ubiquitinyl hydrolase activity(GO:0101005) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0003747 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.1 | GO:0017091 | AU-rich element binding(GO:0017091) |
| 0.0 | 0.5 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0051920 | peroxiredoxin activity(GO:0051920) |
| 0.0 | 0.1 | GO:0016854 | racemase and epimerase activity(GO:0016854) |
| 0.0 | 0.0 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.0 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0001848 | complement binding(GO:0001848) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.7 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 1.8 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.2 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.6 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.1 | PID UPA UPAR PATHWAY | Urokinase-type plasminogen activator (uPA) and uPAR-mediated signaling |
| 0.0 | 0.1 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.1 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.3 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.1 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.1 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.0 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.5 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.2 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.3 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.0 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.4 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.3 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.1 | SIG CHEMOTAXIS | Genes related to chemotaxis |
| 0.0 | 0.1 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.4 | PID IL4 2PATHWAY | IL4-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.1 | 0.5 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.4 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.5 | REACTOME OXYGEN DEPENDENT PROLINE HYDROXYLATION OF HYPOXIA INDUCIBLE FACTOR ALPHA | Genes involved in Oxygen-dependent Proline Hydroxylation of Hypoxia-inducible Factor Alpha |
| 0.0 | 0.4 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.4 | REACTOME G BETA GAMMA SIGNALLING THROUGH PLC BETA | Genes involved in G beta:gamma signalling through PLC beta |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.4 | REACTOME N GLYCAN ANTENNAE ELONGATION IN THE MEDIAL TRANS GOLGI | Genes involved in N-glycan antennae elongation in the medial/trans-Golgi |
| 0.0 | 0.0 | REACTOME INHIBITION OF REPLICATION INITIATION OF DAMAGED DNA BY RB1 E2F1 | Genes involved in Inhibition of replication initiation of damaged DNA by RB1/E2F1 |
| 0.0 | 0.6 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.0 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.5 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.5 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.1 | REACTOME G PROTEIN BETA GAMMA SIGNALLING | Genes involved in G-protein beta:gamma signalling |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.2 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF TGF BETA RECEPTOR SIGNALING | Genes involved in Downregulation of TGF-beta receptor signaling |
| 0.0 | 0.2 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.0 | REACTOME SIGNALING BY ILS | Genes involved in Signaling by Interleukins |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY BRF1 | Genes involved in Destabilization of mRNA by Butyrate Response Factor 1 (BRF1) |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.5 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.4 | REACTOME RNA POL II TRANSCRIPTION PRE INITIATION AND PROMOTER OPENING | Genes involved in RNA Polymerase II Transcription Pre-Initiation And Promoter Opening |
| 0.0 | 0.1 | REACTOME ACTIVATION OF BH3 ONLY PROTEINS | Genes involved in Activation of BH3-only proteins |
| 0.0 | 0.0 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |