Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
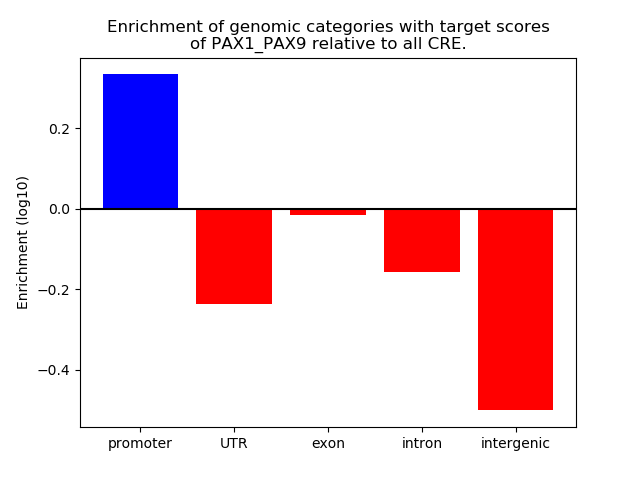
Results for PAX1_PAX9
Z-value: 0.79
Transcription factors associated with PAX1_PAX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
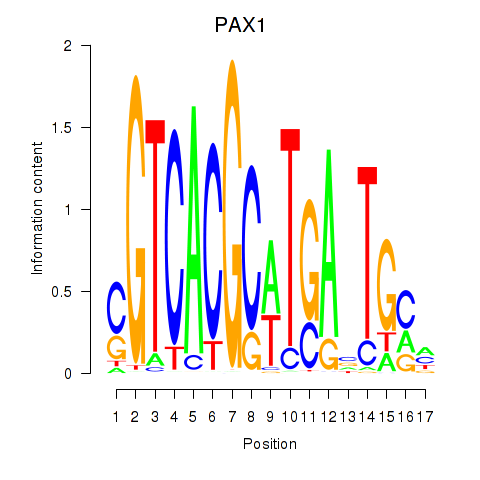
PAX1
|
ENSG00000125813.9 | paired box 1 |
|
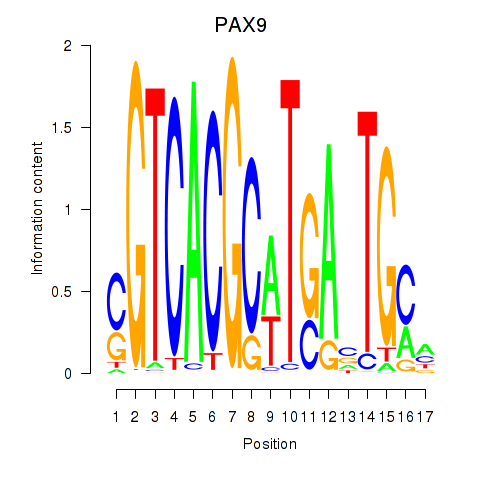
PAX9
|
ENSG00000198807.8 | paired box 9 |
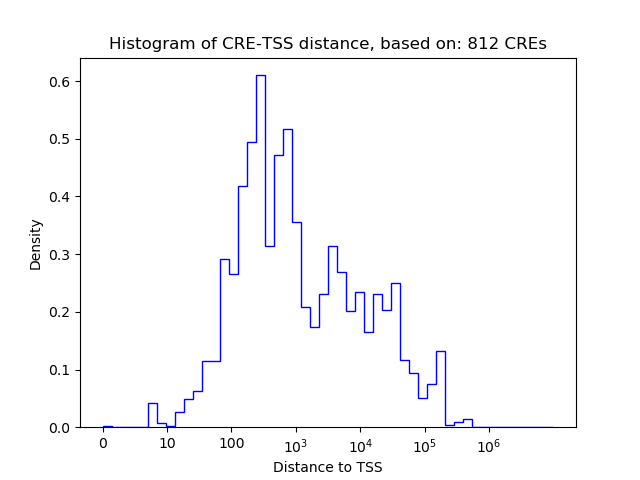
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr20_21961725_21961876 | PAX1 | 274939 | 0.015233 | 0.81 | 8.4e-03 | Click! |
| chr20_21698251_21698402 | PAX1 | 11465 | 0.305732 | -0.78 | 1.3e-02 | Click! |
| chr20_22052219_22052370 | PAX1 | 365433 | 0.008806 | -0.66 | 5.4e-02 | Click! |
| chr20_21662264_21662415 | PAX1 | 23958 | 0.261941 | -0.66 | 5.5e-02 | Click! |
| chr20_21683536_21683700 | PAX1 | 2679 | 0.416922 | -0.56 | 1.2e-01 | Click! |
| chr14_37116502_37116669 | PAX9 | 10188 | 0.177446 | -0.58 | 1.0e-01 | Click! |
| chr14_37131505_37132074 | PAX9 | 10 | 0.976824 | -0.56 | 1.2e-01 | Click! |
| chr14_37144898_37145049 | PAX9 | 13194 | 0.186371 | -0.53 | 1.4e-01 | Click! |
| chr14_37117391_37117614 | PAX9 | 9271 | 0.179851 | -0.46 | 2.1e-01 | Click! |
| chr14_37125375_37125526 | PAX9 | 1323 | 0.404544 | -0.42 | 2.6e-01 | Click! |
Activity of the PAX1_PAX9 motif across conditions
Conditions sorted by the z-value of the PAX1_PAX9 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
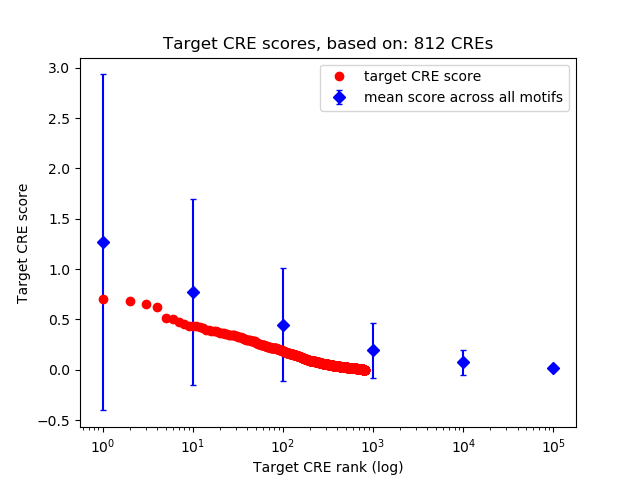
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_198608305_198608501 | 0.70 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
111 |
0.98 |
| chrX_128914480_128914899 | 0.68 |
SASH3 |
SAM and SH3 domain containing 3 |
729 |
0.68 |
| chr17_62097753_62097982 | 0.65 |
ICAM2 |
intercellular adhesion molecule 2 |
44 |
0.97 |
| chr9_137029602_137029922 | 0.63 |
ENSG00000221676 |
. |
76 |
0.97 |
| chr4_36257318_36257571 | 0.52 |
RP11-431M7.3 |
|
696 |
0.72 |
| chr12_102321689_102321840 | 0.50 |
RP11-512N21.3 |
|
3267 |
0.22 |
| chr15_86127194_86127729 | 0.48 |
RP11-815J21.2 |
|
4052 |
0.21 |
| chr2_99953063_99953235 | 0.46 |
TXNDC9 |
thioredoxin domain containing 9 |
224 |
0.81 |
| chr2_207629814_207630047 | 0.44 |
MDH1B |
malate dehydrogenase 1B, NAD (soluble) |
80 |
0.6 |
| chr2_198160747_198160898 | 0.44 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
6421 |
0.18 |
| chr17_79211950_79212101 | 0.43 |
ENTHD2 |
ENTH domain containing 2 |
627 |
0.46 |
| chr20_47437784_47438180 | 0.43 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
6438 |
0.29 |
| chr6_2999439_2999644 | 0.41 |
RP1-90J20.8 |
|
173 |
0.82 |
| chr11_59538510_59538661 | 0.39 |
STX3 |
syntaxin 3 |
15660 |
0.13 |
| chr20_52225525_52225734 | 0.39 |
ZNF217 |
zinc finger protein 217 |
198 |
0.95 |
| chr19_40791502_40791751 | 0.39 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
183 |
0.89 |
| chr16_2689724_2689875 | 0.39 |
PDPK2 |
|
2634 |
0.14 |
| chr21_46529839_46530147 | 0.38 |
PRED58 |
|
4880 |
0.17 |
| chr18_74843066_74843217 | 0.38 |
MBP |
myelin basic protein |
499 |
0.88 |
| chr19_1856293_1856444 | 0.37 |
CTB-31O20.8 |
|
3881 |
0.1 |
| chr10_82223089_82223265 | 0.37 |
TSPAN14 |
tetraspanin 14 |
4119 |
0.25 |
| chr6_41702064_41702224 | 0.36 |
TFEB |
transcription factor EB |
5 |
0.96 |
| chr3_25824543_25824893 | 0.36 |
OXSM |
3-oxoacyl-ACP synthase, mitochondrial |
154 |
0.49 |
| chr10_30723111_30723504 | 0.35 |
MAP3K8 |
mitogen-activated protein kinase kinase kinase 8 |
134 |
0.97 |
| chr5_133747742_133748022 | 0.35 |
CDKN2AIPNL |
CDKN2A interacting protein N-terminal like |
293 |
0.89 |
| chr5_140997567_140997853 | 0.35 |
AC008781.7 |
|
271 |
0.75 |
| chr2_64957643_64958616 | 0.35 |
ENSG00000253082 |
. |
45109 |
0.14 |
| chr17_40400915_40401141 | 0.34 |
RP11-358B23.5 |
|
19577 |
0.1 |
| chr8_101965285_101965580 | 0.34 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
137 |
0.95 |
| chr1_1474981_1475180 | 0.34 |
TMEM240 |
transmembrane protein 240 |
657 |
0.54 |
| chr7_150066210_150066658 | 0.34 |
REPIN1 |
replication initiator 1 |
474 |
0.45 |
| chr3_9885443_9885670 | 0.33 |
RPUSD3 |
RNA pseudouridylate synthase domain containing 3 |
70 |
0.94 |
| chr3_15247783_15248155 | 0.33 |
CAPN7 |
calpain 7 |
219 |
0.93 |
| chr5_36875665_36875984 | 0.32 |
NIPBL |
Nipped-B homolog (Drosophila) |
1037 |
0.69 |
| chr9_129183867_129184018 | 0.31 |
ENSG00000252985 |
. |
5230 |
0.22 |
| chr12_58335952_58336152 | 0.31 |
XRCC6BP1 |
XRCC6 binding protein 1 |
692 |
0.67 |
| chr6_34856993_34857273 | 0.31 |
ANKS1A |
ankyrin repeat and sterile alpha motif domain containing 1A |
53 |
0.96 |
| chr7_147792201_147792352 | 0.31 |
CNTNAP2 |
contactin associated protein-like 2 |
38500 |
0.22 |
| chr20_4794261_4794412 | 0.30 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
1433 |
0.44 |
| chr16_30006409_30006560 | 0.30 |
INO80E |
INO80 complex subunit E |
131 |
0.79 |
| chr18_13612881_13613032 | 0.29 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
338 |
0.8 |
| chr10_93559450_93559702 | 0.29 |
TNKS2 |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase 2 |
1507 |
0.34 |
| chr19_6801178_6801329 | 0.29 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
17124 |
0.13 |
| chr19_52132706_52132857 | 0.29 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
807 |
0.48 |
| chr14_103852201_103853000 | 0.29 |
MARK3 |
MAP/microtubule affinity-regulating kinase 3 |
24 |
0.98 |
| chr5_100236594_100237334 | 0.29 |
ST8SIA4 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
1954 |
0.48 |
| chr2_69921062_69921213 | 0.29 |
ENSG00000238708 |
. |
26646 |
0.17 |
| chr5_177591147_177591316 | 0.29 |
NHP2 |
NHP2 ribonucleoprotein |
10263 |
0.17 |
| chr16_88770281_88770566 | 0.28 |
RNF166 |
ring finger protein 166 |
398 |
0.64 |
| chr6_30585634_30586241 | 0.27 |
MRPS18B |
mitochondrial ribosomal protein S18B |
313 |
0.67 |
| chr5_154137578_154138162 | 0.27 |
LARP1 |
La ribonucleoprotein domain family, member 1 |
1136 |
0.49 |
| chr5_126365438_126366192 | 0.26 |
MARCH3 |
membrane-associated ring finger (C3HC4) 3, E3 ubiquitin protein ligase |
685 |
0.77 |
| chr9_4740769_4741101 | 0.26 |
AK3 |
adenylate kinase 3 |
292 |
0.91 |
| chr7_50727890_50728167 | 0.25 |
GRB10 |
growth factor receptor-bound protein 10 |
33401 |
0.21 |
| chr1_6684874_6685147 | 0.25 |
THAP3 |
THAP domain containing, apoptosis associated protein 3 |
84 |
0.95 |
| chr18_22744503_22745506 | 0.25 |
ZNF521 |
zinc finger protein 521 |
59749 |
0.16 |
| chr12_132413289_132413563 | 0.25 |
PUS1 |
pseudouridylate synthase 1 |
319 |
0.87 |
| chr4_77996688_77997050 | 0.25 |
CCNI |
cyclin I |
273 |
0.93 |
| chr7_3083802_3083953 | 0.25 |
CARD11 |
caspase recruitment domain family, member 11 |
298 |
0.93 |
| chr9_90113946_90114123 | 0.24 |
DAPK1 |
death-associated protein kinase 1 |
114 |
0.98 |
| chr17_39968419_39968821 | 0.24 |
LEPREL4 |
leprecan-like 4 |
169 |
0.65 |
| chr2_74766675_74766942 | 0.24 |
DOK1 |
docking protein 1, 62kDa (downstream of tyrosine kinase 1) |
9345 |
0.07 |
| chr13_67106963_67107114 | 0.24 |
ENSG00000238500 |
. |
97870 |
0.09 |
| chr6_37140750_37140901 | 0.24 |
PIM1 |
pim-1 oncogene |
2846 |
0.25 |
| chr22_24408267_24408637 | 0.24 |
CABIN1 |
calcineurin binding protein 1 |
584 |
0.65 |
| chr8_26435531_26436419 | 0.23 |
DPYSL2 |
dihydropyrimidinase-like 2 |
54 |
0.98 |
| chr21_36901341_36902041 | 0.23 |
ENSG00000211590 |
. |
191322 |
0.03 |
| chr22_39745597_39745764 | 0.23 |
SYNGR1 |
synaptogyrin 1 |
250 |
0.88 |
| chr5_72251895_72252188 | 0.23 |
FCHO2 |
FCH domain only 2 |
143 |
0.97 |
| chr18_33077197_33077498 | 0.23 |
INO80C |
INO80 complex subunit C |
216 |
0.79 |
| chr11_67251391_67251565 | 0.23 |
AIP |
aryl hydrocarbon receptor interacting protein |
966 |
0.31 |
| chrX_79830027_79830462 | 0.23 |
FAM46D |
family with sequence similarity 46, member D |
154279 |
0.04 |
| chr13_110055389_110055540 | 0.22 |
MYO16-AS1 |
MYO16 antisense RNA 1 |
201633 |
0.03 |
| chr14_23388119_23388349 | 0.22 |
RBM23 |
RNA binding motif protein 23 |
104 |
0.8 |
| chr19_47989811_47989962 | 0.22 |
NAPA-AS1 |
NAPA antisense RNA 1 |
2321 |
0.19 |
| chr17_14206700_14206851 | 0.22 |
HS3ST3B1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 3B1 |
2375 |
0.39 |
| chr2_54823751_54824124 | 0.22 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
38406 |
0.14 |
| chr6_108581372_108582140 | 0.22 |
SNX3 |
sorting nexin 3 |
412 |
0.8 |
| chr13_111364022_111364765 | 0.22 |
ING1 |
inhibitor of growth family, member 1 |
690 |
0.72 |
| chr10_27389546_27389885 | 0.21 |
ANKRD26 |
ankyrin repeat domain 26 |
294 |
0.9 |
| chr1_207496793_207497173 | 0.21 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
1847 |
0.46 |
| chr19_52149205_52149440 | 0.21 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
524 |
0.49 |
| chr6_26341940_26342091 | 0.21 |
ENSG00000252399 |
. |
11249 |
0.09 |
| chr6_6588687_6588838 | 0.21 |
LY86 |
lymphocyte antigen 86 |
140 |
0.95 |
| chr16_3074363_3074588 | 0.21 |
HCFC1R1 |
host cell factor C1 regulator 1 (XPO1 dependent) |
188 |
0.65 |
| chr1_202776022_202776519 | 0.21 |
KDM5B |
lysine (K)-specific demethylase 5B |
157 |
0.93 |
| chr7_93753483_93753634 | 0.21 |
BET1 |
Bet1 golgi vesicular membrane trafficking protein |
119864 |
0.06 |
| chr3_18486507_18486658 | 0.21 |
ENSG00000228956 |
. |
66 |
0.7 |
| chr14_62035309_62035460 | 0.21 |
RP11-47I22.3 |
Uncharacterized protein |
1930 |
0.36 |
| chr9_127025877_127026028 | 0.20 |
RP11-121A14.3 |
|
797 |
0.55 |
| chr11_128422978_128423129 | 0.20 |
RP11-1007G5.2 |
|
27016 |
0.2 |
| chr10_76345165_76345892 | 0.20 |
ENSG00000206756 |
. |
34378 |
0.2 |
| chr8_121824505_121824719 | 0.20 |
RP11-713M15.2 |
|
153 |
0.61 |
| chr17_33814121_33814953 | 0.19 |
SLFN12L |
schlafen family member 12-like |
316 |
0.84 |
| chr9_468840_468991 | 0.19 |
RP11-165F24.3 |
|
887 |
0.45 |
| chr2_44222356_44222725 | 0.19 |
LRPPRC |
leucine-rich pentatricopeptide repeat containing |
575 |
0.82 |
| chr2_68383846_68384031 | 0.19 |
WDR92 |
WD repeat domain 92 |
667 |
0.53 |
| chr14_22447214_22447365 | 0.19 |
ENSG00000238634 |
. |
163598 |
0.03 |
| chr14_53618338_53618850 | 0.19 |
DDHD1 |
DDHD domain containing 1 |
1222 |
0.3 |
| chr10_134364208_134364359 | 0.19 |
INPP5A |
inositol polyphosphate-5-phosphatase, 40kDa |
12640 |
0.21 |
| chr12_120700998_120701172 | 0.18 |
PXN |
paxillin |
2478 |
0.18 |
| chr20_62257208_62257699 | 0.18 |
GMEB2 |
glucocorticoid modulatory element binding protein 2 |
941 |
0.32 |
| chr2_240187102_240187645 | 0.18 |
ENSG00000265215 |
. |
39784 |
0.13 |
| chr9_93573094_93573245 | 0.18 |
SYK |
spleen tyrosine kinase |
8960 |
0.33 |
| chr2_98334930_98335281 | 0.17 |
ZAP70 |
zeta-chain (TCR) associated protein kinase 70kDa |
5082 |
0.19 |
| chr16_58034790_58035185 | 0.17 |
USB1 |
U6 snRNA biogenesis 1 |
290 |
0.71 |
| chr9_115571889_115572040 | 0.17 |
SNX30 |
sorting nexin family member 30 |
28901 |
0.18 |
| chr15_73587715_73587866 | 0.17 |
RP11-272D12.2 |
|
39340 |
0.16 |
| chr17_75452866_75453125 | 0.17 |
SEPT9 |
septin 9 |
547 |
0.72 |
| chr2_26726429_26726904 | 0.17 |
OTOF |
otoferlin |
25756 |
0.17 |
| chr19_37177817_37178305 | 0.17 |
ZNF567 |
zinc finger protein 567 |
469 |
0.78 |
| chrX_154032790_154032975 | 0.17 |
MPP1 |
membrane protein, palmitoylated 1, 55kDa |
790 |
0.52 |
| chr7_7679826_7680046 | 0.17 |
RPA3 |
replication protein A3, 14kDa |
279 |
0.94 |
| chr5_179246588_179246964 | 0.17 |
SQSTM1 |
sequestosome 1 |
898 |
0.41 |
| chr11_747907_748726 | 0.17 |
TALDO1 |
transaldolase 1 |
888 |
0.33 |
| chr3_29185407_29185558 | 0.17 |
ENSG00000238470 |
. |
132749 |
0.05 |
| chr1_192777377_192777993 | 0.16 |
RGS2 |
regulator of G-protein signaling 2, 24kDa |
486 |
0.87 |
| chr2_86669408_86669754 | 0.16 |
KDM3A |
lysine (K)-specific demethylase 3A |
410 |
0.86 |
| chr7_18461225_18461376 | 0.16 |
AC010082.2 |
|
9061 |
0.28 |
| chr5_142600508_142600686 | 0.16 |
ARHGAP26 |
Rho GTPase activating protein 26 |
13832 |
0.24 |
| chrX_73512588_73513347 | 0.16 |
ENSG00000199168 |
. |
5775 |
0.15 |
| chr15_76630019_76630254 | 0.16 |
ISL2 |
ISL LIM homeobox 2 |
1071 |
0.52 |
| chr4_100742826_100743086 | 0.16 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
4953 |
0.27 |
| chr15_41316618_41316840 | 0.16 |
RP11-540O11.4 |
|
32 |
0.97 |
| chr19_1088228_1088379 | 0.16 |
POLR2E |
polymerase (RNA) II (DNA directed) polypeptide E, 25kDa |
7050 |
0.1 |
| chr5_149792089_149792429 | 0.16 |
CD74 |
CD74 molecule, major histocompatibility complex, class II invariant chain |
36 |
0.97 |
| chr7_5230948_5231124 | 0.16 |
WIPI2 |
WD repeat domain, phosphoinositide interacting 2 |
1132 |
0.48 |
| chr20_61549533_61549684 | 0.16 |
DIDO1 |
death inducer-obliterator 1 |
3850 |
0.18 |
| chr10_105727511_105727782 | 0.16 |
SLK |
STE20-like kinase |
687 |
0.66 |
| chrX_55186864_55187302 | 0.15 |
FAM104B |
family with sequence similarity 104, member B |
452 |
0.82 |
| chr8_21817389_21817540 | 0.15 |
XPO7 |
exportin 7 |
6285 |
0.19 |
| chr1_26758963_26759170 | 0.15 |
DHDDS |
dehydrodolichyl diphosphate synthase |
172 |
0.93 |
| chr12_54753390_54753672 | 0.15 |
GPR84 |
G protein-coupled receptor 84 |
4727 |
0.09 |
| chr2_109237700_109238010 | 0.15 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
133 |
0.97 |
| chr16_53983720_53983924 | 0.15 |
FTO |
fat mass and obesity associated |
15880 |
0.2 |
| chr6_33393517_33393718 | 0.15 |
SYNGAP1 |
synaptic Ras GTPase activating protein 1 |
5585 |
0.11 |
| chr2_86850787_86850979 | 0.15 |
RNF103 |
ring finger protein 103 |
95 |
0.91 |
| chr12_107350126_107350477 | 0.15 |
C12orf23 |
chromosome 12 open reading frame 23 |
262 |
0.85 |
| chr13_99911468_99911619 | 0.14 |
GPR18 |
G protein-coupled receptor 18 |
861 |
0.62 |
| chr6_15268791_15268942 | 0.14 |
JARID2 |
jumonji, AT rich interactive domain 2 |
19723 |
0.17 |
| chr5_79647576_79647767 | 0.14 |
ENSG00000206774 |
. |
13980 |
0.17 |
| chr17_77817111_77817406 | 0.14 |
CBX4 |
chromobox homolog 4 |
4030 |
0.19 |
| chr11_27695896_27696047 | 0.14 |
BDNF-AS |
BDNF antisense RNA |
9164 |
0.23 |
| chr9_115096689_115097200 | 0.14 |
PTBP3 |
polypyrimidine tract binding protein 3 |
997 |
0.57 |
| chr17_77071312_77071553 | 0.14 |
ENGASE |
endo-beta-N-acetylglucosaminidase |
281 |
0.89 |
| chr4_3533160_3533351 | 0.14 |
LRPAP1 |
low density lipoprotein receptor-related protein associated protein 1 |
1031 |
0.5 |
| chr2_232062733_232062927 | 0.14 |
ARMC9 |
armadillo repeat containing 9 |
430 |
0.86 |
| chr15_40475830_40476133 | 0.14 |
BUB1B |
BUB1 mitotic checkpoint serine/threonine kinase B |
22713 |
0.12 |
| chr8_142275255_142276084 | 0.14 |
RP11-10J21.3 |
Uncharacterized protein |
11005 |
0.14 |
| chr14_64319967_64320824 | 0.13 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
663 |
0.73 |
| chr20_4707737_4707988 | 0.13 |
PRND |
prion protein 2 (dublet) |
5306 |
0.21 |
| chr5_148725091_148725557 | 0.13 |
GRPEL2 |
GrpE-like 2, mitochondrial (E. coli) |
259 |
0.87 |
| chr17_81040457_81040608 | 0.13 |
METRNL |
meteorin, glial cell differentiation regulator-like |
1417 |
0.48 |
| chr17_56410108_56410593 | 0.13 |
MIR142 |
microRNA 142 |
481 |
0.66 |
| chr2_26569037_26569246 | 0.13 |
EPT1 |
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
124 |
0.84 |
| chr21_46973241_46973392 | 0.13 |
SLC19A1 |
solute carrier family 19 (folate transporter), member 1 |
8991 |
0.23 |
| chr1_22109491_22109642 | 0.13 |
USP48 |
ubiquitin specific peptidase 48 |
82 |
0.97 |
| chr5_177632434_177632741 | 0.13 |
HNRNPAB |
heterogeneous nuclear ribonucleoprotein A/B |
1015 |
0.54 |
| chr6_10412096_10412574 | 0.13 |
TFAP2A |
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha) |
272 |
0.9 |
| chr13_51795997_51796228 | 0.13 |
FAM124A |
family with sequence similarity 124A |
391 |
0.9 |
| chr8_121822100_121822251 | 0.12 |
SNTB1 |
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
2208 |
0.3 |
| chr1_235323819_235324540 | 0.12 |
RBM34 |
RNA binding motif protein 34 |
356 |
0.84 |
| chr12_78002829_78002980 | 0.12 |
AC073528.1 |
|
52336 |
0.19 |
| chr17_27229464_27229953 | 0.12 |
DHRS13 |
dehydrogenase/reductase (SDR family) member 13 |
275 |
0.78 |
| chr19_56826578_56826877 | 0.12 |
AC006116.17 |
|
104 |
0.79 |
| chr3_150995573_150995821 | 0.12 |
P2RY14 |
purinergic receptor P2Y, G-protein coupled, 14 |
460 |
0.8 |
| chr13_21140562_21140933 | 0.12 |
IFT88 |
intraflagellar transport 88 homolog (Chlamydomonas) |
162 |
0.96 |
| chr11_67186305_67186456 | 0.11 |
PPP1CA |
protein phosphatase 1, catalytic subunit, alpha isozyme |
2272 |
0.11 |
| chr19_8478353_8478504 | 0.11 |
MARCH2 |
membrane-associated ring finger (C3HC4) 2, E3 ubiquitin protein ligase |
122 |
0.93 |
| chr16_90172947_90173188 | 0.11 |
PRDM7 |
PR domain containing 7 |
30729 |
0.13 |
| chr19_41222549_41222955 | 0.11 |
ADCK4 |
aarF domain containing kinase 4 |
38 |
0.76 |
| chr15_90437690_90437846 | 0.11 |
AP3S2 |
adaptor-related protein complex 3, sigma 2 subunit |
194 |
0.88 |
| chr11_126138529_126138703 | 0.11 |
SRPR |
signal recognition particle receptor (docking protein) |
238 |
0.63 |
| chr6_144461776_144461927 | 0.11 |
STX11 |
syntaxin 11 |
9812 |
0.24 |
| chr1_161903850_161904001 | 0.11 |
OLFML2B |
olfactomedin-like 2B |
51097 |
0.14 |
| chr6_113650155_113650306 | 0.11 |
ENSG00000222677 |
. |
185959 |
0.03 |
| chr15_49102623_49102987 | 0.11 |
CEP152 |
centrosomal protein 152kDa |
14 |
0.57 |
| chr9_93644008_93644159 | 0.11 |
SYK |
spleen tyrosine kinase |
54313 |
0.18 |
| chr19_344917_345170 | 0.11 |
MIER2 |
mesoderm induction early response 1, family member 2 |
250 |
0.9 |
| chr8_28972672_28972823 | 0.11 |
CTD-2647L4.1 |
|
4657 |
0.17 |
| chr7_151038891_151039078 | 0.10 |
NUB1 |
negative regulator of ubiquitin-like proteins 1 |
86 |
0.96 |
| chr16_2318513_2318889 | 0.10 |
RNPS1 |
RNA binding protein S1, serine-rich domain |
288 |
0.64 |
| chr6_21595977_21596233 | 0.10 |
SOX4 |
SRY (sex determining region Y)-box 4 |
2034 |
0.5 |
| chr12_90693182_90693369 | 0.10 |
ENSG00000252823 |
. |
545439 |
0.0 |
| chr14_23387653_23388059 | 0.10 |
RBM23 |
RNA binding motif protein 23 |
482 |
0.48 |
| chr11_47421524_47421704 | 0.10 |
ENSG00000264583 |
. |
907 |
0.36 |
| chr15_20980933_20981246 | 0.10 |
AC012414.1 |
Uncharacterized protein |
23598 |
0.15 |
| chr3_71111322_71111570 | 0.10 |
FOXP1 |
forkhead box P1 |
2631 |
0.43 |
| chr21_46239564_46239715 | 0.10 |
SUMO3 |
small ubiquitin-like modifier 3 |
1595 |
0.28 |
| chr16_68279313_68279519 | 0.10 |
PLA2G15 |
phospholipase A2, group XV |
121 |
0.9 |
| chrX_109247622_109247773 | 0.10 |
TMEM164 |
transmembrane protein 164 |
1354 |
0.53 |
| chr15_34627235_34627720 | 0.09 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
1568 |
0.25 |
| chr6_35268137_35268288 | 0.09 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
2583 |
0.26 |
| chr13_98086771_98087714 | 0.09 |
RAP2A |
RAP2A, member of RAS oncogene family |
766 |
0.75 |
| chr14_99697762_99697913 | 0.09 |
AL109767.1 |
|
31448 |
0.18 |
| chr2_46925496_46926304 | 0.09 |
SOCS5 |
suppressor of cytokine signaling 5 |
191 |
0.95 |
| chr14_60716792_60717657 | 0.09 |
PPM1A |
protein phosphatase, Mg2+/Mn2+ dependent, 1A |
75 |
0.97 |
| chr8_56806111_56806262 | 0.09 |
RP11-318K15.2 |
|
32 |
0.97 |
| chr10_99258721_99258937 | 0.09 |
UBTD1 |
ubiquitin domain containing 1 |
204 |
0.61 |
| chr14_105885619_105886050 | 0.09 |
RP11-521B24.3 |
|
242 |
0.57 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.3 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.1 | 0.5 | GO:0002726 | positive regulation of T cell cytokine production(GO:0002726) |
| 0.1 | 0.3 | GO:2000192 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.1 | 0.2 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.1 | 0.2 | GO:0019886 | antigen processing and presentation of exogenous peptide antigen via MHC class II(GO:0019886) |
| 0.1 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.1 | 0.2 | GO:0034088 | maintenance of sister chromatid cohesion(GO:0034086) maintenance of mitotic sister chromatid cohesion(GO:0034088) |
| 0.1 | 0.1 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.1 | 0.2 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.2 | GO:0051790 | short-chain fatty acid biosynthetic process(GO:0051790) |
| 0.0 | 0.1 | GO:0045425 | positive regulation of granulocyte macrophage colony-stimulating factor biosynthetic process(GO:0045425) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0045625 | regulation of T-helper 1 cell differentiation(GO:0045625) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.6 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.1 | GO:0032212 | positive regulation of telomere maintenance via telomerase(GO:0032212) positive regulation of telomere maintenance via telomere lengthening(GO:1904358) |
| 0.0 | 0.2 | GO:0014049 | positive regulation of glutamate secretion(GO:0014049) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0045003 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.2 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.1 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.1 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.0 | 0.3 | GO:0045577 | regulation of B cell differentiation(GO:0045577) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.2 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0034643 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.1 | GO:0005997 | xylulose metabolic process(GO:0005997) |
| 0.0 | 0.1 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0072393 | microtubule anchoring at microtubule organizing center(GO:0072393) |
| 0.0 | 0.2 | GO:0006303 | double-strand break repair via nonhomologous end joining(GO:0006303) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0019276 | UDP-N-acetylgalactosamine metabolic process(GO:0019276) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0045047 | protein targeting to ER(GO:0045047) establishment of protein localization to endoplasmic reticulum(GO:0072599) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0046184 | aldehyde biosynthetic process(GO:0046184) |
| 0.0 | 0.1 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0090022 | regulation of neutrophil chemotaxis(GO:0090022) regulation of neutrophil migration(GO:1902622) |
| 0.0 | 0.1 | GO:0042119 | neutrophil activation(GO:0042119) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.3 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.0 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.1 | 0.6 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.1 | 0.3 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0005958 | DNA-dependent protein kinase-DNA ligase 4 complex(GO:0005958) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.2 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.0 | GO:0043625 | delta DNA polymerase complex(GO:0043625) |
| 0.0 | 0.1 | GO:0042575 | DNA polymerase complex(GO:0042575) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0005766 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.2 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.2 | GO:0005763 | organellar small ribosomal subunit(GO:0000314) mitochondrial small ribosomal subunit(GO:0005763) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.1 | 0.2 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.1 | 0.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.1 | 0.2 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.1 | 0.2 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.7 | GO:0019198 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.1 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0016615 | malate dehydrogenase activity(GO:0016615) |
| 0.0 | 0.5 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.2 | GO:0004017 | adenylate kinase activity(GO:0004017) |
| 0.0 | 0.1 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.2 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0004312 | fatty acid synthase activity(GO:0004312) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0016744 | transferase activity, transferring aldehyde or ketonic groups(GO:0016744) |
| 0.0 | 0.1 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.0 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.1 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0016778 | diphosphotransferase activity(GO:0016778) |
| 0.0 | 0.0 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.0 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.0 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.0 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.0 | PID PS1 PATHWAY | Presenilin action in Notch and Wnt signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |
| 0.0 | 0.7 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |
| 0.0 | 0.2 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.2 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.2 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |