Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
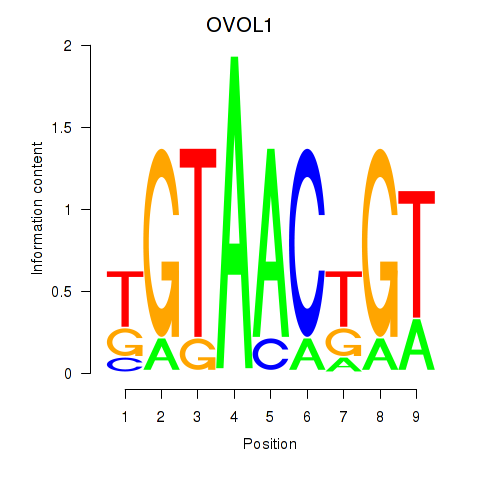
Results for OVOL1
Z-value: 0.90
Transcription factors associated with OVOL1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OVOL1
|
ENSG00000172818.5 | ovo like transcriptional repressor 1 |
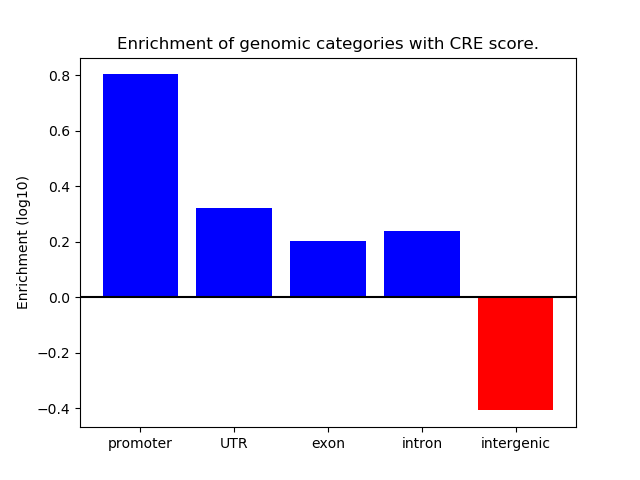
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_65555266_65555920 | OVOL1 | 1100 | 0.277447 | -0.87 | 2.1e-03 | Click! |
| chr11_65553938_65554232 | OVOL1 | 408 | 0.691119 | -0.72 | 3.0e-02 | Click! |
| chr11_65554622_65554896 | OVOL1 | 266 | 0.812927 | -0.69 | 3.8e-02 | Click! |
| chr11_65560364_65560515 | OVOL1 | 741 | 0.426536 | -0.09 | 8.3e-01 | Click! |
Activity of the OVOL1 motif across conditions
Conditions sorted by the z-value of the OVOL1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
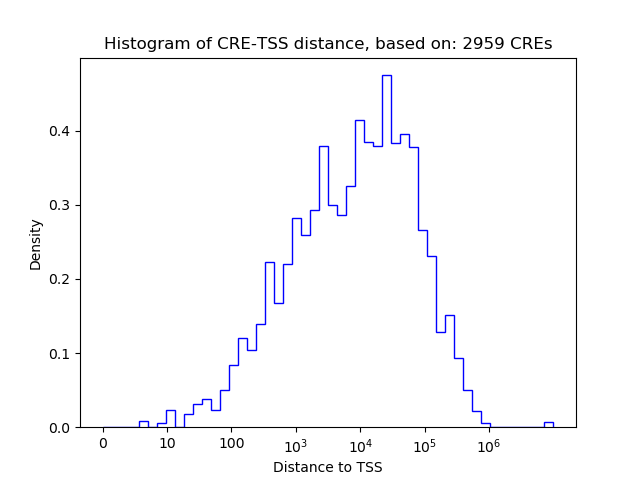
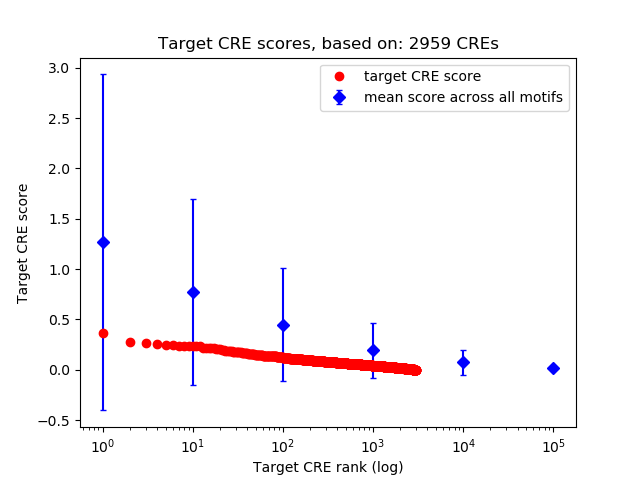
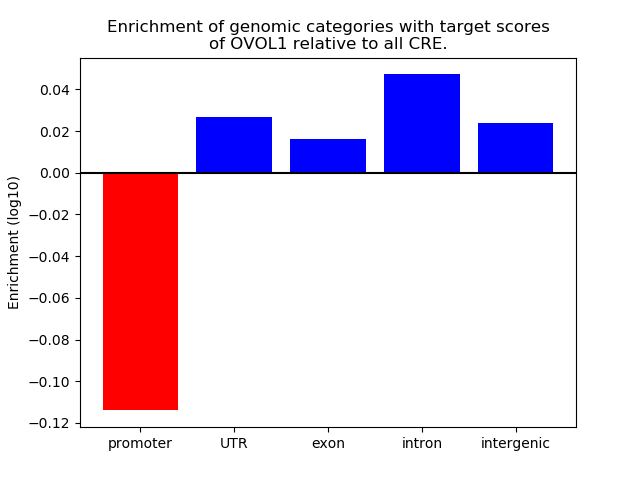
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr17_46177753_46179071 | 0.37 |
CBX1 |
chromobox homolog 1 |
148 |
0.91 |
| chr11_27491201_27491490 | 0.27 |
RP11-159H22.2 |
|
1931 |
0.27 |
| chr5_15510117_15510353 | 0.27 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
8688 |
0.31 |
| chr3_16219029_16219297 | 0.26 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
2947 |
0.33 |
| chr18_5543806_5544379 | 0.24 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
149 |
0.95 |
| chrX_82764698_82764849 | 0.24 |
RP3-326L13.2 |
|
551 |
0.75 |
| chr3_112358958_112359194 | 0.24 |
CCDC80 |
coiled-coil domain containing 80 |
1040 |
0.62 |
| chr20_50178264_50178663 | 0.24 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
705 |
0.79 |
| chr1_98515040_98515403 | 0.24 |
ENSG00000225206 |
. |
3494 |
0.38 |
| chr18_48571368_48571694 | 0.24 |
SMAD4 |
SMAD family member 4 |
202 |
0.96 |
| chr6_154800959_154801329 | 0.23 |
CNKSR3 |
CNKSR family member 3 |
1519 |
0.56 |
| chr6_2789380_2789655 | 0.23 |
WRNIP1 |
Werner helicase interacting protein 1 |
20510 |
0.17 |
| chr18_43335869_43336020 | 0.22 |
RP11-116O18.3 |
|
8764 |
0.2 |
| chr8_20126437_20126669 | 0.22 |
LZTS1 |
leucine zipper, putative tumor suppressor 1 |
13750 |
0.19 |
| chr9_79451100_79451279 | 0.21 |
PRUNE2 |
prune homolog 2 (Drosophila) |
69812 |
0.12 |
| chr3_197099341_197099661 | 0.21 |
ENSG00000238491 |
. |
33079 |
0.17 |
| chr4_126602291_126602442 | 0.21 |
FAT4 |
FAT atypical cadherin 4 |
287275 |
0.01 |
| chr3_174157293_174157469 | 0.21 |
NAALADL2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
1396 |
0.61 |
| chr7_77930640_77930829 | 0.21 |
RP5-1185I7.1 |
|
310997 |
0.01 |
| chr5_71544123_71544318 | 0.20 |
ENSG00000244748 |
. |
66295 |
0.09 |
| chr2_202088333_202088488 | 0.20 |
CASP8 |
caspase 8, apoptosis-related cysteine peptidase |
9756 |
0.18 |
| chr6_18434642_18434820 | 0.20 |
ENSG00000238458 |
. |
32448 |
0.19 |
| chr2_19807945_19808132 | 0.19 |
OSR1 |
odd-skipped related transciption factor 1 |
249624 |
0.02 |
| chrX_33228916_33229133 | 0.19 |
DMD |
dystrophin |
405 |
0.92 |
| chr1_211589992_211590552 | 0.19 |
TRAF5 |
TNF receptor-associated factor 5 |
70566 |
0.1 |
| chr21_17566581_17566826 | 0.18 |
ENSG00000201025 |
. |
90386 |
0.1 |
| chr7_120631344_120631706 | 0.18 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
1849 |
0.35 |
| chr5_87972381_87972532 | 0.18 |
CTC-467M3.1 |
|
420 |
0.85 |
| chr21_36256046_36256553 | 0.18 |
RUNX1 |
runt-related transcription factor 1 |
3181 |
0.38 |
| chr2_17721828_17722425 | 0.18 |
VSNL1 |
visinin-like 1 |
163 |
0.97 |
| chr12_96580571_96580722 | 0.18 |
ELK3 |
ELK3, ETS-domain protein (SRF accessory protein 2) |
7514 |
0.22 |
| chr8_86971534_86971711 | 0.18 |
ATP6V0D2 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d2 |
27930 |
0.23 |
| chr5_124748629_124749002 | 0.18 |
ENSG00000222107 |
. |
61991 |
0.16 |
| chr22_29489752_29489903 | 0.17 |
CTA-747E2.10 |
|
17844 |
0.13 |
| chrX_9435964_9436115 | 0.17 |
TBL1X |
transducin (beta)-like 1X-linked |
2705 |
0.43 |
| chr1_101334241_101334590 | 0.17 |
EXTL2 |
exostosin-like glycosyltransferase 2 |
25805 |
0.15 |
| chr4_16619235_16619484 | 0.17 |
LDB2 |
LIM domain binding 2 |
21861 |
0.28 |
| chr7_84121800_84121951 | 0.17 |
SEMA3A |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A |
10 |
0.99 |
| chr17_41790734_41791669 | 0.17 |
ENSG00000215964 |
. |
34367 |
0.11 |
| chr5_121409447_121409998 | 0.17 |
LOX |
lysyl oxidase |
4258 |
0.27 |
| chr10_99030444_99030681 | 0.16 |
ARHGAP19 |
Rho GTPase activating protein 19 |
141 |
0.95 |
| chr9_13034691_13034857 | 0.16 |
MPDZ |
multiple PDZ domain protein |
101406 |
0.08 |
| chr6_16337793_16338215 | 0.16 |
ENSG00000265642 |
. |
90750 |
0.08 |
| chr19_48216774_48216947 | 0.16 |
EHD2 |
EH-domain containing 2 |
181 |
0.93 |
| chr9_12814147_12814330 | 0.15 |
RP11-3L8.3 |
|
106 |
0.97 |
| chr3_150138037_150138333 | 0.15 |
TSC22D2 |
TSC22 domain family, member 2 |
9389 |
0.28 |
| chr6_131360061_131360448 | 0.15 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
23272 |
0.26 |
| chr10_52567810_52567961 | 0.15 |
RP11-449O16.2 |
|
16400 |
0.22 |
| chr10_21803540_21803691 | 0.15 |
SKIDA1 |
SKI/DACH domain containing 1 |
3233 |
0.17 |
| chr6_117920563_117920770 | 0.15 |
GOPC |
golgi-associated PDZ and coiled-coil motif containing |
2861 |
0.34 |
| chr10_52984247_52984441 | 0.15 |
RP11-40C11.2 |
|
21222 |
0.19 |
| chr20_56827077_56827228 | 0.15 |
PPP4R1L |
protein phosphatase 4, regulatory subunit 1-like |
330 |
0.9 |
| chr7_55001398_55001578 | 0.15 |
ENSG00000252054 |
. |
67977 |
0.12 |
| chr1_29066265_29066416 | 0.15 |
YTHDF2 |
YTH domain family, member 2 |
2863 |
0.24 |
| chr6_1256084_1256235 | 0.15 |
FOXQ1 |
forkhead box Q1 |
56516 |
0.14 |
| chr12_80080022_80080173 | 0.15 |
PAWR |
PRKC, apoptosis, WT1, regulator |
3763 |
0.31 |
| chr1_120217864_120218312 | 0.15 |
ZNF697 |
zinc finger protein 697 |
27692 |
0.16 |
| chr3_58067974_58068125 | 0.14 |
FLNB |
filamin B, beta |
3836 |
0.31 |
| chr16_83847696_83847847 | 0.14 |
HSBP1 |
heat shock factor binding protein 1 |
6177 |
0.18 |
| chr5_55705557_55705708 | 0.14 |
ENSG00000265665 |
. |
47333 |
0.14 |
| chr12_64432896_64433047 | 0.14 |
SRGAP1 |
SLIT-ROBO Rho GTPase activating protein 1 |
56666 |
0.11 |
| chr13_43891097_43891540 | 0.14 |
ENOX1 |
ecto-NOX disulfide-thiol exchanger 1 |
43895 |
0.21 |
| chrX_84307873_84308024 | 0.14 |
APOOL |
apolipoprotein O-like |
49116 |
0.16 |
| chr14_69419003_69419387 | 0.14 |
ACTN1 |
actinin, alpha 1 |
4930 |
0.26 |
| chr2_142369486_142369637 | 0.14 |
ENSG00000265224 |
. |
26852 |
0.28 |
| chr20_30930232_30930398 | 0.14 |
ASXL1 |
additional sex combs like 1 (Drosophila) |
15840 |
0.17 |
| chr3_15724598_15724749 | 0.14 |
ENSG00000264354 |
. |
14205 |
0.18 |
| chr10_12095697_12095857 | 0.14 |
UPF2 |
UPF2 regulator of nonsense transcripts homolog (yeast) |
10754 |
0.17 |
| chr8_123605100_123605251 | 0.14 |
ENSG00000238901 |
. |
78355 |
0.11 |
| chr8_90624271_90624438 | 0.14 |
RIPK2 |
receptor-interacting serine-threonine kinase 2 |
145621 |
0.05 |
| chr7_122314795_122315038 | 0.14 |
RNF133 |
ring finger protein 133 |
24294 |
0.19 |
| chr11_8789155_8789306 | 0.14 |
ST5 |
suppression of tumorigenicity 5 |
971 |
0.37 |
| chr2_196478572_196478732 | 0.14 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
37951 |
0.19 |
| chr5_78712289_78712440 | 0.14 |
HOMER1 |
homer homolog 1 (Drosophila) |
96253 |
0.07 |
| chr5_67780816_67780967 | 0.13 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
192495 |
0.03 |
| chr1_178340504_178341006 | 0.13 |
RASAL2 |
RAS protein activator like 2 |
30149 |
0.22 |
| chr2_45805085_45805236 | 0.13 |
ENSG00000239396 |
. |
8529 |
0.22 |
| chr3_112356424_112356629 | 0.13 |
CCDC80 |
coiled-coil domain containing 80 |
418 |
0.88 |
| chr7_90229787_90229938 | 0.13 |
AC002456.2 |
|
3195 |
0.24 |
| chr8_49834597_49834856 | 0.13 |
SNAI2 |
snail family zinc finger 2 |
427 |
0.91 |
| chr16_4909761_4909912 | 0.13 |
UBN1 |
ubinuclein 1 |
7620 |
0.14 |
| chr16_57226974_57227125 | 0.13 |
RSPRY1 |
ring finger and SPRY domain containing 1 |
6804 |
0.14 |
| chr9_34560276_34560427 | 0.13 |
CNTFR-AS1 |
CNTFR antisense RNA 1 |
7662 |
0.11 |
| chr8_127989678_127989829 | 0.13 |
ENSG00000212451 |
. |
305986 |
0.01 |
| chr6_131290698_131290904 | 0.13 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
822 |
0.76 |
| chr11_106268675_106268826 | 0.13 |
RP11-680E19.1 |
|
133708 |
0.05 |
| chr6_20347614_20347765 | 0.13 |
E2F3 |
E2F transcription factor 3 |
54709 |
0.12 |
| chr10_31603702_31603954 | 0.13 |
ENSG00000237036 |
. |
4033 |
0.23 |
| chr2_72684771_72684922 | 0.13 |
ENSG00000222536 |
. |
264089 |
0.02 |
| chr1_15650263_15650414 | 0.13 |
RP3-467K16.7 |
|
11622 |
0.16 |
| chrX_19688369_19688923 | 0.13 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
166 |
0.97 |
| chr10_59997503_59997654 | 0.13 |
ENSG00000238970 |
. |
943 |
0.61 |
| chr7_119913514_119913995 | 0.13 |
KCND2 |
potassium voltage-gated channel, Shal-related subfamily, member 2 |
32 |
0.99 |
| chrX_130912468_130912619 | 0.13 |
ENSG00000200587 |
. |
160441 |
0.04 |
| chr5_158467035_158467186 | 0.13 |
CTD-2363C16.2 |
|
54296 |
0.13 |
| chr13_26312101_26312591 | 0.13 |
AL138815.1 |
Uncharacterized protein |
129715 |
0.05 |
| chr2_54331076_54331227 | 0.12 |
ACYP2 |
acylphosphatase 2, muscle type |
11389 |
0.25 |
| chrX_51638814_51638965 | 0.12 |
MAGED1 |
melanoma antigen family D, 1 |
2154 |
0.37 |
| chrX_33227444_33227711 | 0.12 |
DMD |
dystrophin |
1852 |
0.53 |
| chr11_27140156_27140307 | 0.12 |
BBOX1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
63467 |
0.13 |
| chr5_151124636_151124864 | 0.12 |
ATOX1 |
antioxidant 1 copper chaperone |
6929 |
0.15 |
| chr18_58201323_58201474 | 0.12 |
ENSG00000253011 |
. |
20978 |
0.26 |
| chr3_189348802_189349006 | 0.12 |
TP63 |
tumor protein p63 |
312 |
0.95 |
| chr9_91609546_91609862 | 0.12 |
S1PR3 |
sphingosine-1-phosphate receptor 3 |
1717 |
0.38 |
| chr3_112931196_112931871 | 0.12 |
BOC |
BOC cell adhesion associated, oncogene regulated |
124 |
0.97 |
| chr12_118793748_118794037 | 0.12 |
TAOK3 |
TAO kinase 3 |
3018 |
0.32 |
| chr22_41613159_41613310 | 0.12 |
RP4-756G23.5 |
|
393 |
0.76 |
| chr16_18796482_18796633 | 0.12 |
RPS15A |
ribosomal protein S15a |
410 |
0.8 |
| chr2_145868497_145868648 | 0.12 |
ENSG00000253036 |
. |
224066 |
0.02 |
| chr11_106031251_106031422 | 0.12 |
RP11-677I18.3 |
|
58634 |
0.12 |
| chr8_91996797_91997429 | 0.12 |
C8orf88 |
chromosome 8 open reading frame 88 |
372 |
0.85 |
| chr2_151439039_151439190 | 0.12 |
RND3 |
Rho family GTPase 3 |
43589 |
0.22 |
| chr7_50859293_50859444 | 0.12 |
GRB10 |
growth factor receptor-bound protein 10 |
1298 |
0.58 |
| chr16_10214963_10215114 | 0.12 |
GRIN2A |
glutamate receptor, ionotropic, N-methyl D-aspartate 2A |
59230 |
0.14 |
| chrX_34828763_34829004 | 0.12 |
FAM47B |
family with sequence similarity 47, member B |
132030 |
0.06 |
| chr8_30095301_30095452 | 0.12 |
RP11-51J9.4 |
|
56972 |
0.12 |
| chr1_119703325_119703476 | 0.12 |
RP11-418J17.1 |
|
14368 |
0.21 |
| chr6_139764774_139764925 | 0.11 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
69092 |
0.13 |
| chr16_88367140_88367361 | 0.11 |
ZNF469 |
zinc finger protein 469 |
126629 |
0.05 |
| chr10_64824310_64824738 | 0.11 |
ENSG00000199446 |
. |
45375 |
0.15 |
| chr4_81186827_81187126 | 0.11 |
FGF5 |
fibroblast growth factor 5 |
777 |
0.72 |
| chr5_53383790_53383941 | 0.11 |
ENSG00000265421 |
. |
12452 |
0.25 |
| chr5_162867024_162867396 | 0.11 |
CCNG1 |
cyclin G1 |
974 |
0.44 |
| chr4_81176376_81176527 | 0.11 |
FGF5 |
fibroblast growth factor 5 |
11302 |
0.24 |
| chr3_62174290_62174618 | 0.11 |
ENSG00000252184 |
. |
70270 |
0.12 |
| chr16_24999768_24999967 | 0.11 |
ARHGAP17 |
Rho GTPase activating protein 17 |
26785 |
0.22 |
| chr12_13351879_13352395 | 0.11 |
EMP1 |
epithelial membrane protein 1 |
2417 |
0.37 |
| chr8_82124094_82124245 | 0.11 |
FABP5 |
fatty acid binding protein 5 (psoriasis-associated) |
68429 |
0.11 |
| chr7_17347048_17347199 | 0.11 |
AC003075.4 |
|
8142 |
0.23 |
| chr3_159483781_159484285 | 0.11 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
1141 |
0.44 |
| chr12_91502482_91502678 | 0.11 |
LUM |
lumican |
3028 |
0.3 |
| chr3_70576201_70576352 | 0.11 |
RP11-231I13.2 |
|
214399 |
0.02 |
| chr11_125497097_125497248 | 0.11 |
CHEK1 |
checkpoint kinase 1 |
529 |
0.7 |
| chr14_45712160_45712311 | 0.11 |
MIS18BP1 |
MIS18 binding protein 1 |
10145 |
0.2 |
| chr7_134469036_134469352 | 0.11 |
CALD1 |
caldesmon 1 |
4765 |
0.32 |
| chr11_108767349_108767500 | 0.11 |
ENSG00000201243 |
. |
108820 |
0.07 |
| chr17_18362132_18362656 | 0.11 |
LGALS9C |
lectin, galactoside-binding, soluble, 9C |
17657 |
0.11 |
| chr4_124705698_124705849 | 0.11 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
384650 |
0.01 |
| chr4_160304105_160304256 | 0.11 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
32800 |
0.22 |
| chr8_144512945_144513817 | 0.11 |
MAFA |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog A |
805 |
0.48 |
| chr3_157639138_157639289 | 0.11 |
ENSG00000251751 |
. |
9301 |
0.31 |
| chrX_23686647_23686860 | 0.11 |
PRDX4 |
peroxiredoxin 4 |
1085 |
0.58 |
| chr4_53580350_53580621 | 0.11 |
ENSG00000212588 |
. |
1069 |
0.38 |
| chr7_23511944_23512095 | 0.11 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
1933 |
0.3 |
| chr2_232326502_232326776 | 0.11 |
ENSG00000202400 |
. |
1488 |
0.22 |
| chr10_57391119_57391436 | 0.11 |
PCDH15 |
protocadherin-related 15 |
3575 |
0.26 |
| chr7_134540602_134540753 | 0.11 |
CALD1 |
caldesmon 1 |
10915 |
0.29 |
| chr7_134473160_134473311 | 0.11 |
CALD1 |
caldesmon 1 |
8806 |
0.29 |
| chr15_49967106_49967257 | 0.10 |
RP11-353B9.1 |
|
22845 |
0.2 |
| chr5_33148047_33148451 | 0.10 |
CTD-2203K17.1 |
|
292476 |
0.01 |
| chr4_88000475_88000743 | 0.10 |
AFF1 |
AF4/FMR2 family, member 1 |
29805 |
0.23 |
| chr13_23412141_23412547 | 0.10 |
ENSG00000253094 |
. |
35060 |
0.19 |
| chr13_80658291_80658484 | 0.10 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
255407 |
0.02 |
| chr12_92488429_92488723 | 0.10 |
C12orf79 |
chromosome 12 open reading frame 79 |
42221 |
0.15 |
| chr4_74962901_74963052 | 0.10 |
CXCL2 |
chemokine (C-X-C motif) ligand 2 |
2034 |
0.28 |
| chr1_157013915_157014350 | 0.10 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
733 |
0.68 |
| chr1_115879628_115879779 | 0.10 |
NGF |
nerve growth factor (beta polypeptide) |
1154 |
0.6 |
| chr7_13960202_13960381 | 0.10 |
ETV1 |
ets variant 1 |
65775 |
0.14 |
| chr6_127498434_127498585 | 0.10 |
RSPO3 |
R-spondin 3 |
58326 |
0.14 |
| chr3_152314614_152314796 | 0.10 |
RP11-362A9.3 |
|
100103 |
0.08 |
| chr2_178453442_178453593 | 0.10 |
AC073834.3 |
|
14300 |
0.18 |
| chr17_35854577_35854849 | 0.10 |
DUSP14 |
dual specificity phosphatase 14 |
3143 |
0.25 |
| chr1_3816010_3816785 | 0.10 |
C1orf174 |
chromosome 1 open reading frame 174 |
452 |
0.8 |
| chr5_75903118_75903290 | 0.10 |
CTD-2236F14.1 |
|
1591 |
0.31 |
| chr5_151056524_151056678 | 0.10 |
CTB-113P19.1 |
|
67 |
0.96 |
| chr6_163951025_163951421 | 0.10 |
QKI |
QKI, KH domain containing, RNA binding |
4889 |
0.36 |
| chr15_71054590_71054766 | 0.10 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
1091 |
0.55 |
| chr6_100381252_100381443 | 0.10 |
MCHR2-AS1 |
MCHR2 antisense RNA 1 |
60473 |
0.14 |
| chr8_48436041_48436263 | 0.10 |
RP11-697N18.4 |
|
3807 |
0.33 |
| chr18_22929163_22929595 | 0.10 |
ZNF521 |
zinc finger protein 521 |
1778 |
0.48 |
| chr3_42066063_42066214 | 0.10 |
ULK4 |
unc-51 like kinase 4 |
62216 |
0.12 |
| chr1_212841074_212841261 | 0.10 |
ENSG00000207491 |
. |
24439 |
0.13 |
| chr5_71127239_71127421 | 0.10 |
CARTPT |
CART prepropeptide |
112340 |
0.07 |
| chr20_50178684_50179923 | 0.10 |
NFATC2 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 |
67 |
0.99 |
| chr13_45330241_45330498 | 0.10 |
ENSG00000238932 |
. |
127560 |
0.05 |
| chr5_5419809_5420018 | 0.10 |
KIAA0947 |
KIAA0947 |
2894 |
0.42 |
| chr18_9619807_9620005 | 0.10 |
ENSG00000212572 |
. |
1836 |
0.31 |
| chr4_183859496_183859668 | 0.10 |
DCTD |
dCMP deaminase |
20493 |
0.24 |
| chr20_9049976_9050220 | 0.10 |
PLCB4 |
phospholipase C, beta 4 |
350 |
0.89 |
| chr18_5500688_5500839 | 0.10 |
EPB41L3 |
erythrocyte membrane protein band 4.1-like 3 |
11394 |
0.2 |
| chr12_115889903_115890336 | 0.10 |
RP11-116D17.1 |
HCG2038717; Uncharacterized protein |
89302 |
0.1 |
| chr7_4885363_4885579 | 0.10 |
RADIL |
Ras association and DIL domains |
7783 |
0.17 |
| chr9_8821970_8822217 | 0.10 |
ENSG00000239102 |
. |
24958 |
0.2 |
| chr1_231964431_231964582 | 0.10 |
DISC1-IT1 |
DISC1 intronic transcript 1 (non-protein coding) |
97074 |
0.08 |
| chr2_87551749_87551924 | 0.10 |
IGKV3OR2-268 |
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
13798 |
0.29 |
| chr6_143447345_143447496 | 0.10 |
AIG1 |
androgen-induced 1 |
33 |
0.98 |
| chr10_125608306_125608596 | 0.10 |
CPXM2 |
carboxypeptidase X (M14 family), member 2 |
42879 |
0.15 |
| chr5_141432956_141433107 | 0.10 |
GNPDA1 |
glucosamine-6-phosphate deaminase 1 |
40425 |
0.13 |
| chr16_70554540_70554691 | 0.10 |
COG4 |
component of oligomeric golgi complex 4 |
2488 |
0.18 |
| chr8_94934276_94934427 | 0.10 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
2244 |
0.3 |
| chr17_70147844_70148239 | 0.10 |
SOX9 |
SRY (sex determining region Y)-box 9 |
30880 |
0.25 |
| chr3_57186762_57186915 | 0.10 |
IL17RD |
interleukin 17 receptor D |
10304 |
0.19 |
| chr17_2827552_2827726 | 0.10 |
CTD-3060P21.1 |
|
41550 |
0.15 |
| chr10_89629938_89630093 | 0.10 |
KLLN |
killin, p53-regulated DNA replication inhibitor |
6821 |
0.14 |
| chr2_157421974_157422125 | 0.10 |
AC011308.1 |
|
69394 |
0.12 |
| chr9_100465936_100466445 | 0.10 |
ENSG00000266608 |
. |
3674 |
0.21 |
| chr14_20812432_20812706 | 0.10 |
RPPH1 |
ribonuclease P RNA component H1 |
725 |
0.32 |
| chr12_51008245_51008396 | 0.10 |
ENSG00000207136 |
. |
18736 |
0.2 |
| chrX_125186493_125186644 | 0.10 |
DCAF12L2 |
DDB1 and CUL4 associated factor 12-like 2 |
113366 |
0.07 |
| chr12_32261770_32262117 | 0.10 |
RP11-843B15.2 |
|
1481 |
0.38 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0048859 | formation of anatomical boundary(GO:0048859) |
| 0.1 | 0.2 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.2 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.5 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.1 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.0 | GO:2000143 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.1 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0033632 | regulation of cell-cell adhesion mediated by integrin(GO:0033632) |
| 0.0 | 0.0 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.0 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.0 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0003078 | obsolete regulation of natriuresis(GO:0003078) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:2000095 | regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0010560 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.0 | GO:0045198 | establishment of epithelial cell apical/basal polarity(GO:0045198) |
| 0.0 | 0.0 | GO:0008215 | spermine metabolic process(GO:0008215) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0090399 | replicative senescence(GO:0090399) |
| 0.0 | 0.0 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.2 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0008300 | isoprenoid catabolic process(GO:0008300) |
| 0.0 | 0.0 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.3 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.1 | 0.3 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.2 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.2 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.3 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.1 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0003696 | satellite DNA binding(GO:0003696) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.0 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.0 | GO:0043175 | RNA polymerase core enzyme binding(GO:0043175) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.0 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |