Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for OTX2_CRX
Z-value: 0.60
Transcription factors associated with OTX2_CRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
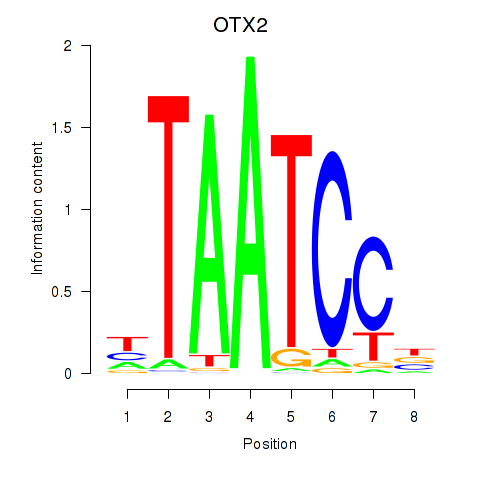
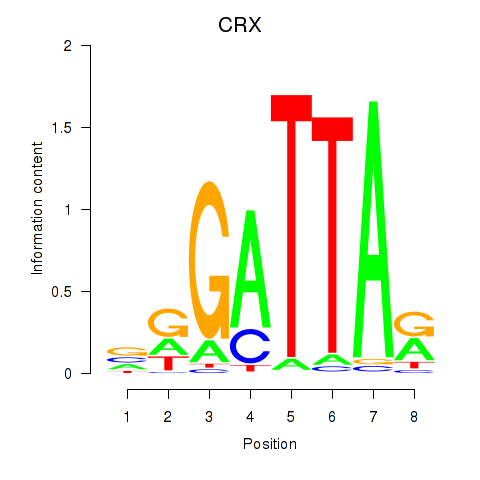
OTX2
|
ENSG00000165588.12 | orthodenticle homeobox 2 |
|
CRX
|
ENSG00000105392.11 | cone-rod homeobox |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr14_57274863_57275150 | OTX2 | 2160 | 0.260373 | 0.64 | 6.3e-02 | Click! |
| chr14_57275358_57275543 | OTX2 | 1716 | 0.309414 | 0.45 | 2.2e-01 | Click! |
| chr14_57264424_57264575 | OTX2 | 7867 | 0.170193 | -0.36 | 3.4e-01 | Click! |
| chr14_57274118_57274269 | OTX2 | 1812 | 0.300379 | 0.28 | 4.6e-01 | Click! |
| chr14_57274481_57274632 | OTX2 | 2175 | 0.260477 | 0.23 | 5.4e-01 | Click! |
Activity of the OTX2_CRX motif across conditions
Conditions sorted by the z-value of the OTX2_CRX motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
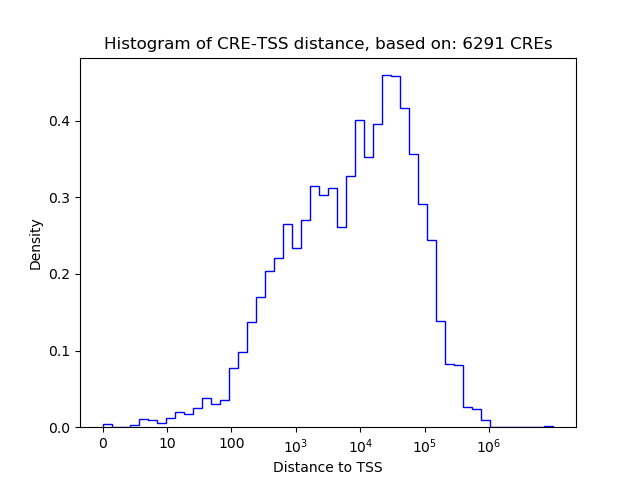
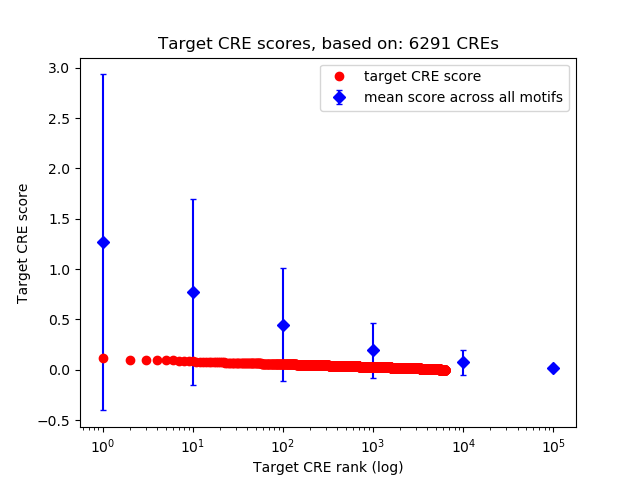
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr15_89914451_89914936 | 0.11 |
ENSG00000207819 |
. |
3445 |
0.23 |
| chrX_34671991_34672311 | 0.10 |
TMEM47 |
transmembrane protein 47 |
3254 |
0.41 |
| chr2_36585229_36585663 | 0.10 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
1832 |
0.5 |
| chr8_58907686_58908288 | 0.10 |
FAM110B |
family with sequence similarity 110, member B |
874 |
0.76 |
| chr3_49169788_49170138 | 0.10 |
LAMB2 |
laminin, beta 2 (laminin S) |
490 |
0.65 |
| chr9_90588787_90589959 | 0.09 |
CDK20 |
cyclin-dependent kinase 20 |
29 |
0.98 |
| chr5_3598562_3598819 | 0.09 |
CTD-2012M11.3 |
|
1612 |
0.42 |
| chr3_123518515_123518699 | 0.09 |
MYLK |
myosin light chain kinase |
5919 |
0.23 |
| chr4_112600114_112600265 | 0.08 |
ENSG00000200963 |
. |
347621 |
0.01 |
| chr2_12842465_12842741 | 0.08 |
TRIB2 |
tribbles pseudokinase 2 |
14412 |
0.26 |
| chrX_99899847_99900028 | 0.08 |
SRPX2 |
sushi-repeat containing protein, X-linked 2 |
722 |
0.65 |
| chr14_27065271_27065422 | 0.08 |
NOVA1 |
neuro-oncological ventral antigen 1 |
65 |
0.96 |
| chr11_130320694_130320845 | 0.08 |
ADAMTS15 |
ADAM metallopeptidase with thrombospondin type 1 motif, 15 |
1900 |
0.41 |
| chr4_75718841_75720131 | 0.08 |
BTC |
betacellulin |
410 |
0.91 |
| chr13_65834890_65835427 | 0.08 |
ENSG00000221685 |
. |
603147 |
0.0 |
| chr14_53301814_53302501 | 0.08 |
FERMT2 |
fermitin family member 2 |
29082 |
0.15 |
| chr6_26575872_26576023 | 0.08 |
ABT1 |
activator of basal transcription 1 |
21233 |
0.14 |
| chr19_6740138_6741070 | 0.07 |
TRIP10 |
thyroid hormone receptor interactor 10 |
325 |
0.81 |
| chr1_114522384_114522566 | 0.07 |
OLFML3 |
olfactomedin-like 3 |
220 |
0.93 |
| chr16_75232194_75232506 | 0.07 |
CTRB2 |
chymotrypsinogen B2 |
7343 |
0.14 |
| chr16_54969072_54969449 | 0.07 |
IRX5 |
iroquois homeobox 5 |
3276 |
0.4 |
| chr3_155109533_155109828 | 0.07 |
ENSG00000272096 |
. |
45932 |
0.16 |
| chr7_73867713_73868217 | 0.07 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
155 |
0.96 |
| chr2_170366040_170366191 | 0.07 |
KLHL41 |
kelch-like family member 41 |
97 |
0.97 |
| chr6_46458168_46458592 | 0.07 |
RCAN2 |
regulator of calcineurin 2 |
719 |
0.62 |
| chr3_87040086_87040946 | 0.07 |
VGLL3 |
vestigial like 3 (Drosophila) |
247 |
0.96 |
| chr13_34250323_34250584 | 0.07 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
141733 |
0.05 |
| chr16_28549300_28549715 | 0.07 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
822 |
0.5 |
| chr8_136478593_136478847 | 0.07 |
KHDRBS3 |
KH domain containing, RNA binding, signal transduction associated 3 |
7770 |
0.25 |
| chr22_31198410_31198813 | 0.07 |
OSBP2 |
oxysterol binding protein 2 |
462 |
0.84 |
| chr20_49307514_49308405 | 0.07 |
FAM65C |
family with sequence similarity 65, member C |
43 |
0.97 |
| chr1_235795999_235796340 | 0.07 |
GNG4 |
guanine nucleotide binding protein (G protein), gamma 4 |
17124 |
0.25 |
| chr5_9546839_9547056 | 0.07 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
760 |
0.6 |
| chr7_93552303_93552503 | 0.07 |
GNG11 |
guanine nucleotide binding protein (G protein), gamma 11 |
1392 |
0.38 |
| chr16_54970178_54971110 | 0.07 |
IRX5 |
iroquois homeobox 5 |
4660 |
0.36 |
| chr15_38547303_38547454 | 0.07 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
1996 |
0.49 |
| chr19_10444919_10445184 | 0.07 |
ICAM3 |
intercellular adhesion molecule 3 |
348 |
0.62 |
| chr8_55294582_55295235 | 0.07 |
ENSG00000244107 |
. |
38120 |
0.17 |
| chr9_89049166_89049463 | 0.07 |
ENSG00000222293 |
. |
11271 |
0.28 |
| chr1_215257188_215257339 | 0.07 |
KCNK2 |
potassium channel, subfamily K, member 2 |
403 |
0.92 |
| chr12_81102183_81102935 | 0.07 |
MYF6 |
myogenic factor 6 (herculin) |
1282 |
0.49 |
| chr10_97049818_97050125 | 0.07 |
PDLIM1 |
PDZ and LIM domain 1 |
810 |
0.7 |
| chr6_35286420_35286751 | 0.07 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
9070 |
0.18 |
| chr18_20746992_20747143 | 0.07 |
CABLES1 |
Cdk5 and Abl enzyme substrate 1 |
11278 |
0.21 |
| chr12_3308931_3310293 | 0.07 |
TSPAN9 |
tetraspanin 9 |
729 |
0.75 |
| chr18_53530407_53530634 | 0.07 |
TCF4 |
transcription factor 4 |
198502 |
0.03 |
| chr10_95848621_95848951 | 0.07 |
PLCE1 |
phospholipase C, epsilon 1 |
44 |
0.97 |
| chr3_28383486_28383637 | 0.07 |
AZI2 |
5-azacytidine induced 2 |
6407 |
0.24 |
| chr10_77159524_77159700 | 0.06 |
ENSG00000237149 |
. |
1665 |
0.33 |
| chr12_1427708_1428104 | 0.06 |
RP5-951N9.2 |
|
67093 |
0.11 |
| chr1_164531408_164531670 | 0.06 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
503 |
0.87 |
| chr1_8772244_8772432 | 0.06 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
9060 |
0.25 |
| chr1_91191218_91191722 | 0.06 |
BARHL2 |
BarH-like homeobox 2 |
8676 |
0.3 |
| chr10_119303944_119304573 | 0.06 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
321 |
0.88 |
| chr3_112359594_112359745 | 0.06 |
CCDC80 |
coiled-coil domain containing 80 |
447 |
0.87 |
| chr14_35179773_35180008 | 0.06 |
CFL2 |
cofilin 2 (muscle) |
3144 |
0.26 |
| chr2_176987899_176988234 | 0.06 |
HOXD9 |
homeobox D9 |
978 |
0.29 |
| chr4_164253766_164254020 | 0.06 |
NPY1R |
neuropeptide Y receptor Y1 |
121 |
0.97 |
| chr12_91576386_91576586 | 0.06 |
DCN |
decorin |
0 |
0.99 |
| chr2_164591967_164592245 | 0.06 |
FIGN |
fidgetin |
411 |
0.92 |
| chr17_762649_762818 | 0.06 |
NXN |
nucleoredoxin |
4604 |
0.18 |
| chr1_39735914_39736147 | 0.06 |
MACF1 |
microtubule-actin crosslinking factor 1 |
1831 |
0.35 |
| chr18_12309515_12309681 | 0.06 |
TUBB6 |
tubulin, beta 6 class V |
1336 |
0.41 |
| chr15_65901005_65901156 | 0.06 |
VWA9 |
von Willebrand factor A domain containing 9 |
2327 |
0.2 |
| chr2_216299709_216300678 | 0.06 |
FN1 |
fibronectin 1 |
597 |
0.55 |
| chr13_30276703_30277107 | 0.06 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
107080 |
0.07 |
| chr8_99963595_99963771 | 0.06 |
OSR2 |
odd-skipped related transciption factor 2 |
3063 |
0.23 |
| chr9_73483972_73484180 | 0.06 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
102 |
0.98 |
| chr20_19974216_19974491 | 0.06 |
NAA20 |
N(alpha)-acetyltransferase 20, NatB catalytic subunit |
23407 |
0.19 |
| chr15_32933967_32934156 | 0.06 |
SCG5 |
secretogranin V (7B2 protein) |
158 |
0.94 |
| chr2_216298025_216298259 | 0.06 |
FN1 |
fibronectin 1 |
2648 |
0.29 |
| chr1_92907603_92907960 | 0.06 |
ENSG00000242764 |
. |
39539 |
0.16 |
| chr13_114792883_114793184 | 0.06 |
RASA3 |
RAS p21 protein activator 3 |
50405 |
0.16 |
| chr2_207307729_207308161 | 0.06 |
ADAM23 |
ADAM metallopeptidase domain 23 |
318 |
0.92 |
| chr6_113953461_113953992 | 0.06 |
ENSG00000266650 |
. |
29609 |
0.21 |
| chr13_36704166_36704317 | 0.06 |
DCLK1 |
doublecortin-like kinase 1 |
1202 |
0.59 |
| chrX_3263508_3263724 | 0.06 |
MXRA5 |
matrix-remodelling associated 5 |
1066 |
0.64 |
| chr7_55089371_55089524 | 0.06 |
EGFR |
epidermal growth factor receptor |
2636 |
0.41 |
| chr6_128977127_128977410 | 0.06 |
ENSG00000238938 |
. |
42922 |
0.16 |
| chr16_55090478_55090815 | 0.06 |
IRX5 |
iroquois homeobox 5 |
124662 |
0.06 |
| chr19_46286294_46286481 | 0.06 |
DMPK |
dystrophia myotonica-protein kinase |
640 |
0.45 |
| chr22_38657243_38657394 | 0.06 |
TMEM184B |
transmembrane protein 184B |
11352 |
0.11 |
| chr15_95217234_95217771 | 0.06 |
MCTP2 |
multiple C2 domains, transmembrane 2 |
318299 |
0.01 |
| chr1_75138923_75139762 | 0.06 |
C1orf173 |
chromosome 1 open reading frame 173 |
80 |
0.98 |
| chr5_9544908_9545592 | 0.06 |
SEMA5A |
sema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A |
937 |
0.56 |
| chr5_95950045_95950381 | 0.06 |
ENSG00000238518 |
. |
24233 |
0.16 |
| chr11_125497097_125497248 | 0.06 |
CHEK1 |
checkpoint kinase 1 |
529 |
0.7 |
| chr3_170132220_170132371 | 0.06 |
CLDN11 |
claudin 11 |
4358 |
0.29 |
| chr4_115520348_115520872 | 0.06 |
UGT8 |
UDP glycosyltransferase 8 |
684 |
0.8 |
| chr3_112961154_112961680 | 0.06 |
BOC |
BOC cell adhesion associated, oncogene regulated |
26219 |
0.19 |
| chr3_123168721_123169253 | 0.06 |
ADCY5 |
adenylate cyclase 5 |
382 |
0.9 |
| chr4_150453103_150453254 | 0.06 |
ENSG00000202331 |
. |
375534 |
0.01 |
| chr12_50636266_50636502 | 0.06 |
LIMA1 |
LIM domain and actin binding 1 |
7292 |
0.13 |
| chr15_35949903_35950054 | 0.06 |
DPH6 |
diphthamine biosynthesis 6 |
111585 |
0.07 |
| chr16_53086342_53086851 | 0.06 |
RP11-467J12.4 |
|
189 |
0.95 |
| chr7_143579344_143579634 | 0.06 |
FAM115A |
family with sequence similarity 115, member A |
487 |
0.8 |
| chr15_23931247_23932124 | 0.06 |
NDN |
necdin, melanoma antigen (MAGE) family member |
765 |
0.7 |
| chr11_19420110_19420332 | 0.06 |
ENSG00000200687 |
. |
3307 |
0.25 |
| chr9_109624363_109624788 | 0.06 |
ZNF462 |
zinc finger protein 462 |
803 |
0.69 |
| chr7_19408295_19408500 | 0.06 |
FERD3L |
Fer3-like bHLH transcription factor |
223353 |
0.02 |
| chr4_152148784_152149033 | 0.06 |
SH3D19 |
SH3 domain containing 19 |
274 |
0.93 |
| chr1_154941803_154941985 | 0.06 |
SHC1 |
SHC (Src homology 2 domain containing) transforming protein 1 |
525 |
0.53 |
| chr2_19557855_19558307 | 0.06 |
OSR1 |
odd-skipped related transciption factor 1 |
333 |
0.92 |
| chr9_89559920_89560071 | 0.06 |
GAS1 |
growth arrest-specific 1 |
2109 |
0.48 |
| chr5_3597166_3597716 | 0.06 |
IRX1 |
iroquois homeobox 1 |
1273 |
0.51 |
| chr1_151953452_151953706 | 0.06 |
S100A10 |
S100 calcium binding protein A10 |
11475 |
0.14 |
| chr8_93115764_93116215 | 0.06 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
475 |
0.9 |
| chr1_180882927_180883078 | 0.06 |
KIAA1614 |
KIAA1614 |
677 |
0.71 |
| chr9_91568737_91568931 | 0.06 |
C9orf47 |
chromosome 9 open reading frame 47 |
36944 |
0.17 |
| chr13_80824030_80824188 | 0.06 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
89685 |
0.1 |
| chr18_2906194_2907483 | 0.06 |
RP11-737O24.5 |
|
14126 |
0.16 |
| chr3_65583777_65583941 | 0.06 |
MAGI1 |
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
298 |
0.95 |
| chr6_3517843_3518042 | 0.06 |
SLC22A23 |
solute carrier family 22, member 23 |
60686 |
0.14 |
| chr1_162582264_162582415 | 0.06 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
18824 |
0.16 |
| chr2_217180593_217180957 | 0.06 |
AC069155.1 |
|
12727 |
0.18 |
| chr13_76361393_76361544 | 0.06 |
LMO7 |
LIM domain 7 |
1506 |
0.51 |
| chr3_55519561_55519953 | 0.06 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
1574 |
0.4 |
| chr1_8433877_8434267 | 0.06 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
9174 |
0.19 |
| chr1_157986336_157986537 | 0.05 |
KIRREL-IT1 |
KIRREL intronic transcript 1 (non-protein coding) |
8904 |
0.22 |
| chr17_78044749_78045392 | 0.05 |
CCDC40 |
coiled-coil domain containing 40 |
12346 |
0.15 |
| chr6_147235797_147235979 | 0.05 |
STXBP5-AS1 |
STXBP5 antisense RNA 1 |
1133 |
0.66 |
| chr2_11051733_11051998 | 0.05 |
KCNF1 |
potassium voltage-gated channel, subfamily F, member 1 |
198 |
0.95 |
| chr10_98272514_98273588 | 0.05 |
TLL2 |
tolloid-like 2 |
617 |
0.79 |
| chr15_62862079_62862274 | 0.05 |
TLN2 |
talin 2 |
8612 |
0.27 |
| chr5_88178054_88178448 | 0.05 |
MEF2C |
myocyte enhancer factor 2C |
713 |
0.52 |
| chr16_88450130_88450579 | 0.05 |
ZNF469 |
zinc finger protein 469 |
43525 |
0.15 |
| chr8_38965103_38965508 | 0.05 |
ADAM32 |
ADAM metallopeptidase domain 32 |
13 |
0.98 |
| chr4_187901481_187901677 | 0.05 |
ENSG00000252382 |
. |
122969 |
0.06 |
| chr7_94536809_94537187 | 0.05 |
PPP1R9A |
protein phosphatase 1, regulatory subunit 9A |
11 |
0.99 |
| chr1_215740105_215740888 | 0.05 |
KCTD3 |
potassium channel tetramerization domain containing 3 |
239 |
0.96 |
| chr1_170851017_170851182 | 0.05 |
MROH9 |
maestro heat-like repeat family member 9 |
53513 |
0.16 |
| chr8_121137253_121137925 | 0.05 |
COL14A1 |
collagen, type XIV, alpha 1 |
237 |
0.96 |
| chr2_235541588_235541739 | 0.05 |
ARL4C |
ADP-ribosylation factor-like 4C |
135966 |
0.05 |
| chr9_109876632_109876800 | 0.05 |
RP11-508N12.2 |
|
11447 |
0.29 |
| chr2_198909706_198909857 | 0.05 |
PLCL1 |
phospholipase C-like 1 |
38755 |
0.22 |
| chr2_175711229_175711408 | 0.05 |
CHN1 |
chimerin 1 |
175 |
0.97 |
| chr14_95949817_95950024 | 0.05 |
SYNE3 |
spectrin repeat containing, nuclear envelope family member 3 |
7747 |
0.18 |
| chr18_8707466_8707765 | 0.05 |
RP11-674N23.1 |
|
4 |
0.73 |
| chr16_10671409_10671697 | 0.05 |
RP11-27M24.2 |
|
487 |
0.78 |
| chr2_240230661_240230844 | 0.05 |
HDAC4 |
histone deacetylase 4 |
155 |
0.95 |
| chr12_91575790_91576058 | 0.05 |
DCN |
decorin |
505 |
0.87 |
| chr6_127839583_127839785 | 0.05 |
SOGA3 |
SOGA family member 3 |
352 |
0.91 |
| chr1_174081654_174081874 | 0.05 |
RABGAP1L |
RAB GTPase activating protein 1-like |
46784 |
0.14 |
| chr10_80009045_80009196 | 0.05 |
ENSG00000201393 |
. |
118144 |
0.06 |
| chr15_40269217_40269368 | 0.05 |
EIF2AK4 |
eukaryotic translation initiation factor 2 alpha kinase 4 |
23116 |
0.14 |
| chr16_121633_122294 | 0.05 |
RHBDF1 |
rhomboid 5 homolog 1 (Drosophila) |
666 |
0.51 |
| chr8_37556330_37556485 | 0.05 |
ZNF703 |
zinc finger protein 703 |
3138 |
0.17 |
| chr3_100852493_100852644 | 0.05 |
ENSG00000238312 |
. |
68108 |
0.12 |
| chr9_97682991_97683218 | 0.05 |
RP11-54O15.3 |
|
14124 |
0.17 |
| chr6_113885726_113885909 | 0.05 |
ENSG00000266650 |
. |
38300 |
0.18 |
| chr17_80970638_80970845 | 0.05 |
RP11-1197K16.2 |
|
31783 |
0.15 |
| chr8_124829147_124829427 | 0.05 |
FER1L6 |
fer-1-like 6 (C. elegans) |
34940 |
0.17 |
| chr11_125365710_125366094 | 0.05 |
FEZ1 |
fasciculation and elongation protein zeta 1 (zygin I) |
116 |
0.91 |
| chr11_121593168_121593875 | 0.05 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
132393 |
0.05 |
| chr8_18941319_18941572 | 0.05 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
795 |
0.76 |
| chr5_34487804_34487985 | 0.05 |
RAI14 |
retinoic acid induced 14 |
168448 |
0.03 |
| chr4_177712658_177713139 | 0.05 |
VEGFC |
vascular endothelial growth factor C |
983 |
0.71 |
| chr7_30952459_30952610 | 0.05 |
AQP1 |
aquaporin 1 (Colton blood group) |
1064 |
0.6 |
| chr7_19136961_19137112 | 0.05 |
AC003986.6 |
|
15061 |
0.16 |
| chr5_150283969_150284454 | 0.05 |
ZNF300 |
zinc finger protein 300 |
140 |
0.96 |
| chr10_17241830_17242242 | 0.05 |
TRDMT1 |
tRNA aspartic acid methyltransferase 1 |
1543 |
0.37 |
| chr11_75272983_75274329 | 0.05 |
SERPINH1 |
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
216 |
0.91 |
| chr3_73652541_73652692 | 0.05 |
ENSG00000239119 |
. |
9463 |
0.21 |
| chr3_189306684_189306989 | 0.05 |
TP63 |
tumor protein p63 |
42380 |
0.21 |
| chr5_171839559_171839710 | 0.05 |
ENSG00000216127 |
. |
27124 |
0.19 |
| chr18_46020104_46020474 | 0.05 |
ENSG00000200872 |
. |
18318 |
0.21 |
| chr1_34630860_34631011 | 0.05 |
CSMD2 |
CUB and Sushi multiple domains 2 |
60 |
0.96 |
| chr5_159305990_159306237 | 0.05 |
ADRA1B |
adrenoceptor alpha 1B |
37677 |
0.2 |
| chr18_56532811_56532962 | 0.05 |
ZNF532 |
zinc finger protein 532 |
100 |
0.97 |
| chr12_309110_309261 | 0.05 |
RP11-283I3.2 |
|
3623 |
0.16 |
| chr1_86859564_86859715 | 0.05 |
ODF2L |
outer dense fiber of sperm tails 2-like |
2021 |
0.38 |
| chrY_14075891_14076042 | 0.05 |
ENSG00000265161 |
. |
128454 |
0.06 |
| chr8_87052179_87052395 | 0.05 |
PSKH2 |
protein serine kinase H2 |
29639 |
0.19 |
| chr1_173835540_173835691 | 0.05 |
ENSG00000234741 |
. |
106 |
0.88 |
| chr22_25348691_25349626 | 0.05 |
KIAA1671 |
KIAA1671 |
461 |
0.83 |
| chr5_86478454_86478741 | 0.05 |
AC008394.1 |
Uncharacterized protein |
56225 |
0.12 |
| chr18_12745437_12745588 | 0.05 |
ENSG00000201466 |
. |
15204 |
0.16 |
| chr21_35133624_35133954 | 0.05 |
ITSN1 |
intersectin 1 (SH3 domain protein) |
26443 |
0.2 |
| chr10_74544262_74544615 | 0.05 |
RP11-354E23.5 |
|
18120 |
0.19 |
| chr2_134325508_134325697 | 0.05 |
NCKAP5 |
NCK-associated protein 5 |
429 |
0.88 |
| chrX_90690422_90690847 | 0.05 |
PABPC5-AS1 |
PABPC5 antisense RNA 1 |
636 |
0.54 |
| chr10_3290980_3291169 | 0.05 |
PITRM1 |
pitrilysin metallopeptidase 1 |
76071 |
0.11 |
| chr4_111397420_111397787 | 0.05 |
ENPEP |
glutamyl aminopeptidase (aminopeptidase A) |
374 |
0.89 |
| chr7_3299969_3300120 | 0.05 |
SDK1 |
sidekick cell adhesion molecule 1 |
41036 |
0.16 |
| chr13_109740818_109740969 | 0.05 |
MYO16-AS2 |
MYO16 antisense RNA 2 |
13374 |
0.3 |
| chr8_71359441_71359951 | 0.05 |
ENSG00000253143 |
. |
33026 |
0.17 |
| chr6_83073574_83074143 | 0.05 |
TPBG |
trophoblast glycoprotein |
103 |
0.98 |
| chr12_8012330_8012481 | 0.05 |
ENSG00000201663 |
. |
8991 |
0.12 |
| chr18_59560344_59560906 | 0.05 |
RNF152 |
ring finger protein 152 |
367 |
0.93 |
| chr4_4862278_4863048 | 0.05 |
MSX1 |
msh homeobox 1 |
1270 |
0.58 |
| chr4_39525757_39525928 | 0.05 |
UGDH |
UDP-glucose 6-dehydrogenase |
2335 |
0.24 |
| chr10_52833938_52834229 | 0.05 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
149 |
0.97 |
| chr5_169230726_169230933 | 0.05 |
CTB-37A13.1 |
|
24460 |
0.22 |
| chr7_555957_556150 | 0.05 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
2092 |
0.33 |
| chr11_101453394_101453891 | 0.05 |
TRPC6 |
transient receptor potential cation channel, subfamily C, member 6 |
592 |
0.84 |
| chr1_214722280_214722431 | 0.05 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
2211 |
0.42 |
| chr8_39966106_39966460 | 0.05 |
C8orf4 |
chromosome 8 open reading frame 4 |
44706 |
0.18 |
| chr3_114477226_114477377 | 0.05 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
486 |
0.86 |
| chr17_1081882_1082496 | 0.05 |
ABR |
active BCR-related |
889 |
0.58 |
| chr5_150181131_150181300 | 0.05 |
AC010441.1 |
|
23348 |
0.14 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0072081 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.0 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.0 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.0 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.0 | GO:1902170 | cellular response to nitric oxide(GO:0071732) cellular response to reactive nitrogen species(GO:1902170) |
| 0.0 | 0.0 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.1 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.0 | GO:0002866 | positive regulation of acute inflammatory response to antigenic stimulus(GO:0002866) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.0 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0043260 | laminin-11 complex(GO:0043260) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.0 | GO:0018995 | host(GO:0018995) host cell part(GO:0033643) host intracellular part(GO:0033646) intracellular region of host(GO:0043656) host cell(GO:0043657) other organism(GO:0044215) other organism cell(GO:0044216) other organism part(GO:0044217) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.0 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.4 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.0 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0001228 | transcription factor activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0000982) transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific binding(GO:0001077) transcriptional activator activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001228) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME PI 3K CASCADE | Genes involved in PI-3K cascade |
| 0.0 | 0.0 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.0 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |