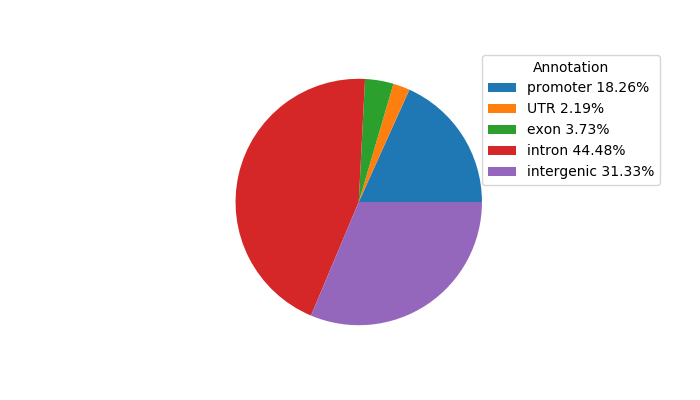
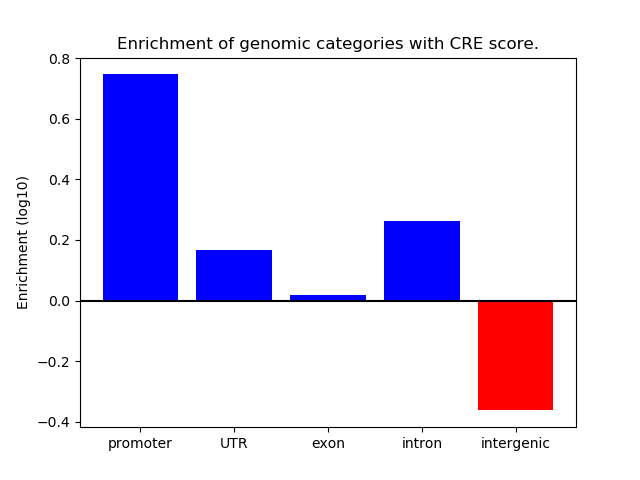
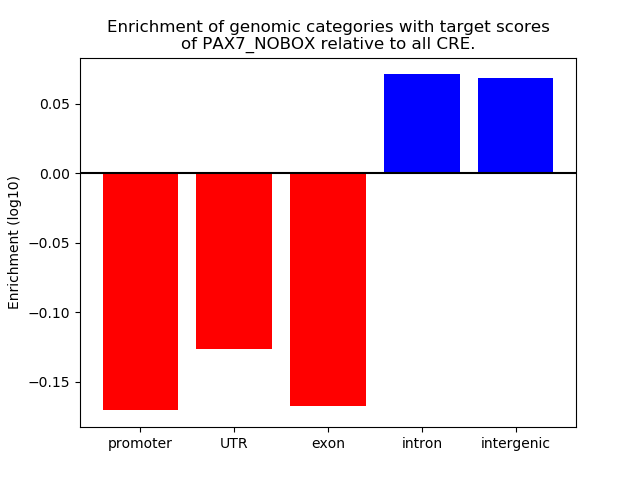
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for PAX7_NOBOX
Z-value: 0.74
Transcription factors associated with PAX7_NOBOX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
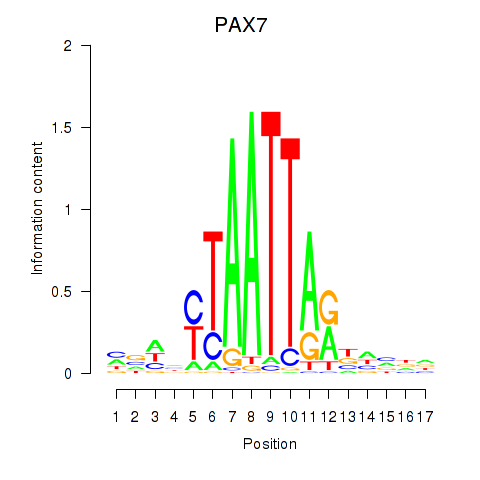
PAX7
|
ENSG00000009709.7 | paired box 7 |
|
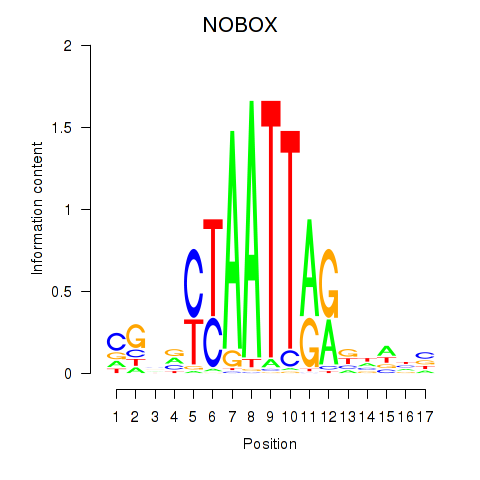
NOBOX
|
ENSG00000106410.10 | NOBOX oogenesis homeobox |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr7_144104551_144104702 | NOBOX | 2694 | 0.244758 | 0.82 | 6.4e-03 | Click! |
| chr7_144113392_144113543 | NOBOX | 6147 | 0.187773 | 0.82 | 7.1e-03 | Click! |
| chr7_144104809_144104960 | NOBOX | 2436 | 0.260426 | 0.64 | 6.3e-02 | Click! |
| chr7_144113151_144113302 | NOBOX | 5906 | 0.189144 | 0.63 | 6.6e-02 | Click! |
| chr7_144081989_144082140 | NOBOX | 18722 | 0.133433 | 0.54 | 1.4e-01 | Click! |
| chr1_18957647_18957941 | PAX7 | 221 | 0.963424 | 0.83 | 6.1e-03 | Click! |
| chr1_18957463_18957614 | PAX7 | 38 | 0.987288 | 0.81 | 8.0e-03 | Click! |
| chr1_19020395_19020546 | PAX7 | 62452 | 0.134235 | 0.80 | 9.9e-03 | Click! |
| chr1_18970260_18970411 | PAX7 | 12317 | 0.285708 | 0.68 | 4.3e-02 | Click! |
| chr1_18970063_18970214 | PAX7 | 12120 | 0.286284 | 0.67 | 4.7e-02 | Click! |
Activity of the PAX7_NOBOX motif across conditions
Conditions sorted by the z-value of the PAX7_NOBOX motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
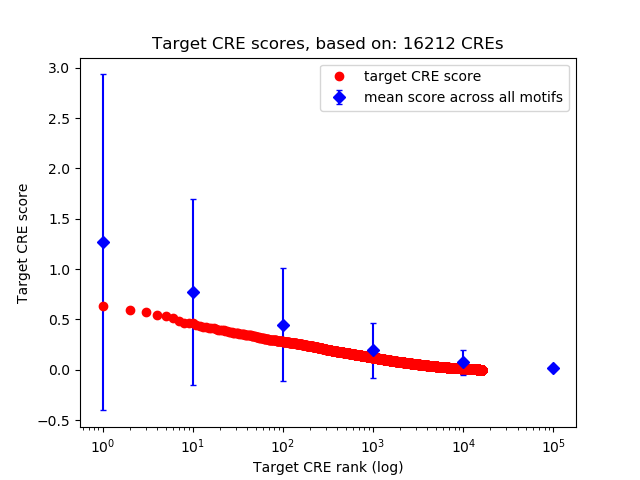
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_115135696_115136278 | 0.63 |
TBX3 |
T-box 3 |
14018 |
0.21 |
| chr20_14315841_14316127 | 0.59 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
2270 |
0.36 |
| chr1_45083000_45083241 | 0.58 |
RNF220 |
ring finger protein 220 |
8878 |
0.17 |
| chr5_3597777_3597928 | 0.55 |
IRX1 |
iroquois homeobox 1 |
1684 |
0.41 |
| chr6_56528559_56528710 | 0.54 |
DST |
dystonin |
20840 |
0.27 |
| chr3_67581837_67581988 | 0.52 |
SUCLG2 |
succinate-CoA ligase, GDP-forming, beta subunit |
3266 |
0.41 |
| chr1_200865204_200865355 | 0.48 |
C1orf106 |
chromosome 1 open reading frame 106 |
1330 |
0.42 |
| chr2_23603304_23603524 | 0.46 |
KLHL29 |
kelch-like family member 29 |
4674 |
0.35 |
| chr11_131780111_131780399 | 0.46 |
NTM |
neurotrimin |
642 |
0.77 |
| chr6_125855681_125855832 | 0.46 |
RP11-735G4.1 |
|
160286 |
0.04 |
| chr4_90581488_90581639 | 0.44 |
RP11-115D19.1 |
|
21100 |
0.27 |
| chr1_159111648_159111897 | 0.43 |
AIM2 |
absent in melanoma 2 |
1474 |
0.37 |
| chrX_135231057_135231247 | 0.43 |
FHL1 |
four and a half LIM domains 1 |
415 |
0.86 |
| chr5_73839186_73839374 | 0.42 |
HEXB |
hexosaminidase B (beta polypeptide) |
96568 |
0.07 |
| chr11_69454777_69455129 | 0.42 |
CCND1 |
cyclin D1 |
902 |
0.63 |
| chr5_36591027_36591178 | 0.42 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
15355 |
0.27 |
| chr1_230223080_230223231 | 0.41 |
GALNT2 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 2 (GalNAc-T2) |
20137 |
0.23 |
| chr22_18575359_18575510 | 0.41 |
XXbac-B476C20.9 |
|
13784 |
0.14 |
| chr7_80377911_80378104 | 0.40 |
CD36 |
CD36 molecule (thrombospondin receptor) |
75315 |
0.12 |
| chr7_70060715_70061143 | 0.39 |
AUTS2 |
autism susceptibility candidate 2 |
133196 |
0.06 |
| chr9_9701186_9701337 | 0.39 |
ENSG00000265735 |
. |
259201 |
0.02 |
| chr13_37397601_37397752 | 0.39 |
RFXAP |
regulatory factor X-associated protein |
4315 |
0.24 |
| chr9_98188423_98189086 | 0.39 |
PTCH1 |
patched 1 |
54013 |
0.12 |
| chr1_1099465_1099616 | 0.39 |
ENSG00000207730 |
. |
2944 |
0.11 |
| chr7_32929941_32930760 | 0.38 |
KBTBD2 |
kelch repeat and BTB (POZ) domain containing 2 |
415 |
0.87 |
| chr2_192176773_192176924 | 0.38 |
MYO1B |
myosin IB |
35237 |
0.19 |
| chr11_24518160_24518510 | 0.37 |
LUZP2 |
leucine zipper protein 2 |
389 |
0.91 |
| chr6_33589286_33590289 | 0.37 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
626 |
0.65 |
| chr4_57984994_57985389 | 0.37 |
IGFBP7 |
insulin-like growth factor binding protein 7 |
8640 |
0.18 |
| chr9_72745702_72745853 | 0.37 |
MAMDC2-AS1 |
MAMDC2 antisense RNA 1 |
17001 |
0.22 |
| chr19_13127591_13127981 | 0.36 |
CTC-239J10.1 |
|
2461 |
0.15 |
| chr6_27342407_27343330 | 0.36 |
ZNF391 |
zinc finger protein 391 |
13629 |
0.22 |
| chr14_74724486_74725063 | 0.36 |
VSX2 |
visual system homeobox 2 |
18599 |
0.14 |
| chr4_27219592_27219774 | 0.35 |
ENSG00000222206 |
. |
5366 |
0.35 |
| chr11_119572371_119572522 | 0.35 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
26817 |
0.16 |
| chr2_113958350_113958501 | 0.35 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
4568 |
0.16 |
| chr2_163171316_163171467 | 0.35 |
IFIH1 |
interferon induced with helicase C domain 1 |
3803 |
0.25 |
| chr4_23975239_23975390 | 0.35 |
PPARGC1A |
peroxisome proliferator-activated receptor gamma, coactivator 1 alpha |
83614 |
0.11 |
| chr18_61555988_61556228 | 0.35 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
1115 |
0.49 |
| chr8_59027426_59027596 | 0.34 |
FAM110B |
family with sequence similarity 110, member B |
120398 |
0.07 |
| chr13_23092547_23092698 | 0.34 |
ENSG00000253094 |
. |
284662 |
0.01 |
| chr22_46929020_46929201 | 0.34 |
CELSR1 |
cadherin, EGF LAG seven-pass G-type receptor 1 |
2081 |
0.33 |
| chr11_37525950_37526107 | 0.34 |
ENSG00000251838 |
. |
197742 |
0.03 |
| chr20_51273685_51273836 | 0.34 |
TSHZ2 |
teashirt zinc finger homeobox 2 |
315186 |
0.01 |
| chr10_75938020_75938413 | 0.34 |
ADK |
adenosine kinase |
1695 |
0.42 |
| chr13_110757726_110757877 | 0.34 |
ENSG00000265885 |
. |
7303 |
0.31 |
| chr22_34248307_34248458 | 0.34 |
LARGE |
like-glycosyltransferase |
9230 |
0.27 |
| chr14_81789932_81790083 | 0.34 |
STON2 |
stonin 2 |
46730 |
0.17 |
| chr3_54897831_54897982 | 0.33 |
CACNA2D3-AS1 |
CACNA2D3 antisense RNA 1 |
37376 |
0.21 |
| chr4_160357796_160357947 | 0.33 |
ENSG00000251979 |
. |
70309 |
0.12 |
| chr12_29716327_29716478 | 0.32 |
TMTC1 |
transmembrane and tetratricopeptide repeat containing 1 |
40755 |
0.16 |
| chr5_96000350_96000501 | 0.32 |
CAST |
calpastatin |
1668 |
0.37 |
| chr18_59619337_59619488 | 0.32 |
RNF152 |
ring finger protein 152 |
57948 |
0.16 |
| chr15_99297407_99297593 | 0.32 |
ENSG00000264480 |
. |
30155 |
0.19 |
| chr7_1443631_1443991 | 0.32 |
MICALL2 |
MICAL-like 2 |
55151 |
0.11 |
| chr4_113331922_113332073 | 0.32 |
RP11-402J6.1 |
|
104544 |
0.06 |
| chr13_39327903_39328054 | 0.32 |
ENSG00000252795 |
. |
25583 |
0.19 |
| chrX_3264180_3264467 | 0.32 |
MXRA5 |
matrix-remodelling associated 5 |
359 |
0.92 |
| chr15_93999319_93999470 | 0.32 |
ENSG00000212063 |
. |
169842 |
0.04 |
| chr10_126315472_126315623 | 0.32 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
76647 |
0.09 |
| chr2_113539557_113539708 | 0.31 |
IL1A |
interleukin 1, alpha |
2535 |
0.25 |
| chr13_30415572_30415723 | 0.31 |
UBL3 |
ubiquitin-like 3 |
9174 |
0.3 |
| chr8_70622919_70623070 | 0.31 |
RP11-102F4.2 |
|
2631 |
0.31 |
| chr11_14375529_14375805 | 0.31 |
RRAS2 |
related RAS viral (r-ras) oncogene homolog 2 |
379 |
0.91 |
| chr15_98835948_98836124 | 0.30 |
FAM169B |
family with sequence similarity 169, member B |
193171 |
0.03 |
| chr16_66851667_66851818 | 0.30 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
7140 |
0.13 |
| chr9_124846791_124846942 | 0.30 |
TTLL11 |
tubulin tyrosine ligase-like family, member 11 |
9019 |
0.2 |
| chr4_139635357_139635508 | 0.30 |
ENSG00000238971 |
. |
97488 |
0.08 |
| chr7_100492646_100493161 | 0.30 |
ACHE |
acetylcholinesterase (Yt blood group) |
579 |
0.57 |
| chr8_112438072_112438223 | 0.30 |
ENSG00000222146 |
. |
722907 |
0.0 |
| chr1_181050920_181051071 | 0.30 |
IER5 |
immediate early response 5 |
6643 |
0.23 |
| chr1_97033525_97033676 | 0.30 |
ENSG00000241992 |
. |
15165 |
0.29 |
| chr20_1892625_1892776 | 0.30 |
SIRPA |
signal-regulatory protein alpha |
16758 |
0.21 |
| chr8_76807046_76807197 | 0.30 |
ENSG00000238595 |
. |
1982 |
0.49 |
| chr11_26604890_26605185 | 0.30 |
MUC15 |
mucin 15, cell surface associated |
11257 |
0.3 |
| chr5_100168975_100169126 | 0.30 |
ENSG00000221263 |
. |
16781 |
0.26 |
| chr3_151034381_151034532 | 0.30 |
GPR87 |
G protein-coupled receptor 87 |
284 |
0.9 |
| chr9_92032170_92032321 | 0.29 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
1503 |
0.49 |
| chr22_19794405_19794556 | 0.29 |
C22orf29 |
chromosome 22 open reading frame 29 |
47859 |
0.1 |
| chrX_150695771_150695922 | 0.29 |
PASD1 |
PAS domain containing 1 |
36248 |
0.2 |
| chr15_101899572_101899723 | 0.29 |
PCSK6 |
proprotein convertase subtilisin/kexin type 6 |
6820 |
0.2 |
| chr7_121585085_121585236 | 0.29 |
PTPRZ1 |
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
71745 |
0.12 |
| chr10_6763782_6764322 | 0.29 |
PRKCQ |
protein kinase C, theta |
141789 |
0.05 |
| chr10_108111933_108112084 | 0.29 |
SORCS1 |
sortilin-related VPS10 domain containing receptor 1 |
259737 |
0.02 |
| chr6_41472777_41473161 | 0.29 |
RP11-328M4.2 |
|
40843 |
0.13 |
| chr3_49447935_49448923 | 0.29 |
RHOA |
ras homolog family member A |
932 |
0.32 |
| chr9_89952337_89952906 | 0.29 |
ENSG00000212421 |
. |
77256 |
0.11 |
| chr15_99449448_99449599 | 0.29 |
RP11-654A16.1 |
|
12759 |
0.21 |
| chr12_56917781_56918152 | 0.29 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
2183 |
0.23 |
| chr7_98970111_98970529 | 0.29 |
ARPC1B |
actin related protein 2/3 complex, subunit 1B, 41kDa |
1552 |
0.28 |
| chr12_125179754_125179913 | 0.29 |
SCARB1 |
scavenger receptor class B, member 1 |
122420 |
0.06 |
| chr20_14316163_14316462 | 0.29 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
1942 |
0.4 |
| chr14_94641841_94642049 | 0.28 |
PPP4R4 |
protein phosphatase 4, regulatory subunit 4 |
147 |
0.96 |
| chr18_30310087_30311054 | 0.28 |
AC012123.1 |
Uncharacterized protein |
39188 |
0.17 |
| chr3_79028408_79028559 | 0.28 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
39211 |
0.19 |
| chr3_88191159_88191414 | 0.28 |
ZNF654 |
zinc finger protein 654 |
3032 |
0.24 |
| chr15_96903910_96904291 | 0.28 |
AC087477.1 |
Uncharacterized protein |
387 |
0.85 |
| chr17_12569246_12569498 | 0.28 |
MYOCD |
myocardin |
66 |
0.98 |
| chr2_36589291_36589442 | 0.28 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
5752 |
0.33 |
| chr8_143757309_143757460 | 0.28 |
PSCA |
prostate stem cell antigen |
4490 |
0.13 |
| chr4_77067485_77067636 | 0.28 |
NUP54 |
nucleoporin 54kDa |
1990 |
0.3 |
| chr9_22239467_22239618 | 0.28 |
CDKN2B-AS1 |
CDKN2B antisense RNA 1 |
125865 |
0.06 |
| chr17_40700695_40700889 | 0.28 |
HSD17B1 |
hydroxysteroid (17-beta) dehydrogenase 1 |
440 |
0.63 |
| chr8_79469123_79469274 | 0.28 |
PKIA |
protein kinase (cAMP-dependent, catalytic) inhibitor alpha |
34268 |
0.19 |
| chr15_36905175_36905326 | 0.28 |
C15orf41 |
chromosome 15 open reading frame 41 |
18149 |
0.26 |
| chr2_119188658_119188833 | 0.28 |
INSIG2 |
insulin induced gene 2 |
342695 |
0.01 |
| chr9_75771174_75771814 | 0.28 |
ANXA1 |
annexin A1 |
1076 |
0.66 |
| chr13_79182716_79183828 | 0.27 |
POU4F1 |
POU class 4 homeobox 1 |
5599 |
0.19 |
| chr6_132610665_132610816 | 0.27 |
MOXD1 |
monooxygenase, DBH-like 1 |
85464 |
0.09 |
| chr1_30417491_30417642 | 0.27 |
ENSG00000222787 |
. |
59817 |
0.17 |
| chr13_74606153_74606483 | 0.27 |
KLF12 |
Kruppel-like factor 12 |
37132 |
0.24 |
| chr4_173311851_173312002 | 0.27 |
GALNTL6 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 |
338330 |
0.01 |
| chr7_92254665_92254816 | 0.27 |
FAM133B |
family with sequence similarity 133, member B |
35032 |
0.17 |
| chr10_24754741_24755189 | 0.27 |
KIAA1217 |
KIAA1217 |
495 |
0.84 |
| chr2_36597223_36597374 | 0.27 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
13684 |
0.29 |
| chr3_170527527_170527678 | 0.27 |
ENSG00000222411 |
. |
32359 |
0.19 |
| chr14_53275157_53275308 | 0.27 |
GNPNAT1 |
glucosamine-phosphate N-acetyltransferase 1 |
16846 |
0.16 |
| chr14_61445912_61446063 | 0.27 |
TRMT5 |
tRNA methyltransferase 5 |
1782 |
0.29 |
| chr4_184961314_184961487 | 0.27 |
STOX2 |
storkhead box 2 |
38922 |
0.18 |
| chr1_40471048_40471199 | 0.27 |
CAP1 |
CAP, adenylate cyclase-associated protein 1 (yeast) |
34782 |
0.13 |
| chr2_196281841_196281992 | 0.27 |
ENSG00000202206 |
. |
96844 |
0.09 |
| chr4_68636095_68636246 | 0.27 |
GNRHR |
gonadotropin-releasing hormone receptor |
16092 |
0.17 |
| chr15_83951840_83952048 | 0.27 |
BNC1 |
basonuclin 1 |
127 |
0.97 |
| chr4_77611462_77611724 | 0.27 |
AC107072.2 |
|
52817 |
0.12 |
| chr8_39259417_39259568 | 0.27 |
ENSG00000252176 |
. |
158964 |
0.04 |
| chr1_246034788_246034939 | 0.27 |
RP11-83A16.1 |
|
161990 |
0.04 |
| chr10_119302142_119302346 | 0.27 |
EMX2 |
empty spiracles homeobox 2 |
265 |
0.64 |
| chr1_209761749_209761900 | 0.27 |
CAMK1G |
calcium/calmodulin-dependent protein kinase IG |
4750 |
0.21 |
| chr5_44235116_44235267 | 0.27 |
FGF10-AS1 |
FGF10 antisense RNA 1 |
153643 |
0.04 |
| chr5_140864017_140864278 | 0.27 |
PCDHGC4 |
protocadherin gamma subfamily C, 4 |
594 |
0.53 |
| chr9_23769338_23769489 | 0.27 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
9960 |
0.33 |
| chr8_31771478_31771629 | 0.26 |
NRG1-IT1 |
NRG1 intronic transcript 1 (non-protein coding) |
112182 |
0.07 |
| chr10_114552786_114553214 | 0.26 |
RP11-57H14.4 |
|
30249 |
0.21 |
| chr14_55272428_55272636 | 0.26 |
SAMD4A |
sterile alpha motif domain containing 4A |
50981 |
0.13 |
| chr4_153272766_153273041 | 0.26 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
1220 |
0.52 |
| chr4_3387991_3388142 | 0.26 |
RGS12 |
regulator of G-protein signaling 12 |
5 |
0.98 |
| chr6_106972751_106973424 | 0.26 |
AIM1 |
absent in melanoma 1 |
13357 |
0.2 |
| chr8_62566222_62566373 | 0.26 |
ASPH |
aspartate beta-hydroxylase |
6931 |
0.24 |
| chr3_31575511_31576107 | 0.26 |
STT3B |
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
1527 |
0.55 |
| chr2_184434080_184434231 | 0.26 |
ENSG00000238306 |
. |
380008 |
0.01 |
| chr3_100660155_100660398 | 0.26 |
ENSG00000201642 |
. |
11195 |
0.24 |
| chr4_75174481_75174674 | 0.26 |
EPGN |
epithelial mitogen |
237 |
0.93 |
| chr7_139047352_139047690 | 0.26 |
C7orf55-LUC7L2 |
C7orf55-LUC7L2 readthrough |
2867 |
0.19 |
| chr10_134261699_134261957 | 0.26 |
C10orf91 |
chromosome 10 open reading frame 91 |
3135 |
0.24 |
| chr12_95511000_95511151 | 0.26 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
326 |
0.91 |
| chr17_79315981_79316442 | 0.26 |
TMEM105 |
transmembrane protein 105 |
11737 |
0.13 |
| chr4_183455582_183455733 | 0.26 |
ENSG00000252343 |
. |
14155 |
0.26 |
| chr3_120277024_120277175 | 0.26 |
NDUFB4 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa |
38057 |
0.18 |
| chr21_17909068_17909746 | 0.26 |
ENSG00000207638 |
. |
2002 |
0.33 |
| chr3_189509857_189510008 | 0.26 |
TP63 |
tumor protein p63 |
2342 |
0.39 |
| chr2_21444769_21444920 | 0.26 |
TDRD15 |
tudor domain containing 15 |
98055 |
0.09 |
| chr16_86817661_86817812 | 0.25 |
FOXL1 |
forkhead box L1 |
205621 |
0.02 |
| chr3_60921693_60921844 | 0.25 |
ENSG00000212211 |
. |
79491 |
0.12 |
| chr4_166361643_166362016 | 0.25 |
CPE |
carboxypeptidase E |
22451 |
0.23 |
| chr7_79953980_79954131 | 0.25 |
ENSG00000240347 |
. |
6486 |
0.23 |
| chr12_125624965_125625116 | 0.25 |
AACS |
acetoacetyl-CoA synthetase |
11948 |
0.29 |
| chr14_99713575_99713726 | 0.25 |
AL109767.1 |
|
15635 |
0.21 |
| chr11_106239153_106239583 | 0.25 |
RP11-680E19.1 |
|
104326 |
0.08 |
| chr1_198250905_198251056 | 0.25 |
NEK7 |
NIMA-related kinase 7 |
61051 |
0.16 |
| chr2_119989374_119989591 | 0.25 |
STEAP3 |
STEAP family member 3, metalloreductase |
8053 |
0.2 |
| chr5_39058731_39058980 | 0.25 |
RICTOR |
RPTOR independent companion of MTOR, complex 2 |
15636 |
0.24 |
| chr12_50665639_50665877 | 0.25 |
LIMA1 |
LIM domain and actin binding 1 |
11509 |
0.13 |
| chr3_129383548_129383699 | 0.25 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
8054 |
0.22 |
| chr1_63212144_63212320 | 0.25 |
ATG4C |
autophagy related 4C, cysteine peptidase |
37574 |
0.17 |
| chr16_59637744_59637895 | 0.25 |
ENSG00000200062 |
. |
63789 |
0.15 |
| chr1_3003277_3003428 | 0.25 |
PRDM16 |
PR domain containing 16 |
17577 |
0.18 |
| chr2_207998546_207999225 | 0.25 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
24 |
0.98 |
| chr6_64239754_64239905 | 0.25 |
PTP4A1 |
protein tyrosine phosphatase type IVA, member 1 |
42088 |
0.17 |
| chr5_180523773_180523999 | 0.25 |
OR2V1 |
olfactory receptor, family 2, subfamily V, member 1 |
28418 |
0.12 |
| chr14_52534801_52535890 | 0.25 |
NID2 |
nidogen 2 (osteonidogen) |
367 |
0.89 |
| chr17_18901833_18902110 | 0.25 |
FAM83G |
family with sequence similarity 83, member G |
5518 |
0.14 |
| chr10_74436092_74436294 | 0.25 |
MCU |
mitochondrial calcium uniporter |
15696 |
0.15 |
| chr2_55810508_55810659 | 0.25 |
ENSG00000212175 |
. |
17744 |
0.15 |
| chr15_67390777_67391884 | 0.25 |
SMAD3 |
SMAD family member 3 |
313 |
0.93 |
| chr6_117804579_117805184 | 0.25 |
DCBLD1 |
discoidin, CUB and LCCL domain containing 1 |
1056 |
0.49 |
| chr18_47385264_47385415 | 0.24 |
MYO5B |
myosin VB |
9123 |
0.15 |
| chr2_188418561_188419245 | 0.24 |
AC007319.1 |
|
27 |
0.69 |
| chr4_26344236_26344574 | 0.24 |
RBPJ |
recombination signal binding protein for immunoglobulin kappa J region |
357 |
0.93 |
| chr11_131170059_131170210 | 0.24 |
NTM |
neurotrimin |
70239 |
0.12 |
| chr2_95689867_95690018 | 0.24 |
MAL |
mal, T-cell differentiation protein |
1487 |
0.32 |
| chr10_29165095_29165348 | 0.24 |
ENSG00000199402 |
. |
1785 |
0.43 |
| chr2_511397_511548 | 0.24 |
TMEM18 |
transmembrane protein 18 |
164303 |
0.03 |
| chr1_112491209_112491360 | 0.24 |
KCND3 |
potassium voltage-gated channel, Shal-related subfamily, member 3 |
34134 |
0.17 |
| chr22_46476561_46476775 | 0.24 |
FLJ27365 |
hsa-mir-4763 |
476 |
0.66 |
| chr8_87188438_87188625 | 0.24 |
CTD-3118D11.3 |
|
4116 |
0.24 |
| chr20_35830256_35830797 | 0.24 |
MROH8 |
maestro heat-like repeat family member 8 |
22535 |
0.17 |
| chr4_77506894_77507309 | 0.24 |
ENSG00000265314 |
. |
10397 |
0.17 |
| chr7_19141522_19141673 | 0.24 |
AC003986.6 |
|
10500 |
0.16 |
| chr15_47476270_47477144 | 0.24 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
409 |
0.87 |
| chr14_29243474_29243665 | 0.24 |
C14orf23 |
chromosome 14 open reading frame 23 |
1568 |
0.33 |
| chr1_198393639_198393891 | 0.24 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
116039 |
0.07 |
| chr2_149863937_149864088 | 0.24 |
LYPD6B |
LY6/PLAUR domain containing 6B |
30969 |
0.22 |
| chr7_70048822_70048973 | 0.24 |
AUTS2 |
autism susceptibility candidate 2 |
145228 |
0.05 |
| chr7_81040159_81040512 | 0.24 |
AC005008.2 |
Uncharacterized protein |
223229 |
0.02 |
| chr13_97927308_97927620 | 0.24 |
MBNL2 |
muscleblind-like splicing regulator 2 |
994 |
0.69 |
| chr18_6730820_6730971 | 0.24 |
ARHGAP28 |
Rho GTPase activating protein 28 |
853 |
0.47 |
| chr17_69531749_69531900 | 0.24 |
ENSG00000222563 |
. |
224515 |
0.02 |
| chr8_17657715_17658703 | 0.24 |
MTUS1 |
microtubule associated tumor suppressor 1 |
32 |
0.93 |
| chr5_158036732_158037177 | 0.24 |
CTD-2363C16.1 |
|
373060 |
0.01 |
| chr15_96878465_96878729 | 0.24 |
ENSG00000222651 |
. |
2107 |
0.24 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.5 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.1 | 0.5 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.4 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.3 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.5 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.1 | 0.3 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.2 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.1 | 0.3 | GO:0060197 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.1 | 0.3 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.7 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.2 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.1 | 0.3 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.2 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.1 | 0.3 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.1 | 0.1 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.1 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.1 | 0.2 | GO:0044413 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.2 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.2 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.2 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 | 0.1 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.2 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.2 | GO:1900619 | acetylcholine metabolic process(GO:0008291) acetate ester metabolic process(GO:1900619) |
| 0.0 | 0.1 | GO:0060013 | righting reflex(GO:0060013) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.3 | GO:0031579 | membrane raft organization(GO:0031579) |
| 0.0 | 0.1 | GO:0002085 | inhibition of neuroepithelial cell differentiation(GO:0002085) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.1 | GO:0061117 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.1 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.0 | 0.0 | GO:0060462 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.4 | GO:0043616 | keratinocyte proliferation(GO:0043616) |
| 0.0 | 0.2 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.5 | GO:0008038 | neuron recognition(GO:0008038) |
| 0.0 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.1 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.4 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0071421 | manganese ion transmembrane transport(GO:0071421) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.1 | GO:0032353 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.0 | GO:0072071 | mesangial cell differentiation(GO:0072007) kidney interstitial fibroblast differentiation(GO:0072071) renal interstitial fibroblast development(GO:0072141) mesangial cell development(GO:0072143) pericyte cell differentiation(GO:1904238) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.0 | GO:0035112 | genitalia morphogenesis(GO:0035112) |
| 0.0 | 0.1 | GO:0010659 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.3 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.2 | GO:0033144 | negative regulation of intracellular steroid hormone receptor signaling pathway(GO:0033144) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.1 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0021830 | interneuron migration from the subpallium to the cortex(GO:0021830) cerebral cortex GABAergic interneuron migration(GO:0021853) cerebral cortex GABAergic interneuron development(GO:0021894) interneuron migration(GO:1904936) |
| 0.0 | 0.1 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.2 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.1 | GO:0002420 | natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002420) natural killer cell mediated immune response to tumor cell(GO:0002423) regulation of natural killer cell mediated immune response to tumor cell(GO:0002855) positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002858) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.1 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.5 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.0 | GO:0060033 | Mullerian duct regression(GO:0001880) anatomical structure regression(GO:0060033) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.2 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.1 | GO:0060438 | trachea development(GO:0060438) |
| 0.0 | 0.1 | GO:0010792 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.0 | GO:0010002 | cardioblast differentiation(GO:0010002) |
| 0.0 | 0.1 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.0 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.0 | GO:0060057 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.0 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.0 | GO:0055094 | response to lipoprotein particle(GO:0055094) |
| 0.0 | 0.1 | GO:0051446 | positive regulation of meiotic nuclear division(GO:0045836) positive regulation of meiotic cell cycle(GO:0051446) |
| 0.0 | 0.0 | GO:0072193 | ureter development(GO:0072189) ureter smooth muscle development(GO:0072191) ureter smooth muscle cell differentiation(GO:0072193) |
| 0.0 | 0.1 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.4 | GO:0009954 | proximal/distal pattern formation(GO:0009954) |
| 0.0 | 0.2 | GO:0051963 | regulation of synapse assembly(GO:0051963) |
| 0.0 | 0.1 | GO:0032933 | SREBP signaling pathway(GO:0032933) cellular response to sterol depletion(GO:0071501) |
| 0.0 | 0.1 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.0 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0097094 | cranial suture morphogenesis(GO:0060363) craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.5 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.2 | GO:0010633 | negative regulation of epithelial cell migration(GO:0010633) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.1 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.0 | GO:0060124 | positive regulation of growth hormone secretion(GO:0060124) |
| 0.0 | 0.1 | GO:0010894 | negative regulation of steroid biosynthetic process(GO:0010894) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0060134 | prepulse inhibition(GO:0060134) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0048535 | lymph node development(GO:0048535) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0071392 | cellular response to estradiol stimulus(GO:0071392) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0060179 | male courtship behavior(GO:0008049) male mating behavior(GO:0060179) |
| 0.0 | 0.0 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0043457 | regulation of cellular respiration(GO:0043457) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.0 | GO:0000089 | mitotic metaphase(GO:0000089) |
| 0.0 | 0.0 | GO:0006533 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.3 | GO:0090090 | negative regulation of canonical Wnt signaling pathway(GO:0090090) |
| 0.0 | 0.1 | GO:0050872 | white fat cell differentiation(GO:0050872) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0033599 | regulation of mammary gland epithelial cell proliferation(GO:0033599) |
| 0.0 | 0.0 | GO:0071428 | rRNA-containing ribonucleoprotein complex export from nucleus(GO:0071428) |
| 0.0 | 0.0 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0006344 | maintenance of chromatin silencing(GO:0006344) |
| 0.0 | 0.0 | GO:0045747 | positive regulation of Notch signaling pathway(GO:0045747) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.1 | GO:0044065 | regulation of respiratory system process(GO:0044065) |
| 0.0 | 0.1 | GO:0000768 | syncytium formation by plasma membrane fusion(GO:0000768) myoblast fusion(GO:0007520) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0060992 | response to fungicide(GO:0060992) |
| 0.0 | 0.0 | GO:0060605 | tube lumen cavitation(GO:0060605) salivary gland cavitation(GO:0060662) |
| 0.0 | 0.1 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.0 | GO:0006089 | lactate metabolic process(GO:0006089) |
| 0.0 | 0.1 | GO:0001885 | endothelial cell development(GO:0001885) |
| 0.0 | 0.0 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.2 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.0 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.0 | GO:0071288 | cellular response to mercury ion(GO:0071288) |
| 0.0 | 0.0 | GO:0051968 | positive regulation of synaptic transmission, glutamatergic(GO:0051968) |
| 0.0 | 0.1 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.0 | GO:0090102 | cochlea development(GO:0090102) |
| 0.0 | 0.0 | GO:0032261 | purine nucleotide salvage(GO:0032261) |
| 0.0 | 0.0 | GO:0043438 | acetoacetic acid metabolic process(GO:0043438) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.1 | GO:0046339 | diacylglycerol metabolic process(GO:0046339) |
| 0.0 | 0.1 | GO:0047496 | vesicle transport along microtubule(GO:0047496) vesicle cytoskeletal trafficking(GO:0099518) |
| 0.0 | 0.0 | GO:0060754 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.0 | 1.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.0 | GO:0061213 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.0 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.0 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.0 | 0.2 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.4 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.0 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.0 | GO:0032075 | positive regulation of nuclease activity(GO:0032075) |
| 0.0 | 0.1 | GO:0021952 | central nervous system projection neuron axonogenesis(GO:0021952) |
| 0.0 | 0.1 | GO:0071157 | negative regulation of cell cycle arrest(GO:0071157) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.0 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.0 | GO:0043276 | anoikis(GO:0043276) |
| 0.0 | 0.0 | GO:0003097 | renal water transport(GO:0003097) |
| 0.0 | 0.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.0 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.0 | GO:0090493 | dopamine uptake involved in synaptic transmission(GO:0051583) catecholamine uptake involved in synaptic transmission(GO:0051934) catecholamine uptake(GO:0090493) dopamine uptake(GO:0090494) |
| 0.0 | 0.0 | GO:0042670 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.1 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 0.1 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0042640 | anagen(GO:0042640) |
| 0.0 | 0.1 | GO:0030826 | regulation of cGMP biosynthetic process(GO:0030826) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.4 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.2 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.3 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.3 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.4 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0005883 | neurofilament(GO:0005883) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.1 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0031088 | platelet dense granule membrane(GO:0031088) |
| 0.0 | 0.1 | GO:0045178 | basal part of cell(GO:0045178) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.6 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.0 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.2 | GO:0005665 | DNA-directed RNA polymerase II, core complex(GO:0005665) |
| 0.0 | 0.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.3 | GO:0098636 | integrin complex(GO:0008305) protein complex involved in cell adhesion(GO:0098636) |
| 0.0 | 0.1 | GO:0001950 | obsolete plasma membrane enriched fraction(GO:0001950) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.2 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.0 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0001741 | XY body(GO:0001741) |
| 0.0 | 0.2 | GO:0030057 | desmosome(GO:0030057) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.4 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.4 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.1 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.1 | 0.4 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.2 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.1 | 0.2 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.1 | 0.2 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.1 | 0.2 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.5 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.3 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.3 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0050897 | cobalt ion binding(GO:0050897) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.4 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.2 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.3 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.9 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0008035 | high-density lipoprotein particle binding(GO:0008035) |
| 0.0 | 0.1 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.0 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0016423 | tRNA (guanine) methyltransferase activity(GO:0016423) |
| 0.0 | 0.1 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.0 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0016538 | cyclin-dependent protein serine/threonine kinase regulator activity(GO:0016538) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0060229 | lipase activator activity(GO:0060229) |
| 0.0 | 0.0 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.2 | GO:0004835 | tubulin-tyrosine ligase activity(GO:0004835) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.0 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.0 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0004303 | estradiol 17-beta-dehydrogenase activity(GO:0004303) |
| 0.0 | 0.0 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.0 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.0 | GO:0034617 | tetrahydrobiopterin binding(GO:0034617) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0030249 | guanylate cyclase regulator activity(GO:0030249) |
| 0.0 | 0.1 | GO:0047760 | butyrate-CoA ligase activity(GO:0047760) |
| 0.0 | 0.0 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0032052 | bile acid binding(GO:0032052) |
| 0.0 | 0.0 | GO:0031708 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.9 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.4 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.9 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.3 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.0 | PID HIV NEF PATHWAY | HIV-1 Nef: Negative effector of Fas and TNF-alpha |
| 0.0 | 0.7 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.4 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.1 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.9 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.3 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.4 | REACTOME GRB2 EVENTS IN ERBB2 SIGNALING | Genes involved in GRB2 events in ERBB2 signaling |
| 0.0 | 1.1 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.3 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.0 | REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | Genes involved in Class A/1 (Rhodopsin-like receptors) |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.0 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.0 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.0 | REACTOME SHC1 EVENTS IN EGFR SIGNALING | Genes involved in SHC1 events in EGFR signaling |
| 0.0 | 0.2 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.1 | REACTOME SIGNALING BY NOTCH3 | Genes involved in Signaling by NOTCH3 |
| 0.0 | 0.2 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING ACTIVATES SMADS | Genes involved in TGF-beta receptor signaling activates SMADs |
| 0.0 | 0.1 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.1 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.1 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |