Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
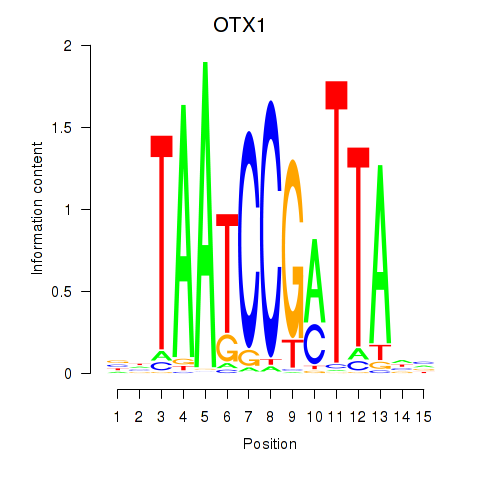
Results for OTX1
Z-value: 1.41
Transcription factors associated with OTX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTX1
|
ENSG00000115507.5 | orthodenticle homeobox 1 |
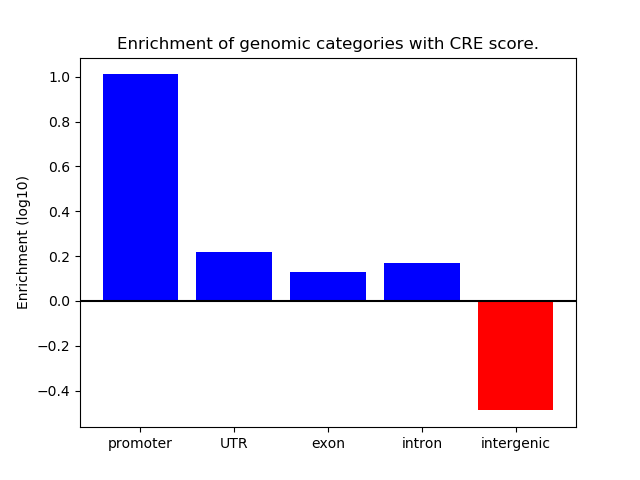
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_63276750_63277690 | OTX1 | 28 | 0.972223 | 0.72 | 2.8e-02 | Click! |
| chr2_63278835_63278986 | OTX1 | 973 | 0.593722 | 0.61 | 7.9e-02 | Click! |
| chr2_63279236_63279503 | OTX1 | 1432 | 0.458319 | 0.54 | 1.3e-01 | Click! |
| chr2_63278511_63278672 | OTX1 | 654 | 0.734523 | 0.51 | 1.6e-01 | Click! |
| chr2_63287244_63287598 | OTX1 | 9484 | 0.232922 | -0.47 | 2.0e-01 | Click! |
Activity of the OTX1 motif across conditions
Conditions sorted by the z-value of the OTX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
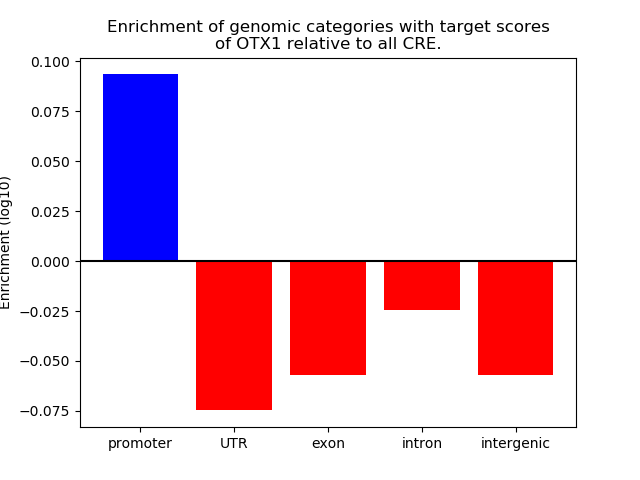
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_122050649_122050906 | 0.60 |
ENSG00000207994 |
. |
27761 |
0.16 |
| chr1_162532316_162532688 | 0.50 |
UAP1 |
UDP-N-acteylglucosamine pyrophosphorylase 1 |
1179 |
0.36 |
| chr8_94930403_94930714 | 0.49 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
580 |
0.72 |
| chr3_98619475_98619750 | 0.48 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
403 |
0.8 |
| chr2_151334124_151334616 | 0.48 |
RND3 |
Rho family GTPase 3 |
7526 |
0.34 |
| chr4_90581488_90581639 | 0.46 |
RP11-115D19.1 |
|
21100 |
0.27 |
| chr11_79150433_79150655 | 0.46 |
TENM4 |
teneurin transmembrane protein 4 |
1151 |
0.54 |
| chr5_98111445_98111873 | 0.45 |
RGMB |
repulsive guidance molecule family member b |
2320 |
0.3 |
| chr5_171708618_171708769 | 0.43 |
UBTD2 |
ubiquitin domain containing 2 |
2382 |
0.24 |
| chr15_45570927_45571126 | 0.42 |
CTD-2651B20.3 |
|
399 |
0.8 |
| chr22_44722319_44722530 | 0.41 |
KIAA1644 |
KIAA1644 |
13693 |
0.24 |
| chr1_119529068_119529220 | 0.41 |
TBX15 |
T-box 15 |
1284 |
0.57 |
| chr5_78712583_78712734 | 0.40 |
HOMER1 |
homer homolog 1 (Drosophila) |
95959 |
0.08 |
| chr3_30659362_30659513 | 0.39 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
11344 |
0.29 |
| chr6_84761983_84762373 | 0.38 |
MRAP2 |
melanocortin 2 receptor accessory protein 2 |
18703 |
0.28 |
| chr19_47756874_47757099 | 0.37 |
CCDC9 |
coiled-coil domain containing 9 |
2730 |
0.19 |
| chr2_9815352_9815503 | 0.37 |
YWHAQ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
44284 |
0.14 |
| chr2_9376393_9376544 | 0.35 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
29574 |
0.23 |
| chr5_72715364_72715530 | 0.34 |
FOXD1 |
forkhead box D1 |
28905 |
0.18 |
| chr3_197022890_197023199 | 0.34 |
DLG1 |
discs, large homolog 1 (Drosophila) |
1031 |
0.42 |
| chr10_114714022_114714173 | 0.34 |
RP11-57H14.2 |
|
2463 |
0.3 |
| chr15_36693921_36694243 | 0.34 |
C15orf41 |
chromosome 15 open reading frame 41 |
177730 |
0.03 |
| chr1_23449014_23449165 | 0.33 |
LUZP1 |
leucine zipper protein 1 |
11083 |
0.14 |
| chr7_156791129_156791280 | 0.33 |
MNX1-AS2 |
MNX1 antisense RNA 2 |
7797 |
0.17 |
| chr8_117698220_117698371 | 0.33 |
EIF3H |
eukaryotic translation initiation factor 3, subunit H |
69728 |
0.11 |
| chr1_171614800_171614951 | 0.33 |
MYOC |
myocilin, trabecular meshwork inducible glucocorticoid response |
6948 |
0.24 |
| chr3_25513057_25513453 | 0.33 |
RARB |
retinoic acid receptor, beta |
13454 |
0.26 |
| chr11_16946573_16946796 | 0.33 |
PLEKHA7 |
pleckstrin homology domain containing, family A member 7 |
43779 |
0.12 |
| chr10_114552786_114553214 | 0.33 |
RP11-57H14.4 |
|
30249 |
0.21 |
| chr3_156561167_156561375 | 0.32 |
LEKR1 |
leucine, glutamate and lysine rich 1 |
17123 |
0.27 |
| chr10_95518290_95518441 | 0.32 |
LGI1 |
leucine-rich, glioma inactivated 1 |
687 |
0.76 |
| chr14_53773167_53773351 | 0.32 |
RP11-547D23.1 |
|
153187 |
0.04 |
| chr5_177631147_177631350 | 0.32 |
HNRNPAB |
heterogeneous nuclear ribonucleoprotein A/B |
260 |
0.92 |
| chr2_237477915_237478284 | 0.32 |
ACKR3 |
atypical chemokine receptor 3 |
185 |
0.97 |
| chr4_124267046_124267197 | 0.31 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
50829 |
0.19 |
| chr1_193154702_193155811 | 0.31 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
528 |
0.8 |
| chr14_68466898_68467139 | 0.31 |
CTD-2566J3.1 |
|
129895 |
0.05 |
| chr6_147236233_147236440 | 0.31 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
685 |
0.82 |
| chr4_177711668_177712016 | 0.31 |
VEGFC |
vascular endothelial growth factor C |
2039 |
0.48 |
| chr8_129873562_129874058 | 0.30 |
ENSG00000221351 |
. |
41770 |
0.22 |
| chr2_163157606_163157781 | 0.30 |
IFIH1 |
interferon induced with helicase C domain 1 |
17501 |
0.19 |
| chr8_101859554_101859783 | 0.30 |
ENSG00000222795 |
. |
15580 |
0.19 |
| chr7_94273794_94273945 | 0.30 |
SGCE |
sarcoglycan, epsilon |
11541 |
0.21 |
| chr2_204398750_204399136 | 0.30 |
RAPH1 |
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1 |
787 |
0.76 |
| chr5_72146808_72146959 | 0.29 |
TNPO1 |
transportin 1 |
2872 |
0.27 |
| chr20_34471632_34471916 | 0.29 |
ENSG00000201221 |
. |
3807 |
0.18 |
| chr12_31522871_31523058 | 0.29 |
ENSG00000207477 |
. |
3742 |
0.2 |
| chr3_149374923_149375174 | 0.29 |
WWTR1 |
WW domain containing transcription regulator 1 |
174 |
0.54 |
| chr7_42275396_42275690 | 0.29 |
GLI3 |
GLI family zinc finger 3 |
1069 |
0.7 |
| chr22_36232996_36233711 | 0.29 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
2912 |
0.37 |
| chr15_31276777_31276928 | 0.29 |
MTMR10 |
myotubularin related protein 10 |
6766 |
0.17 |
| chr5_145207148_145207299 | 0.28 |
PRELID2 |
PRELI domain containing 2 |
7625 |
0.27 |
| chrX_139584639_139584790 | 0.28 |
SOX3 |
SRY (sex determining region Y)-box 3 |
2511 |
0.41 |
| chr13_107182142_107182293 | 0.28 |
EFNB2 |
ephrin-B2 |
5245 |
0.29 |
| chr9_2655572_2655791 | 0.28 |
RP11-125B21.2 |
|
14286 |
0.23 |
| chr1_12675825_12676469 | 0.28 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
1590 |
0.37 |
| chr4_86417012_86417203 | 0.28 |
ARHGAP24 |
Rho GTPase activating protein 24 |
20716 |
0.29 |
| chr6_106546757_106547354 | 0.28 |
PRDM1 |
PR domain containing 1, with ZNF domain |
240 |
0.87 |
| chr12_4967146_4967345 | 0.27 |
RP3-377H17.2 |
|
19124 |
0.2 |
| chr6_132180195_132180361 | 0.27 |
ENSG00000200895 |
. |
38804 |
0.14 |
| chr3_40428700_40429091 | 0.27 |
ENTPD3 |
ectonucleoside triphosphate diphosphohydrolase 3 |
159 |
0.96 |
| chr1_213500454_213500722 | 0.26 |
RPS6KC1 |
ribosomal protein S6 kinase, 52kDa, polypeptide 1 |
275984 |
0.01 |
| chr7_103969727_103969878 | 0.26 |
LHFPL3 |
lipoma HMGIC fusion partner-like 3 |
654 |
0.83 |
| chr14_85998679_85999082 | 0.26 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
2308 |
0.36 |
| chr3_4624343_4624494 | 0.26 |
ITPR1 |
inositol 1,4,5-trisphosphate receptor, type 1 |
66242 |
0.12 |
| chr6_27722042_27722205 | 0.26 |
HIST1H2BL |
histone cluster 1, H2bl |
53586 |
0.06 |
| chr5_80256040_80256245 | 0.26 |
RASGRF2 |
Ras protein-specific guanine nucleotide-releasing factor 2 |
349 |
0.67 |
| chr11_131780111_131780399 | 0.26 |
NTM |
neurotrimin |
642 |
0.77 |
| chr11_125497097_125497248 | 0.26 |
CHEK1 |
checkpoint kinase 1 |
529 |
0.7 |
| chr1_1927919_1928070 | 0.26 |
C1orf222 |
chromosome 1 open reading frame 222 |
5626 |
0.14 |
| chr3_29325145_29325351 | 0.26 |
RBMS3 |
RNA binding motif, single stranded interacting protein 3 |
2141 |
0.37 |
| chr7_42420275_42420426 | 0.26 |
GLI3 |
GLI family zinc finger 3 |
143692 |
0.05 |
| chr17_14028855_14029146 | 0.25 |
ENSG00000252305 |
. |
51448 |
0.12 |
| chr8_62496427_62496578 | 0.25 |
ENSG00000222898 |
. |
48069 |
0.16 |
| chr18_61555988_61556228 | 0.25 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
1115 |
0.49 |
| chr6_170597728_170597879 | 0.25 |
DLL1 |
delta-like 1 (Drosophila) |
1758 |
0.29 |
| chr14_55119300_55119828 | 0.25 |
SAMD4A |
sterile alpha motif domain containing 4A |
84927 |
0.09 |
| chr10_93684687_93684838 | 0.25 |
BTAF1 |
BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa |
1236 |
0.47 |
| chr5_148926669_148926913 | 0.25 |
CSNK1A1 |
casein kinase 1, alpha 1 |
3071 |
0.22 |
| chr18_35145104_35145551 | 0.25 |
CELF4 |
CUGBP, Elav-like family member 4 |
277 |
0.95 |
| chr1_86048991_86049142 | 0.25 |
CYR61 |
cysteine-rich, angiogenic inducer, 61 |
2622 |
0.26 |
| chr7_123389891_123390042 | 0.25 |
RP11-390E23.6 |
|
844 |
0.4 |
| chr8_38899834_38899985 | 0.25 |
ENSG00000207199 |
. |
23707 |
0.15 |
| chr10_70096270_70096993 | 0.25 |
PBLD |
phenazine biosynthesis-like protein domain containing |
3825 |
0.2 |
| chr3_135741763_135741914 | 0.25 |
PPP2R3A |
protein phosphatase 2, regulatory subunit B'', alpha |
261 |
0.95 |
| chr16_57849974_57850125 | 0.25 |
CTD-2600O9.1 |
uncharacterized protein LOC388282 |
2365 |
0.22 |
| chr18_21851017_21851223 | 0.25 |
OSBPL1A |
oxysterol binding protein-like 1A |
1076 |
0.43 |
| chr19_47776409_47776560 | 0.25 |
PRR24 |
proline rich 24 |
1658 |
0.28 |
| chr13_25745649_25746102 | 0.24 |
AMER2 |
APC membrane recruitment protein 2 |
18 |
0.98 |
| chr13_114567217_114567395 | 0.24 |
GAS6 |
growth arrest-specific 6 |
260 |
0.94 |
| chr3_111673457_111673608 | 0.24 |
ABHD10 |
abhydrolase domain containing 10 |
24325 |
0.17 |
| chr17_46664636_46664902 | 0.24 |
HOXB3 |
homeobox B3 |
2828 |
0.09 |
| chr2_27487055_27487432 | 0.24 |
SLC30A3 |
solute carrier family 30 (zinc transporter), member 3 |
239 |
0.85 |
| chr8_31134922_31135073 | 0.24 |
RP11-363L24.3 |
|
100314 |
0.08 |
| chr9_73979405_73979556 | 0.24 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
82271 |
0.11 |
| chr1_224545541_224545743 | 0.24 |
CNIH4 |
cornichon family AMPA receptor auxiliary protein 4 |
1047 |
0.45 |
| chr2_120518344_120518699 | 0.24 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
721 |
0.76 |
| chr2_129315776_129315927 | 0.24 |
ENSG00000238379 |
. |
113079 |
0.07 |
| chr8_49830829_49831037 | 0.24 |
SNAI2 |
snail family zinc finger 2 |
3055 |
0.4 |
| chr2_216948558_216948709 | 0.24 |
TMEM169 |
transmembrane protein 169 |
1991 |
0.27 |
| chr19_2984028_2984281 | 0.23 |
TLE6 |
transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) |
6576 |
0.14 |
| chr17_43971204_43971355 | 0.23 |
MAPT |
microtubule-associated protein tau |
469 |
0.73 |
| chr2_102720712_102721707 | 0.23 |
IL1R1 |
interleukin 1 receptor, type I |
180 |
0.96 |
| chr9_21558481_21558945 | 0.23 |
MIR31HG |
MIR31 host gene (non-protein coding) |
955 |
0.56 |
| chr8_67977563_67977802 | 0.23 |
COPS5 |
COP9 signalosome subunit 5 |
504 |
0.63 |
| chr4_33952877_33953028 | 0.23 |
ENSG00000251755 |
. |
166395 |
0.04 |
| chr5_73926204_73926355 | 0.23 |
HEXB |
hexosaminidase B (beta polypeptide) |
9569 |
0.21 |
| chr1_201605392_201605543 | 0.23 |
NAV1 |
neuron navigator 1 |
11910 |
0.15 |
| chr3_128892906_128893057 | 0.23 |
CNBP |
CCHC-type zinc finger, nucleic acid binding protein |
9761 |
0.13 |
| chr8_135790318_135790469 | 0.23 |
ENSG00000207582 |
. |
22457 |
0.21 |
| chr13_44842345_44842496 | 0.23 |
SERP2 |
stress-associated endoplasmic reticulum protein family member 2 |
105558 |
0.06 |
| chr6_156349604_156349763 | 0.23 |
ENSG00000221456 |
. |
81752 |
0.12 |
| chr7_121944988_121945139 | 0.23 |
FEZF1-AS1 |
FEZF1 antisense RNA 1 |
59 |
0.89 |
| chr7_83946098_83946249 | 0.23 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
121412 |
0.07 |
| chr15_38545953_38546360 | 0.23 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
774 |
0.79 |
| chr3_185651354_185651505 | 0.23 |
TRA2B |
transformer 2 beta homolog (Drosophila) |
4372 |
0.23 |
| chr2_208027525_208027676 | 0.23 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
2695 |
0.3 |
| chr10_33620140_33620362 | 0.23 |
NRP1 |
neuropilin 1 |
3059 |
0.34 |
| chr3_192958571_192958786 | 0.22 |
HRASLS |
HRAS-like suppressor |
236 |
0.96 |
| chr5_171115385_171115536 | 0.22 |
ENSG00000264303 |
. |
82074 |
0.09 |
| chr13_110320110_110320261 | 0.22 |
LINC00676 |
long intergenic non-protein coding RNA 676 |
60444 |
0.15 |
| chr5_125760300_125760451 | 0.22 |
GRAMD3 |
GRAM domain containing 3 |
1225 |
0.57 |
| chr16_86888410_86888561 | 0.22 |
FOXL1 |
forkhead box L1 |
276370 |
0.01 |
| chr9_73483972_73484180 | 0.22 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
102 |
0.98 |
| chr2_102327994_102328145 | 0.22 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
13077 |
0.29 |
| chr17_56013006_56013279 | 0.22 |
CUEDC1 |
CUE domain containing 1 |
19476 |
0.15 |
| chr19_46965021_46965172 | 0.22 |
PNMAL1 |
paraneoplastic Ma antigen family-like 1 |
9568 |
0.14 |
| chr11_2404011_2404162 | 0.22 |
CD81 |
CD81 molecule |
396 |
0.75 |
| chr6_148593838_148594147 | 0.22 |
SASH1 |
SAM and SH3 domain containing 1 |
552 |
0.78 |
| chr14_55077622_55077773 | 0.22 |
SAMD4A |
sterile alpha motif domain containing 4A |
43060 |
0.17 |
| chr4_160173723_160173874 | 0.22 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
14235 |
0.23 |
| chr9_95298468_95298698 | 0.22 |
ECM2 |
extracellular matrix protein 2, female organ and adipocyte specific |
246 |
0.92 |
| chr9_21558983_21559308 | 0.22 |
MIR31HG |
MIR31 host gene (non-protein coding) |
523 |
0.78 |
| chr2_219828438_219828589 | 0.22 |
AC097468.7 |
|
1888 |
0.18 |
| chr7_28682052_28682203 | 0.22 |
CREB5 |
cAMP responsive element binding protein 5 |
33449 |
0.24 |
| chr3_8808825_8808976 | 0.22 |
OXTR |
oxytocin receptor |
1463 |
0.34 |
| chr14_77490003_77490186 | 0.22 |
IRF2BPL |
interferon regulatory factor 2 binding protein-like |
4940 |
0.22 |
| chr8_67343981_67344272 | 0.22 |
ADHFE1 |
alcohol dehydrogenase, iron containing, 1 |
592 |
0.73 |
| chrX_17431758_17432043 | 0.22 |
ENSG00000265465 |
. |
12104 |
0.22 |
| chr6_28806796_28807125 | 0.22 |
ENSG00000265764 |
. |
43148 |
0.11 |
| chr3_173302379_173302636 | 0.22 |
NLGN1 |
neuroligin 1 |
162 |
0.98 |
| chr12_113375085_113375236 | 0.22 |
OAS3 |
2'-5'-oligoadenylate synthetase 3, 100kDa |
997 |
0.54 |
| chr13_30276703_30277107 | 0.22 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
107080 |
0.07 |
| chr2_159251872_159252065 | 0.22 |
CCDC148 |
coiled-coil domain containing 148 |
14496 |
0.23 |
| chr6_11230027_11230357 | 0.22 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
2699 |
0.33 |
| chr10_14051003_14051266 | 0.22 |
FRMD4A |
FERM domain containing 4A |
602 |
0.78 |
| chr10_33620428_33620906 | 0.21 |
NRP1 |
neuropilin 1 |
2643 |
0.37 |
| chr3_58983787_58983938 | 0.21 |
C3orf67 |
chromosome 3 open reading frame 67 |
51855 |
0.18 |
| chr8_93069724_93069875 | 0.21 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
5392 |
0.33 |
| chr13_35517642_35517793 | 0.21 |
NBEA |
neurobeachin |
920 |
0.75 |
| chr10_63809082_63810748 | 0.21 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
945 |
0.69 |
| chr6_27636067_27636272 | 0.21 |
ENSG00000238648 |
. |
36981 |
0.14 |
| chr2_206610963_206611114 | 0.21 |
AC007362.3 |
|
17639 |
0.25 |
| chr11_108724912_108725630 | 0.21 |
ENSG00000201243 |
. |
150973 |
0.04 |
| chr7_18550520_18550717 | 0.21 |
HDAC9 |
histone deacetylase 9 |
1682 |
0.51 |
| chr9_24326132_24326283 | 0.21 |
IZUMO3 |
IZUMO family member 3 |
219467 |
0.02 |
| chr12_90272914_90273065 | 0.21 |
ENSG00000252823 |
. |
125153 |
0.06 |
| chr2_113916305_113916585 | 0.21 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
15103 |
0.13 |
| chr1_6269211_6269420 | 0.21 |
RPL22 |
ribosomal protein L22 |
11 |
0.96 |
| chr1_68414425_68414576 | 0.21 |
GNG12-AS1 |
GNG12 antisense RNA 1 |
6588 |
0.28 |
| chr13_107185720_107185941 | 0.21 |
EFNB2 |
ephrin-B2 |
1632 |
0.49 |
| chr19_48614158_48614373 | 0.21 |
PLA2G4C |
phospholipase A2, group IVC (cytosolic, calcium-independent) |
191 |
0.91 |
| chr5_114713190_114713401 | 0.21 |
CCDC112 |
coiled-coil domain containing 112 |
80767 |
0.09 |
| chr7_130596201_130596683 | 0.21 |
ENSG00000226380 |
. |
34144 |
0.19 |
| chr17_27135559_27135731 | 0.21 |
FAM222B |
family with sequence similarity 222, member B |
3597 |
0.1 |
| chr16_22038297_22038549 | 0.21 |
C16orf52 |
chromosome 16 open reading frame 52 |
18819 |
0.15 |
| chr2_71205181_71205355 | 0.21 |
ANKRD53 |
ankyrin repeat domain 53 |
242 |
0.83 |
| chr6_134211072_134211606 | 0.21 |
TCF21 |
transcription factor 21 |
866 |
0.5 |
| chr2_201248196_201249000 | 0.20 |
ENSG00000264798 |
. |
3300 |
0.23 |
| chr3_61913363_61913514 | 0.20 |
ENSG00000252420 |
. |
188535 |
0.03 |
| chr4_85503338_85503489 | 0.20 |
CDS1 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 1 |
719 |
0.78 |
| chr20_48565386_48565537 | 0.20 |
ENSG00000239157 |
. |
8524 |
0.13 |
| chr9_18475768_18475931 | 0.20 |
ADAMTSL1 |
ADAMTS-like 1 |
1618 |
0.53 |
| chrX_33573585_33573736 | 0.20 |
DMD |
dystrophin |
216102 |
0.02 |
| chr3_54674397_54674548 | 0.20 |
ESRG |
embryonic stem cell related (non-protein coding) |
588 |
0.86 |
| chr19_32828053_32828204 | 0.20 |
ZNF507 |
zinc finger protein 507 |
8372 |
0.19 |
| chr1_214279767_214279918 | 0.20 |
RP11-53A1.2 |
|
75733 |
0.11 |
| chr2_176997573_176997736 | 0.20 |
HOXD8 |
homeobox D8 |
2569 |
0.11 |
| chr1_79115791_79116542 | 0.20 |
IFI44 |
interferon-induced protein 44 |
649 |
0.75 |
| chr10_10834897_10835048 | 0.20 |
CELF2 |
CUGBP, Elav-like family member 2 |
212287 |
0.02 |
| chr6_3844247_3844398 | 0.20 |
FAM50B |
family with sequence similarity 50, member B |
5298 |
0.22 |
| chr12_13079095_13079275 | 0.20 |
RP11-392P7.6 |
|
1475 |
0.3 |
| chr10_65282301_65282503 | 0.20 |
REEP3 |
receptor accessory protein 3 |
1279 |
0.55 |
| chr6_142360108_142360259 | 0.20 |
RP11-137J7.2 |
|
49187 |
0.15 |
| chr7_132260236_132260613 | 0.20 |
PLXNA4 |
plexin A4 |
805 |
0.75 |
| chr8_42748827_42749465 | 0.20 |
ENSG00000266044 |
. |
2272 |
0.19 |
| chr19_51308481_51308642 | 0.20 |
C19orf48 |
chromosome 19 open reading frame 48 |
375 |
0.65 |
| chr6_116693696_116693937 | 0.20 |
DSE |
dermatan sulfate epimerase |
1706 |
0.38 |
| chr12_76740590_76740888 | 0.20 |
BBS10 |
Bardet-Biedl syndrome 10 |
1483 |
0.48 |
| chr14_65801219_65801685 | 0.20 |
ENSG00000266740 |
. |
449 |
0.88 |
| chr21_18348624_18348775 | 0.20 |
ENSG00000239023 |
. |
257382 |
0.02 |
| chr3_104156196_104156347 | 0.20 |
ENSG00000265076 |
. |
382631 |
0.01 |
| chr6_106547367_106547905 | 0.20 |
RP1-134E15.3 |
|
379 |
0.74 |
| chrY_2620247_2620578 | 0.20 |
ENSG00000251841 |
. |
32378 |
0.19 |
| chr16_4368544_4368842 | 0.20 |
GLIS2 |
GLIS family zinc finger 2 |
3931 |
0.16 |
| chr2_70140665_70140816 | 0.20 |
ENSG00000238465 |
. |
913 |
0.4 |
| chr8_69879858_69880009 | 0.20 |
ENSG00000238808 |
. |
142876 |
0.05 |
| chr1_97850069_97850220 | 0.19 |
DPYD-IT1 |
DPYD intronic transcript 1 (non-protein coding) |
35553 |
0.24 |
| chr6_19803841_19803992 | 0.19 |
ID4 |
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
33701 |
0.2 |
| chr22_31692523_31692674 | 0.19 |
PIK3IP1 |
phosphoinositide-3-kinase interacting protein 1 |
4078 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 0.3 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.1 | 0.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.2 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.1 | 0.2 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.1 | 0.2 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.1 | 0.2 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.1 | 0.3 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.1 | 0.2 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.2 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.2 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.2 | GO:0060753 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.4 | GO:0021903 | rostrocaudal neural tube patterning(GO:0021903) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:0046337 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.5 | GO:0050812 | acetyl-CoA biosynthetic process from pyruvate(GO:0006086) regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.1 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.1 | GO:0070669 | response to interleukin-2(GO:0070669) |
| 0.0 | 0.0 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.2 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.2 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.2 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.2 | GO:0001553 | luteinization(GO:0001553) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 | 0.1 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.2 | GO:0045217 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0001955 | blood vessel maturation(GO:0001955) |
| 0.0 | 0.1 | GO:0051176 | positive regulation of sulfur metabolic process(GO:0051176) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.2 | GO:0000959 | mitochondrial RNA metabolic process(GO:0000959) transcription from mitochondrial promoter(GO:0006390) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0032875 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0030952 | establishment or maintenance of cytoskeleton polarity(GO:0030952) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.2 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.0 | GO:0060423 | foregut regionalization(GO:0060423) lung field specification(GO:0060424) lung induction(GO:0060492) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.1 | GO:0021955 | central nervous system projection neuron axonogenesis(GO:0021952) central nervous system neuron axonogenesis(GO:0021955) |
| 0.0 | 0.1 | GO:0048069 | eye pigmentation(GO:0048069) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.1 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.0 | GO:0006925 | inflammatory cell apoptotic process(GO:0006925) |
| 0.0 | 0.0 | GO:0010957 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0050651 | dermatan sulfate proteoglycan biosynthetic process(GO:0050651) |
| 0.0 | 0.1 | GO:0097354 | protein prenylation(GO:0018342) prenylation(GO:0097354) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.2 | GO:0018214 | peptidyl-glutamic acid carboxylation(GO:0017187) protein carboxylation(GO:0018214) |
| 0.0 | 0.1 | GO:0009296 | obsolete flagellum assembly(GO:0009296) |
| 0.0 | 0.2 | GO:0033146 | regulation of intracellular estrogen receptor signaling pathway(GO:0033146) |
| 0.0 | 0.1 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0033630 | positive regulation of cell adhesion mediated by integrin(GO:0033630) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0034227 | tRNA wobble base modification(GO:0002097) tRNA wobble uridine modification(GO:0002098) tRNA thio-modification(GO:0034227) |
| 0.0 | 0.2 | GO:0006103 | 2-oxoglutarate metabolic process(GO:0006103) |
| 0.0 | 0.0 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.1 | GO:0021520 | spinal cord motor neuron cell fate specification(GO:0021520) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.3 | GO:0048286 | lung alveolus development(GO:0048286) |
| 0.0 | 0.1 | GO:0044091 | membrane biogenesis(GO:0044091) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.3 | GO:0048701 | embryonic cranial skeleton morphogenesis(GO:0048701) |
| 0.0 | 0.0 | GO:0010523 | negative regulation of calcium ion transport into cytosol(GO:0010523) |
| 0.0 | 0.0 | GO:0021514 | ventral spinal cord interneuron differentiation(GO:0021514) ventral spinal cord interneuron fate commitment(GO:0060579) cell fate commitment involved in pattern specification(GO:0060581) |
| 0.0 | 0.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.0 | GO:0051307 | resolution of meiotic recombination intermediates(GO:0000712) meiotic chromosome separation(GO:0051307) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.2 | GO:0006376 | mRNA splice site selection(GO:0006376) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.2 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.0 | GO:0046325 | negative regulation of glucose import(GO:0046325) |
| 0.0 | 0.0 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.2 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.1 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0090025 | regulation of monocyte chemotaxis(GO:0090025) positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0090080 | positive regulation of MAPKKK cascade by fibroblast growth factor receptor signaling pathway(GO:0090080) |
| 0.0 | 0.1 | GO:0071027 | RNA surveillance(GO:0071025) nuclear RNA surveillance(GO:0071027) nuclear mRNA surveillance(GO:0071028) |
| 0.0 | 0.2 | GO:0010506 | regulation of autophagy(GO:0010506) |
| 0.0 | 0.0 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0046349 | amino sugar biosynthetic process(GO:0046349) |
| 0.0 | 0.0 | GO:1900078 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.2 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.1 | GO:0042178 | xenobiotic catabolic process(GO:0042178) |
| 0.0 | 0.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 0.0 | GO:0045992 | negative regulation of embryonic development(GO:0045992) |
| 0.0 | 0.0 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.1 | GO:0032464 | positive regulation of protein homooligomerization(GO:0032464) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0061077 | chaperone-mediated protein folding(GO:0061077) |
| 0.0 | 0.0 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0019720 | Mo-molybdopterin cofactor biosynthetic process(GO:0006777) Mo-molybdopterin cofactor metabolic process(GO:0019720) molybdopterin cofactor biosynthetic process(GO:0032324) molybdopterin cofactor metabolic process(GO:0043545) prosthetic group metabolic process(GO:0051189) |
| 0.0 | 0.1 | GO:0090114 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0060045 | positive regulation of cardiac muscle tissue growth(GO:0055023) positive regulation of cardiac muscle tissue development(GO:0055025) positive regulation of cardiac muscle cell proliferation(GO:0060045) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0035058 | nonmotile primary cilium assembly(GO:0035058) |
| 0.0 | 0.2 | GO:0035088 | establishment or maintenance of apical/basal cell polarity(GO:0035088) establishment or maintenance of bipolar cell polarity(GO:0061245) |
| 0.0 | 0.1 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.1 | GO:0045070 | positive regulation of viral genome replication(GO:0045070) |
| 0.0 | 0.0 | GO:0034447 | very-low-density lipoprotein particle clearance(GO:0034447) |
| 0.0 | 0.0 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.0 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0032897 | negative regulation of viral transcription(GO:0032897) |
| 0.0 | 0.1 | GO:0060710 | chorio-allantoic fusion(GO:0060710) |
| 0.0 | 0.1 | GO:0051593 | response to folic acid(GO:0051593) |
| 0.0 | 0.0 | GO:0017085 | response to insecticide(GO:0017085) |
| 0.0 | 0.0 | GO:0043615 | astrocyte cell migration(GO:0043615) |
| 0.0 | 0.0 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.0 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.0 | GO:0072191 | positive regulation of smooth muscle cell differentiation(GO:0051152) ureter development(GO:0072189) ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) kidney smooth muscle tissue development(GO:0072194) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.2 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.4 | GO:0007163 | establishment or maintenance of cell polarity(GO:0007163) |
| 0.0 | 0.2 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0048194 | vesicle coating(GO:0006901) COPI-coated vesicle budding(GO:0035964) Golgi vesicle budding(GO:0048194) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) single-organism membrane budding(GO:1902591) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.0 | GO:0001779 | natural killer cell differentiation(GO:0001779) |
| 0.0 | 0.0 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0031290 | retinal ganglion cell axon guidance(GO:0031290) |
| 0.0 | 0.1 | GO:0045648 | positive regulation of erythrocyte differentiation(GO:0045648) |
| 0.0 | 0.0 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.1 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.2 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.0 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0005852 | eukaryotic translation initiation factor 3 complex(GO:0005852) |
| 0.0 | 0.1 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.0 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0017119 | Golgi transport complex(GO:0017119) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.4 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.0 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.2 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.1 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.1 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.0 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.5 | GO:1903293 | protein serine/threonine phosphatase complex(GO:0008287) phosphatase complex(GO:1903293) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.0 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.4 | GO:0022625 | cytosolic large ribosomal subunit(GO:0022625) |
| 0.0 | 0.0 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.0 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.1 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.1 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.1 | 0.4 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0034187 | obsolete apolipoprotein E binding(GO:0034187) |
| 0.1 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.3 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.2 | GO:0004908 | interleukin-1 receptor activity(GO:0004908) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.3 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.1 | GO:0032139 | dinucleotide insertion or deletion binding(GO:0032139) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0004128 | cytochrome-b5 reductase activity, acting on NAD(P)H(GO:0004128) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.2 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.0 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.2 | GO:0016668 | oxidoreductase activity, acting on a sulfur group of donors, NAD(P) as acceptor(GO:0016668) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0043398 | HLH domain binding(GO:0043398) |
| 0.0 | 0.0 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.0 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.0 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0016671 | oxidoreductase activity, acting on a sulfur group of donors, disulfide as acceptor(GO:0016671) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.2 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.0 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.0 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.0 | GO:0043734 | DNA demethylase activity(GO:0035514) DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.0 | GO:0004694 | eukaryotic translation initiation factor 2alpha kinase activity(GO:0004694) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.0 | GO:0015037 | peptide disulfide oxidoreductase activity(GO:0015037) |
| 0.0 | 0.0 | GO:0005350 | pyrimidine nucleobase transmembrane transporter activity(GO:0005350) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0008312 | 7S RNA binding(GO:0008312) |
| 0.0 | 0.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.6 | GO:0003743 | translation initiation factor activity(GO:0003743) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.1 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.2 | GO:0030331 | estrogen receptor binding(GO:0030331) |
| 0.0 | 0.1 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.0 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.2 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.0 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.0 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.0 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.0 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.3 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID IGF1 PATHWAY | IGF1 pathway |
| 0.0 | 0.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.4 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.3 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.1 | ST INTERFERON GAMMA PATHWAY | Interferon gamma pathway. |
| 0.0 | 0.0 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.0 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.5 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.3 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.0 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.2 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY TRISTETRAPROLIN TTP | Genes involved in Destabilization of mRNA by Tristetraprolin (TTP) |
| 0.0 | 0.3 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.0 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.1 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.2 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.1 | REACTOME COPI MEDIATED TRANSPORT | Genes involved in COPI Mediated Transport |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |