Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
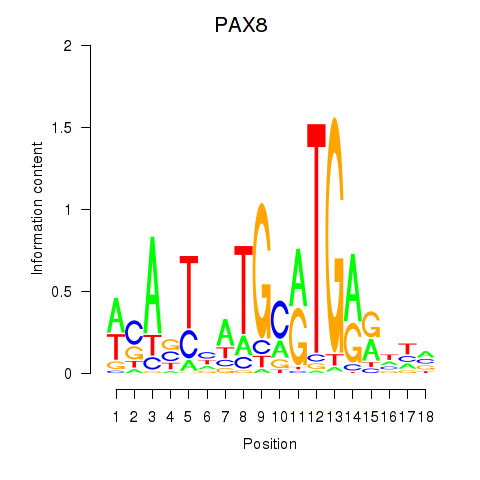
Results for PAX8
Z-value: 2.16
Transcription factors associated with PAX8
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX8
|
ENSG00000125618.12 | paired box 8 |
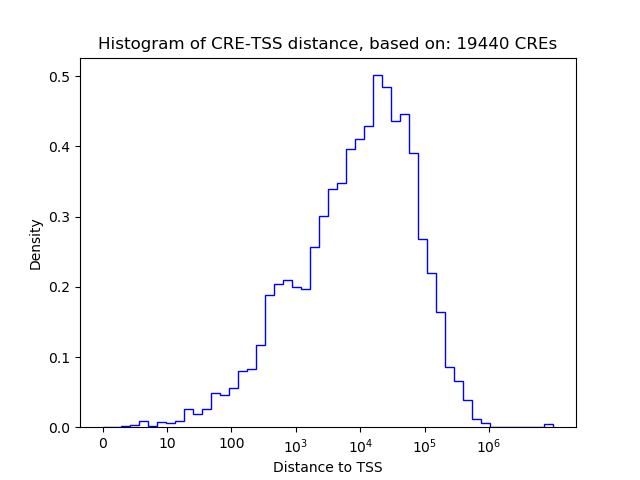
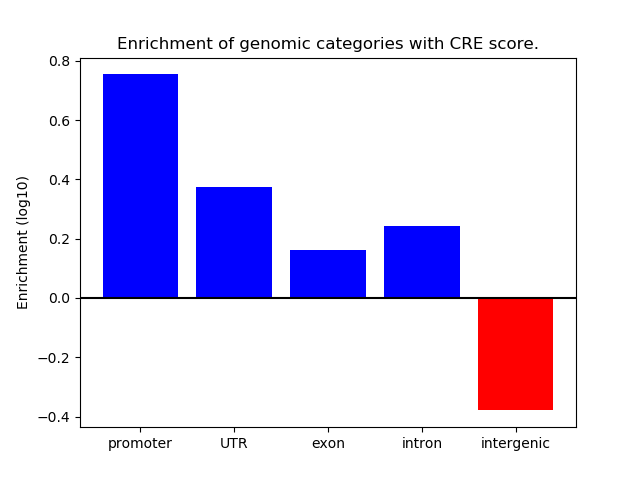
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_113992675_113993110 | PAX8 | 128 | 0.615076 | -0.78 | 1.3e-02 | Click! |
| chr2_114035393_114035544 | PAX8 | 503 | 0.774500 | -0.66 | 5.4e-02 | Click! |
| chr2_114082607_114082820 | PAX8 | 46186 | 0.113986 | -0.60 | 8.4e-02 | Click! |
| chr2_114035017_114035267 | PAX8 | 829 | 0.592173 | -0.52 | 1.5e-01 | Click! |
| chr2_114034526_114034677 | PAX8 | 1370 | 0.399257 | -0.48 | 1.9e-01 | Click! |
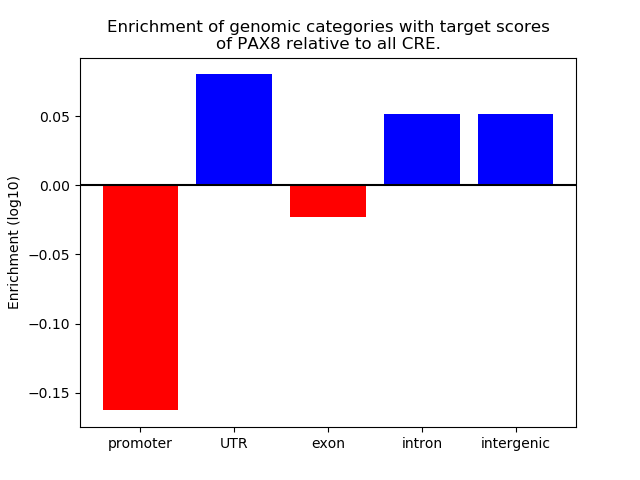
Activity of the PAX8 motif across conditions
Conditions sorted by the z-value of the PAX8 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
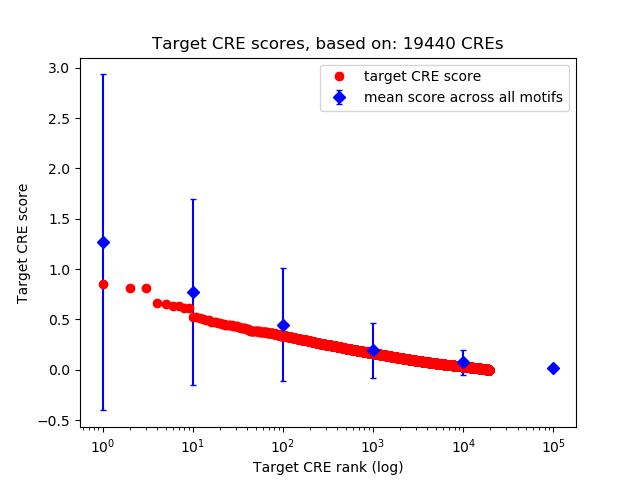
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_84854349_84854509 | 0.86 |
CRISPLD2 |
cysteine-rich secretory protein LCCL domain containing 2 |
818 |
0.61 |
| chrX_153595297_153595831 | 0.81 |
FLNA |
filamin A, alpha |
4087 |
0.1 |
| chr4_71570546_71572213 | 0.81 |
RUFY3 |
RUN and FYVE domain containing 3 |
861 |
0.36 |
| chr3_71293724_71294002 | 0.66 |
FOXP1 |
forkhead box P1 |
453 |
0.89 |
| chr1_9882638_9884014 | 0.65 |
CLSTN1 |
calsyntenin 1 |
716 |
0.65 |
| chr8_9005889_9006259 | 0.64 |
PPP1R3B |
protein phosphatase 1, regulatory subunit 3B |
2132 |
0.26 |
| chr1_26238885_26239088 | 0.63 |
ENSG00000266763 |
. |
746 |
0.47 |
| chr3_45685861_45686013 | 0.62 |
LIMD1-AS1 |
LIMD1 antisense RNA 1 |
44437 |
0.11 |
| chr2_30454378_30455881 | 0.62 |
LBH |
limb bud and heart development |
83 |
0.98 |
| chr16_85793045_85793254 | 0.53 |
C16orf74 |
chromosome 16 open reading frame 74 |
8414 |
0.11 |
| chr10_21462508_21463481 | 0.52 |
NEBL-AS1 |
NEBL antisense RNA 1 |
51 |
0.59 |
| chr8_83170405_83170556 | 0.51 |
SNX16 |
sorting nexin 16 |
415379 |
0.01 |
| chr7_79820122_79820273 | 0.50 |
GNAI1 |
guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 1 |
3822 |
0.32 |
| chr18_51780819_51781029 | 0.50 |
POLI |
polymerase (DNA directed) iota |
14850 |
0.19 |
| chr20_17643261_17643428 | 0.49 |
RRBP1 |
ribosome binding protein 1 |
2192 |
0.33 |
| chr1_168440138_168440500 | 0.48 |
XCL2 |
chemokine (C motif) ligand 2 |
72916 |
0.1 |
| chr4_160169630_160170007 | 0.47 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
18215 |
0.23 |
| chr15_44484891_44485042 | 0.47 |
FRMD5 |
FERM domain containing 5 |
1678 |
0.47 |
| chr16_3333511_3334475 | 0.47 |
ZNF263 |
zinc finger protein 263 |
38 |
0.95 |
| chr5_139525265_139525476 | 0.47 |
IGIP |
IgA-inducing protein |
19849 |
0.12 |
| chr14_100260565_100260825 | 0.46 |
EML1 |
echinoderm microtubule associated protein like 1 |
923 |
0.67 |
| chr12_44130303_44130454 | 0.45 |
RP11-210N13.1 |
|
2254 |
0.3 |
| chrX_9344812_9344963 | 0.45 |
TBL1X |
transducin (beta)-like 1X-linked |
86448 |
0.1 |
| chr15_81074624_81074775 | 0.44 |
KIAA1199 |
KIAA1199 |
2987 |
0.3 |
| chr7_26564902_26565053 | 0.44 |
KIAA0087 |
KIAA0087 |
13430 |
0.27 |
| chr14_90526445_90527696 | 0.44 |
KCNK13 |
potassium channel, subfamily K, member 13 |
1039 |
0.63 |
| chr7_94044432_94044583 | 0.44 |
COL1A2 |
collagen, type I, alpha 2 |
20634 |
0.24 |
| chr7_19154419_19154620 | 0.44 |
AC003986.6 |
|
2422 |
0.22 |
| chr9_134277901_134278052 | 0.43 |
PRRC2B |
proline-rich coiled-coil 2B |
8496 |
0.22 |
| chr3_115511210_115511361 | 0.43 |
ENSG00000243359 |
. |
45430 |
0.2 |
| chr5_169231050_169231345 | 0.43 |
CTB-37A13.1 |
|
24828 |
0.22 |
| chr5_15510117_15510353 | 0.43 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
8688 |
0.31 |
| chr14_104024819_104024970 | 0.42 |
KLC1 |
kinesin light chain 1 |
3339 |
0.09 |
| chr11_75479321_75480435 | 0.42 |
DGAT2 |
diacylglycerol O-acyltransferase 2 |
21 |
0.69 |
| chr8_13371594_13371816 | 0.42 |
DLC1 |
deleted in liver cancer 1 |
569 |
0.79 |
| chr12_59312115_59312531 | 0.42 |
LRIG3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
1004 |
0.56 |
| chr1_234754657_234754906 | 0.41 |
IRF2BP2 |
interferon regulatory factor 2 binding protein 2 |
9510 |
0.19 |
| chr1_247506079_247506341 | 0.41 |
ZNF496 |
zinc finger protein 496 |
11165 |
0.18 |
| chr3_8493679_8493830 | 0.41 |
LMCD1-AS1 |
LMCD1 antisense RNA 1 (head to head) |
49530 |
0.14 |
| chr4_114679710_114679861 | 0.40 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
2439 |
0.44 |
| chr4_146841607_146841966 | 0.40 |
ZNF827 |
zinc finger protein 827 |
17837 |
0.22 |
| chr1_109200139_109200290 | 0.39 |
HENMT1 |
HEN1 methyltransferase homolog 1 (Arabidopsis) |
3463 |
0.24 |
| chr6_138864777_138865078 | 0.39 |
NHSL1 |
NHS-like 1 |
1921 |
0.45 |
| chr4_40844059_40844210 | 0.39 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
399 |
0.83 |
| chr16_67203881_67204335 | 0.39 |
NOL3 |
nucleolar protein 3 (apoptosis repressor with CARD domain) |
51 |
0.91 |
| chr17_13965313_13965500 | 0.39 |
ENSG00000236088 |
. |
7369 |
0.23 |
| chr11_84566576_84566727 | 0.39 |
DLG2 |
discs, large homolog 2 (Drosophila) |
67573 |
0.13 |
| chr1_78836324_78836475 | 0.39 |
ENSG00000212308 |
. |
4554 |
0.3 |
| chr21_17942611_17942934 | 0.39 |
ENSG00000207863 |
. |
19785 |
0.19 |
| chr2_143828948_143829099 | 0.39 |
ARHGAP15 |
Rho GTPase activating protein 15 |
19908 |
0.24 |
| chr7_18832949_18833100 | 0.38 |
ENSG00000222164 |
. |
14878 |
0.28 |
| chr6_144980278_144980429 | 0.38 |
UTRN |
utrophin |
627 |
0.85 |
| chr4_100777669_100777820 | 0.38 |
LAMTOR3 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 3 |
37789 |
0.15 |
| chr19_7744721_7744872 | 0.38 |
TRAPPC5 |
trafficking protein particle complex 5 |
933 |
0.29 |
| chr16_86866838_86867009 | 0.38 |
FOXL1 |
forkhead box L1 |
254808 |
0.02 |
| chr12_120678523_120678674 | 0.38 |
PXN |
paxillin |
9366 |
0.12 |
| chr21_28314071_28314222 | 0.38 |
ENSG00000266133 |
. |
12134 |
0.22 |
| chr18_10593857_10594008 | 0.38 |
NAPG |
N-ethylmaleimide-sensitive factor attachment protein, gamma |
67523 |
0.1 |
| chr15_101611274_101611425 | 0.38 |
RP11-505E24.2 |
|
14922 |
0.21 |
| chr12_109013932_109014083 | 0.37 |
RP11-689B22.2 |
|
8456 |
0.13 |
| chr12_78336292_78336638 | 0.37 |
NAV3 |
neuron navigator 3 |
23591 |
0.28 |
| chr6_152700147_152700338 | 0.37 |
SYNE1-AS1 |
SYNE1 antisense RNA 1 |
1439 |
0.39 |
| chr15_50140285_50140436 | 0.37 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
28532 |
0.19 |
| chr3_13592611_13592832 | 0.37 |
FBLN2 |
fibulin 2 |
2040 |
0.39 |
| chr6_35187804_35187955 | 0.37 |
SCUBE3 |
signal peptide, CUB domain, EGF-like 3 |
5683 |
0.22 |
| chr16_86619409_86619937 | 0.37 |
FOXL1 |
forkhead box L1 |
7558 |
0.18 |
| chr16_51180566_51180717 | 0.37 |
AC009166.5 |
|
2509 |
0.33 |
| chr20_44098779_44099661 | 0.37 |
WFDC2 |
WAP four-disulfide core domain 2 |
824 |
0.44 |
| chr14_102094512_102094862 | 0.37 |
DIO3 |
deiodinase, iodothyronine, type III |
66999 |
0.09 |
| chr17_643901_644052 | 0.37 |
FAM57A |
family with sequence similarity 57, member A |
7611 |
0.14 |
| chr15_76825935_76826086 | 0.37 |
ENSG00000266449 |
. |
52978 |
0.16 |
| chr6_143221298_143221665 | 0.36 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
44857 |
0.18 |
| chr12_6937518_6938631 | 0.36 |
LEPREL2 |
leprecan-like 2 |
502 |
0.55 |
| chr10_103699577_103699728 | 0.36 |
ENSG00000222430 |
. |
12809 |
0.2 |
| chr1_186646717_186646868 | 0.36 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
2767 |
0.4 |
| chr14_33532042_33532240 | 0.36 |
NPAS3 |
neuronal PAS domain protein 3 |
123618 |
0.06 |
| chr15_48351886_48352188 | 0.36 |
SLC24A5 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 5 |
61132 |
0.12 |
| chr10_34827521_34827675 | 0.36 |
PARD3 |
par-3 family cell polarity regulator |
111939 |
0.07 |
| chr6_2857258_2857409 | 0.36 |
ENSG00000266750 |
. |
2992 |
0.24 |
| chr14_95962099_95962345 | 0.36 |
SYNE3 |
spectrin repeat containing, nuclear envelope family member 3 |
20049 |
0.14 |
| chr10_11645069_11645220 | 0.36 |
RP11-138I18.1 |
|
8160 |
0.23 |
| chr8_20060548_20060978 | 0.36 |
ATP6V1B2 |
ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 |
5813 |
0.2 |
| chr4_126311427_126311944 | 0.36 |
FAT4 |
FAT atypical cadherin 4 |
3406 |
0.33 |
| chr3_105104777_105104928 | 0.35 |
ALCAM |
activated leukocyte cell adhesion molecule |
18665 |
0.31 |
| chr11_43945051_43945202 | 0.35 |
C11orf96 |
chromosome 11 open reading frame 96 |
1766 |
0.25 |
| chr21_45006172_45006433 | 0.35 |
HSF2BP |
heat shock transcription factor 2 binding protein |
71723 |
0.1 |
| chr2_242295729_242296527 | 0.35 |
FARP2 |
FERM, RhoGEF and pleckstrin domain protein 2 |
416 |
0.79 |
| chr11_72975754_72976333 | 0.35 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
232 |
0.91 |
| chr1_180881914_180882730 | 0.35 |
KIAA1614 |
KIAA1614 |
3 |
0.98 |
| chr7_120630679_120630951 | 0.35 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
1139 |
0.51 |
| chr8_61821706_61823076 | 0.34 |
RP11-33I11.2 |
|
100226 |
0.08 |
| chr6_36257165_36257316 | 0.34 |
PNPLA1 |
patatin-like phospholipase domain containing 1 |
19003 |
0.17 |
| chr17_36861833_36863162 | 0.34 |
CTB-58E17.3 |
|
17 |
0.9 |
| chr3_30124388_30124539 | 0.34 |
ENSG00000264178 |
. |
55276 |
0.15 |
| chr15_59157116_59157678 | 0.34 |
RNF111 |
ring finger protein 111 |
23 |
0.63 |
| chr1_87858152_87858303 | 0.34 |
ENSG00000199318 |
. |
60829 |
0.14 |
| chr2_20649479_20649630 | 0.34 |
RHOB |
ras homolog family member B |
2719 |
0.28 |
| chr12_50560122_50561166 | 0.34 |
CERS5 |
ceramide synthase 5 |
447 |
0.74 |
| chr1_54200875_54201026 | 0.34 |
GLIS1 |
GLIS family zinc finger 1 |
1073 |
0.53 |
| chr11_19736868_19737019 | 0.34 |
NAV2 |
neuron navigator 2 |
1800 |
0.43 |
| chr1_68258421_68258776 | 0.34 |
ENSG00000238778 |
. |
20262 |
0.19 |
| chr4_13329866_13330017 | 0.34 |
RAB28 |
RAB28, member RAS oncogene family |
53277 |
0.18 |
| chr18_55989081_55989359 | 0.34 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
1332 |
0.5 |
| chr1_110282307_110283361 | 0.34 |
GSTM3 |
glutathione S-transferase mu 3 (brain) |
304 |
0.83 |
| chrX_3629951_3631442 | 0.33 |
PRKX |
protein kinase, X-linked |
953 |
0.63 |
| chr15_49098304_49098486 | 0.33 |
CEP152 |
centrosomal protein 152kDa |
4396 |
0.19 |
| chr6_53934757_53935103 | 0.33 |
MLIP-AS1 |
MLIP antisense RNA 1 |
9594 |
0.22 |
| chr6_111739256_111739407 | 0.33 |
REV3L-IT1 |
REV3L intronic transcript 1 (non-protein coding) |
56521 |
0.11 |
| chr3_66350036_66350187 | 0.33 |
ENSG00000206759 |
. |
6467 |
0.24 |
| chr3_31563579_31563768 | 0.33 |
STT3B |
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
10609 |
0.3 |
| chr16_86542600_86543872 | 0.33 |
FOXF1 |
forkhead box F1 |
897 |
0.62 |
| chr17_67056911_67057062 | 0.33 |
ABCA9 |
ATP-binding cassette, sub-family A (ABC1), member 9 |
61 |
0.98 |
| chr2_160918347_160919586 | 0.33 |
PLA2R1 |
phospholipase A2 receptor 1, 180kDa |
155 |
0.98 |
| chr12_62726310_62726461 | 0.33 |
ENSG00000200814 |
. |
1593 |
0.43 |
| chr1_113070155_113070306 | 0.33 |
RP4-671G15.2 |
|
9167 |
0.18 |
| chr2_182239735_182239886 | 0.33 |
ENSG00000266705 |
. |
69431 |
0.12 |
| chr12_16110077_16110228 | 0.33 |
DERA |
deoxyribose-phosphate aldolase (putative) |
630 |
0.8 |
| chrX_114828631_114828846 | 0.33 |
PLS3 |
plastin 3 |
873 |
0.64 |
| chr1_243687933_243688084 | 0.32 |
RP11-269F20.1 |
|
20826 |
0.27 |
| chrX_117654917_117655068 | 0.32 |
DOCK11 |
dedicator of cytokinesis 11 |
25120 |
0.21 |
| chr7_38885898_38886049 | 0.32 |
VPS41 |
vacuolar protein sorting 41 homolog (S. cerevisiae) |
15027 |
0.28 |
| chr1_9953446_9953936 | 0.32 |
CTNNBIP1 |
catenin, beta interacting protein 1 |
396 |
0.8 |
| chr20_49527457_49527608 | 0.32 |
ADNP |
activity-dependent neuroprotector homeobox |
1204 |
0.39 |
| chr18_34274407_34274743 | 0.32 |
FHOD3 |
formin homology 2 domain containing 3 |
23945 |
0.22 |
| chr15_83240603_83240843 | 0.32 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
105 |
0.95 |
| chr1_12290222_12291451 | 0.32 |
VPS13D |
vacuolar protein sorting 13 homolog D (S. cerevisiae) |
712 |
0.66 |
| chr9_21081854_21082005 | 0.32 |
IFNB1 |
interferon, beta 1, fibroblast |
3986 |
0.21 |
| chr1_32168571_32169014 | 0.32 |
COL16A1 |
collagen, type XVI, alpha 1 |
976 |
0.48 |
| chr1_46378692_46378869 | 0.32 |
MAST2 |
microtubule associated serine/threonine kinase 2 |
480 |
0.87 |
| chr20_33871790_33873092 | 0.32 |
EIF6 |
eukaryotic translation initiation factor 6 |
77 |
0.88 |
| chr15_46147938_46148235 | 0.32 |
SQRDL |
sulfide quinone reductase-like (yeast) |
173348 |
0.03 |
| chr17_56744273_56744616 | 0.32 |
ENSG00000199426 |
. |
386 |
0.76 |
| chr10_78771414_78771608 | 0.31 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
67771 |
0.12 |
| chr16_75600298_75601136 | 0.31 |
GABARAPL2 |
GABA(A) receptor-associated protein-like 2 |
221 |
0.9 |
| chr11_74041520_74041671 | 0.31 |
P4HA3 |
prolyl 4-hydroxylase, alpha polypeptide III |
18893 |
0.16 |
| chr6_72595878_72596126 | 0.31 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
404 |
0.92 |
| chr4_127721162_127721313 | 0.31 |
ENSG00000199862 |
. |
228137 |
0.02 |
| chr3_578313_578483 | 0.31 |
CHL1 |
cell adhesion molecule L1-like |
147270 |
0.05 |
| chr10_63402177_63402609 | 0.31 |
C10orf107 |
chromosome 10 open reading frame 107 |
20326 |
0.24 |
| chr12_91487813_91488090 | 0.31 |
LUM |
lumican |
17657 |
0.21 |
| chr4_114357002_114357251 | 0.31 |
ENSG00000206820 |
. |
15647 |
0.2 |
| chr14_94859853_94860004 | 0.31 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
2898 |
0.25 |
| chr5_159510080_159510300 | 0.31 |
PWWP2A |
PWWP domain containing 2A |
36239 |
0.14 |
| chr2_108202365_108202516 | 0.31 |
RGPD4 |
RANBP2-like and GRIP domain containing 4 |
240948 |
0.02 |
| chr21_29814935_29815086 | 0.31 |
ENSG00000251894 |
. |
300520 |
0.01 |
| chr1_27830414_27830820 | 0.31 |
WASF2 |
WAS protein family, member 2 |
13948 |
0.16 |
| chr10_30574213_30574364 | 0.31 |
ENSG00000200887 |
. |
14314 |
0.19 |
| chr5_135315117_135315268 | 0.31 |
LECT2 |
leukocyte cell-derived chemotaxin 2 |
24469 |
0.18 |
| chr2_95881225_95881376 | 0.31 |
ZNF2 |
zinc finger protein 2 |
49738 |
0.11 |
| chr2_40681805_40681956 | 0.30 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
1302 |
0.63 |
| chr4_159093168_159093728 | 0.30 |
RP11-597D13.9 |
|
281 |
0.64 |
| chr11_65158052_65158759 | 0.30 |
FRMD8 |
FERM domain containing 8 |
4006 |
0.13 |
| chr10_88125313_88125533 | 0.30 |
GRID1 |
glutamate receptor, ionotropic, delta 1 |
812 |
0.7 |
| chr13_76362962_76363113 | 0.30 |
LMO7 |
LIM domain 7 |
63 |
0.98 |
| chr9_115170481_115170632 | 0.30 |
HSDL2 |
hydroxysteroid dehydrogenase like 2 |
28164 |
0.18 |
| chr17_77767116_77767731 | 0.30 |
CBX8 |
chromobox homolog 8 |
3492 |
0.16 |
| chr3_71826949_71827100 | 0.30 |
PROK2 |
prokineticin 2 |
7188 |
0.2 |
| chr2_157912582_157912749 | 0.30 |
ENSG00000263848 |
. |
40896 |
0.21 |
| chr12_29396535_29396686 | 0.30 |
FAR2 |
fatty acyl CoA reductase 2 |
19932 |
0.23 |
| chr4_138787929_138788176 | 0.30 |
ENSG00000250033 |
. |
190566 |
0.03 |
| chr14_73929626_73929971 | 0.30 |
ENSG00000251393 |
. |
669 |
0.64 |
| chr17_1994078_1994475 | 0.30 |
RP11-667K14.5 |
|
1301 |
0.3 |
| chr6_113786265_113786416 | 0.30 |
ENSG00000222677 |
. |
49849 |
0.18 |
| chr1_84630171_84630563 | 0.30 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
3 |
0.99 |
| chr1_4794555_4794706 | 0.30 |
AJAP1 |
adherens junctions associated protein 1 |
79525 |
0.12 |
| chr3_25470358_25470509 | 0.30 |
RARB |
retinoic acid receptor, beta |
631 |
0.81 |
| chr4_166220745_166220896 | 0.30 |
MSMO1 |
methylsterol monooxygenase 1 |
27955 |
0.16 |
| chr20_5142036_5142305 | 0.30 |
CDS2 |
CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 |
17298 |
0.15 |
| chr12_66290552_66290845 | 0.30 |
RP11-366L20.2 |
Uncharacterized protein |
14751 |
0.17 |
| chr4_20196394_20196545 | 0.30 |
SLIT2 |
slit homolog 2 (Drosophila) |
58414 |
0.17 |
| chr9_19161139_19161290 | 0.30 |
PLIN2 |
perilipin 2 |
11938 |
0.17 |
| chr3_32915013_32915164 | 0.30 |
TRIM71 |
tripartite motif containing 71, E3 ubiquitin protein ligase |
55578 |
0.13 |
| chr19_58488964_58489115 | 0.30 |
C19orf18 |
chromosome 19 open reading frame 18 |
3137 |
0.15 |
| chr11_12864005_12864295 | 0.30 |
RP11-47J17.1 |
|
6192 |
0.2 |
| chr12_107631141_107631292 | 0.30 |
BTBD11 |
BTB (POZ) domain containing 11 |
80974 |
0.1 |
| chr1_52132272_52132423 | 0.29 |
OSBPL9 |
oxysterol binding protein-like 9 |
2876 |
0.29 |
| chr16_11663438_11663589 | 0.29 |
LITAF |
lipopolysaccharide-induced TNF factor |
16716 |
0.17 |
| chr19_5036988_5037627 | 0.29 |
KDM4B |
lysine (K)-specific demethylase 4B |
20971 |
0.2 |
| chr6_73905097_73905248 | 0.29 |
KHDC1P1 |
KH homology domain containing 1 pseudogene 1 |
14631 |
0.15 |
| chr4_62239698_62239849 | 0.29 |
LPHN3 |
latrophilin 3 |
123066 |
0.07 |
| chr10_124614021_124614212 | 0.29 |
CUZD1 |
CUB and zona pellucida-like domains 1 |
3807 |
0.21 |
| chr7_7606179_7606870 | 0.29 |
MIOS |
missing oocyte, meiosis regulator, homolog (Drosophila) |
18 |
0.98 |
| chr4_438020_438299 | 0.29 |
ENSG00000252150 |
. |
23266 |
0.14 |
| chr10_24737837_24737988 | 0.29 |
KIAA1217 |
KIAA1217 |
450 |
0.87 |
| chr6_11581235_11581386 | 0.29 |
TMEM170B |
transmembrane protein 170B |
42799 |
0.18 |
| chr18_67507667_67507818 | 0.29 |
CD226 |
CD226 molecule |
106913 |
0.07 |
| chr15_79049667_79049910 | 0.29 |
RP11-160C18.4 |
|
4332 |
0.2 |
| chr17_40157049_40157200 | 0.29 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
6169 |
0.12 |
| chr2_217416160_217416311 | 0.29 |
RPL37A |
ribosomal protein L37a |
52372 |
0.1 |
| chr11_48072208_48072359 | 0.29 |
AC103828.1 |
|
34876 |
0.16 |
| chr6_144471049_144472508 | 0.29 |
STX11 |
syntaxin 11 |
115 |
0.98 |
| chr18_32925570_32925721 | 0.29 |
ZNF24 |
zinc finger protein 24 |
199 |
0.95 |
| chr4_157872609_157873145 | 0.29 |
PDGFC |
platelet derived growth factor C |
19178 |
0.21 |
| chr3_197083218_197083369 | 0.29 |
ENSG00000238491 |
. |
16871 |
0.2 |
| chr15_81773129_81773280 | 0.28 |
TMC3 |
transmembrane channel-like 3 |
106650 |
0.07 |
| chr6_3208796_3209079 | 0.28 |
TUBB2B |
tubulin, beta 2B class IIb |
19032 |
0.15 |
| chr16_28546392_28546543 | 0.28 |
NUPR1 |
nuclear protein, transcriptional regulator, 1 |
3862 |
0.15 |
| chr11_128435928_128436079 | 0.28 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
21450 |
0.21 |
| chr19_47393595_47393746 | 0.28 |
ARHGAP35 |
Rho GTPase activating protein 35 |
28263 |
0.14 |
| chr4_16629429_16629735 | 0.28 |
LDB2 |
LIM domain binding 2 |
32084 |
0.25 |
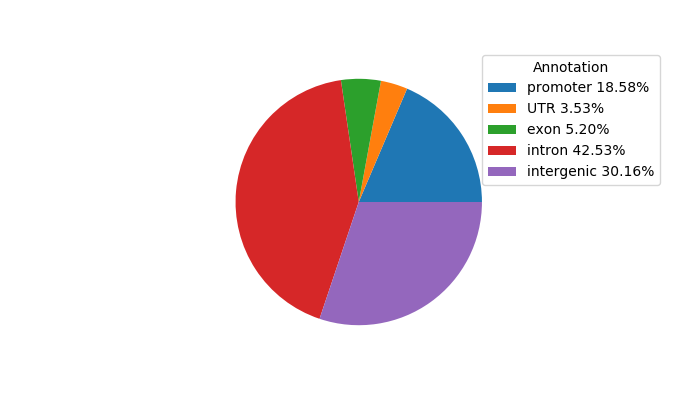
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.2 | 0.8 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.1 | 0.5 | GO:0031394 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.1 | 0.5 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.1 | 0.1 | GO:0060502 | epithelial cell proliferation involved in lung morphogenesis(GO:0060502) |
| 0.1 | 0.5 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.1 | 0.4 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.1 | 0.3 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.3 | GO:0072201 | negative regulation of mesenchymal cell proliferation(GO:0072201) |
| 0.1 | 0.3 | GO:0052033 | pathogen-associated molecular pattern dependent induction by symbiont of host innate immune response(GO:0052033) positive regulation by symbiont of host innate immune response(GO:0052166) modulation by symbiont of host innate immune response(GO:0052167) pathogen-associated molecular pattern dependent modulation by symbiont of host innate immune response(GO:0052169) pathogen-associated molecular pattern dependent induction by organism of innate immune response of other organism involved in symbiotic interaction(GO:0052257) positive regulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052305) modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052306) pathogen-associated molecular pattern dependent modulation by organism of innate immune response in other organism involved in symbiotic interaction(GO:0052308) |
| 0.1 | 0.2 | GO:0032413 | negative regulation of ion transmembrane transporter activity(GO:0032413) |
| 0.1 | 0.5 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.1 | 0.4 | GO:0043570 | maintenance of DNA repeat elements(GO:0043570) |
| 0.1 | 0.3 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.1 | 0.3 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.1 | 0.2 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.1 | 0.2 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.1 | 0.3 | GO:0090400 | stress-induced premature senescence(GO:0090400) |
| 0.1 | 0.2 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.1 | 0.1 | GO:0007195 | adenylate cyclase-inhibiting dopamine receptor signaling pathway(GO:0007195) |
| 0.1 | 0.1 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.1 | 0.2 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.3 | GO:1903672 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 | 0.5 | GO:1901532 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.1 | 0.1 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.1 | 0.3 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.1 | 0.2 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 0.3 | GO:0060536 | trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.1 | 0.2 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.1 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.2 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.3 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.2 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.1 | 0.2 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.1 | 0.1 | GO:0090009 | primitive streak formation(GO:0090009) |
| 0.1 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.1 | 0.3 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.1 | 0.2 | GO:0060430 | lung saccule development(GO:0060430) Type II pneumocyte differentiation(GO:0060510) |
| 0.1 | 0.2 | GO:0035461 | vitamin transmembrane transport(GO:0035461) |
| 0.1 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.1 | 0.1 | GO:0060394 | negative regulation of pathway-restricted SMAD protein phosphorylation(GO:0060394) |
| 0.1 | 0.3 | GO:0071549 | cellular response to dexamethasone stimulus(GO:0071549) |
| 0.1 | 0.3 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.3 | GO:0042090 | interleukin-12 biosynthetic process(GO:0042090) regulation of interleukin-12 biosynthetic process(GO:0045075) |
| 0.0 | 0.1 | GO:0010587 | miRNA metabolic process(GO:0010586) miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.2 | GO:0045630 | positive regulation of T-helper 2 cell differentiation(GO:0045630) |
| 0.0 | 0.2 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0070779 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.1 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.2 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.2 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.0 | GO:2000542 | negative regulation of gastrulation(GO:2000542) |
| 0.0 | 0.1 | GO:0035610 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.3 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.1 | GO:2000647 | negative regulation of stem cell proliferation(GO:2000647) |
| 0.0 | 0.0 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.2 | GO:0002138 | retinoic acid biosynthetic process(GO:0002138) diterpenoid biosynthetic process(GO:0016102) |
| 0.0 | 0.2 | GO:0008354 | germ cell migration(GO:0008354) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.2 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.2 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.2 | GO:0060259 | regulation of feeding behavior(GO:0060259) |
| 0.0 | 0.1 | GO:0048755 | branching morphogenesis of a nerve(GO:0048755) |
| 0.0 | 0.8 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.1 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.1 | GO:0007500 | mesodermal cell fate determination(GO:0007500) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.3 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.0 | GO:0032226 | positive regulation of synaptic transmission, dopaminergic(GO:0032226) positive regulation of dopamine secretion(GO:0033603) |
| 0.0 | 0.0 | GO:0001957 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0021932 | hindbrain radial glia guided cell migration(GO:0021932) |
| 0.0 | 0.1 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.0 | GO:0046136 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0032048 | phosphatidylglycerol biosynthetic process(GO:0006655) cardiolipin metabolic process(GO:0032048) cardiolipin biosynthetic process(GO:0032049) phosphatidylglycerol metabolic process(GO:0046471) |
| 0.0 | 0.1 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0007619 | courtship behavior(GO:0007619) |
| 0.0 | 0.1 | GO:0072311 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0019348 | polyprenol metabolic process(GO:0016093) dolichol metabolic process(GO:0019348) |
| 0.0 | 0.2 | GO:0016322 | neuron remodeling(GO:0016322) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0000052 | citrulline metabolic process(GO:0000052) |
| 0.0 | 0.1 | GO:2000050 | regulation of non-canonical Wnt signaling pathway(GO:2000050) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.1 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) chloride ion homeostasis(GO:0055064) |
| 0.0 | 0.3 | GO:0045600 | positive regulation of fat cell differentiation(GO:0045600) |
| 0.0 | 0.1 | GO:0090083 | regulation of inclusion body assembly(GO:0090083) |
| 0.0 | 0.2 | GO:0030238 | male sex determination(GO:0030238) |
| 0.0 | 0.4 | GO:0035136 | forelimb morphogenesis(GO:0035136) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.1 | GO:0048263 | determination of dorsal identity(GO:0048263) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.0 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) apoptotic process involved in luteolysis(GO:0061364) |
| 0.0 | 0.1 | GO:0001878 | response to yeast(GO:0001878) |
| 0.0 | 0.1 | GO:0031269 | pseudopodium assembly(GO:0031269) regulation of pseudopodium assembly(GO:0031272) positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.1 | GO:1901021 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of calcium ion transmembrane transporter activity(GO:1901021) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) positive regulation of cation channel activity(GO:2001259) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0060677 | ureteric bud elongation(GO:0060677) |
| 0.0 | 0.2 | GO:0032781 | positive regulation of ATPase activity(GO:0032781) |
| 0.0 | 0.1 | GO:0042693 | muscle cell fate commitment(GO:0042693) |
| 0.0 | 0.1 | GO:0043312 | neutrophil activation involved in immune response(GO:0002283) neutrophil degranulation(GO:0043312) |
| 0.0 | 0.0 | GO:0002042 | cell migration involved in sprouting angiogenesis(GO:0002042) |
| 0.0 | 0.2 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.0 | GO:0042637 | catagen(GO:0042637) |
| 0.0 | 0.0 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.0 | GO:1900078 | positive regulation of insulin receptor signaling pathway(GO:0046628) positive regulation of cellular response to insulin stimulus(GO:1900078) |
| 0.0 | 0.0 | GO:0003207 | cardiac chamber formation(GO:0003207) cardiac ventricle formation(GO:0003211) |
| 0.0 | 0.1 | GO:0048867 | stem cell fate commitment(GO:0048865) stem cell fate determination(GO:0048867) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:1902692 | positive regulation of neuroblast proliferation(GO:0002052) regulation of neuroblast proliferation(GO:1902692) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.0 | GO:0045064 | T-helper 2 cell differentiation(GO:0045064) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0090047 | obsolete positive regulation of transcription regulator activity(GO:0090047) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0048715 | negative regulation of oligodendrocyte differentiation(GO:0048715) |
| 0.0 | 0.5 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.1 | GO:1903393 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.1 | GO:1903312 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.1 | GO:0045897 | regulation of transcription during mitosis(GO:0045896) positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.0 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0033148 | positive regulation of intracellular estrogen receptor signaling pathway(GO:0033148) |
| 0.0 | 0.1 | GO:0007571 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.1 | GO:0007413 | axonal fasciculation(GO:0007413) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0031017 | exocrine pancreas development(GO:0031017) |
| 0.0 | 0.1 | GO:0002063 | chondrocyte development(GO:0002063) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:0002830 | positive regulation of type 2 immune response(GO:0002830) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0046856 | phosphatidylinositol dephosphorylation(GO:0046856) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.1 | GO:0048625 | myoblast fate commitment(GO:0048625) |
| 0.0 | 0.1 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.1 | GO:0033962 | cytoplasmic mRNA processing body assembly(GO:0033962) |
| 0.0 | 0.0 | GO:0010944 | negative regulation of transcription by competitive promoter binding(GO:0010944) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0009649 | entrainment of circadian clock(GO:0009649) |
| 0.0 | 0.1 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.0 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.1 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.3 | GO:0042255 | ribosome assembly(GO:0042255) |
| 0.0 | 0.1 | GO:0033194 | response to hydroperoxide(GO:0033194) |
| 0.0 | 0.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0043537 | negative regulation of blood vessel endothelial cell migration(GO:0043537) |
| 0.0 | 0.1 | GO:0048566 | embryonic digestive tract development(GO:0048566) |
| 0.0 | 0.1 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.1 | GO:0051898 | negative regulation of protein kinase B signaling(GO:0051898) |
| 0.0 | 0.3 | GO:0050771 | negative regulation of axonogenesis(GO:0050771) |
| 0.0 | 0.1 | GO:0032352 | positive regulation of hormone metabolic process(GO:0032352) positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.1 | GO:0071731 | response to nitric oxide(GO:0071731) |
| 0.0 | 0.1 | GO:0045746 | negative regulation of Notch signaling pathway(GO:0045746) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.1 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.1 | GO:0002024 | diet induced thermogenesis(GO:0002024) adaptive thermogenesis(GO:1990845) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.2 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.1 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.1 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.0 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.0 | 0.0 | GO:0010665 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.0 | 0.1 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.0 | GO:0048853 | forebrain morphogenesis(GO:0048853) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0070266 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.0 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.0 | GO:0043049 | otic placode formation(GO:0043049) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.2 | GO:0045987 | positive regulation of smooth muscle contraction(GO:0045987) |
| 0.0 | 0.0 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:0016242 | negative regulation of macroautophagy(GO:0016242) |
| 0.0 | 0.0 | GO:0006931 | substrate-dependent cell migration, cell attachment to substrate(GO:0006931) |
| 0.0 | 0.3 | GO:0001755 | neural crest cell migration(GO:0001755) |
| 0.0 | 0.1 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.0 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:0030422 | production of siRNA involved in RNA interference(GO:0030422) |
| 0.0 | 0.0 | GO:0090197 | chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.0 | GO:0014834 | skeletal muscle satellite cell maintenance involved in skeletal muscle regeneration(GO:0014834) |
| 0.0 | 0.0 | GO:0043129 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.3 | GO:0009620 | response to fungus(GO:0009620) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.0 | GO:0010919 | regulation of inositol phosphate biosynthetic process(GO:0010919) |
| 0.0 | 0.1 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.0 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:0006154 | adenosine catabolic process(GO:0006154) |
| 0.0 | 0.1 | GO:0090231 | regulation of spindle checkpoint(GO:0090231) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.1 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0016079 | synaptic vesicle exocytosis(GO:0016079) |
| 0.0 | 0.0 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
| 0.0 | 0.0 | GO:0032823 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.1 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.1 | GO:2000258 | negative regulation of complement activation(GO:0045916) negative regulation of protein activation cascade(GO:2000258) |
| 0.0 | 0.0 | GO:0060405 | regulation of penile erection(GO:0060405) positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.0 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.0 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.0 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.0 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.0 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.0 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.0 | GO:0033147 | negative regulation of intracellular estrogen receptor signaling pathway(GO:0033147) |
| 0.0 | 0.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0060840 | artery development(GO:0060840) |
| 0.0 | 0.0 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.0 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.1 | GO:0002027 | regulation of heart rate(GO:0002027) |
| 0.0 | 0.2 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.0 | GO:0060297 | regulation of sarcomere organization(GO:0060297) |
| 0.0 | 0.0 | GO:0060900 | embryonic camera-type eye formation(GO:0060900) |
| 0.0 | 0.0 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 1.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.0 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.2 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.1 | GO:0005513 | detection of calcium ion(GO:0005513) |
| 0.0 | 0.1 | GO:0016264 | gap junction assembly(GO:0016264) |
| 0.0 | 0.0 | GO:0042538 | hyperosmotic salinity response(GO:0042538) |
| 0.0 | 0.0 | GO:0006999 | nuclear pore organization(GO:0006999) |
| 0.0 | 0.0 | GO:0006435 | threonyl-tRNA aminoacylation(GO:0006435) |
| 0.0 | 0.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.0 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.0 | GO:1901976 | regulation of cell cycle checkpoint(GO:1901976) |
| 0.0 | 0.0 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.0 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.0 | GO:0046185 | aldehyde catabolic process(GO:0046185) |
| 0.0 | 0.1 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.0 | GO:0052510 | modulation by symbiont of host defense response(GO:0052031) modulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052255) positive regulation by symbiont of host defense response(GO:0052509) positive regulation by organism of defense response of other organism involved in symbiotic interaction(GO:0052510) modulation by organism of immune response of other organism involved in symbiotic interaction(GO:0052552) modulation by symbiont of host immune response(GO:0052553) |
| 0.0 | 0.0 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.2 | GO:0042597 | outer membrane-bounded periplasmic space(GO:0030288) periplasmic space(GO:0042597) |
| 0.1 | 0.2 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.1 | 0.2 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.2 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.1 | 0.3 | GO:0071437 | invadopodium(GO:0071437) |
| 0.1 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.3 | GO:0005593 | FACIT collagen trimer(GO:0005593) |
| 0.0 | 0.1 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.3 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.7 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.3 | GO:0098651 | basement membrane collagen trimer(GO:0098651) |
| 0.0 | 0.2 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0031512 | motile primary cilium(GO:0031512) |
| 0.0 | 0.3 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.3 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.2 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.2 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 0.7 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.3 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.3 | GO:0000421 | autophagosome membrane(GO:0000421) |
| 0.0 | 0.1 | GO:0030904 | retromer complex(GO:0030904) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.0 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.0 | GO:0000805 | X chromosome(GO:0000805) |
| 0.0 | 0.1 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.2 | GO:0016581 | NuRD complex(GO:0016581) CHD-type complex(GO:0090545) nuclear transcriptional repressor complex(GO:0090568) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0005858 | axonemal dynein complex(GO:0005858) |
| 0.0 | 0.1 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0032391 | photoreceptor connecting cilium(GO:0032391) |
| 0.0 | 0.0 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0060293 | P granule(GO:0043186) pole plasm(GO:0045495) germ plasm(GO:0060293) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.2 | GO:0005891 | voltage-gated calcium channel complex(GO:0005891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.4 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.2 | GO:0043559 | insulin binding(GO:0043559) |
| 0.1 | 0.4 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.1 | 0.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.1 | 0.5 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.3 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.2 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.1 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.1 | 0.3 | GO:0005000 | vasopressin receptor activity(GO:0005000) |
| 0.1 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.3 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.2 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.1 | 0.2 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.2 | GO:0008518 | reduced folate carrier activity(GO:0008518) |
| 0.1 | 0.2 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.4 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.2 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.1 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.2 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.2 | GO:0070696 | transmembrane receptor protein serine/threonine kinase binding(GO:0070696) |
| 0.0 | 0.2 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.3 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.2 | GO:0004331 | fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.3 | GO:0043548 | phosphatidylinositol 3-kinase binding(GO:0043548) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.3 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.0 | 0.3 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.0 | 0.1 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0097493 | extracellular matrix constituent conferring elasticity(GO:0030023) structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.2 | GO:0004954 | prostanoid receptor activity(GO:0004954) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.0 | 0.1 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.2 | GO:0022821 | potassium ion antiporter activity(GO:0022821) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0004022 | alcohol dehydrogenase (NAD) activity(GO:0004022) |
| 0.0 | 0.4 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0016832 | aldehyde-lyase activity(GO:0016832) |
| 0.0 | 0.7 | GO:0005080 | protein kinase C binding(GO:0005080) |
| 0.0 | 0.1 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.4 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.0 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.1 | GO:0008504 | monoamine transmembrane transporter activity(GO:0008504) |
| 0.0 | 0.1 | GO:0008331 | high voltage-gated calcium channel activity(GO:0008331) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.3 | GO:0005003 | ephrin receptor activity(GO:0005003) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0047115 | trans-1,2-dihydrobenzene-1,2-diol dehydrogenase activity(GO:0047115) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.0 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 1.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.0 | 0.0 | GO:0004645 | phosphorylase activity(GO:0004645) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.1 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.1 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.3 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.1 | GO:0031545 | procollagen-proline 4-dioxygenase activity(GO:0004656) peptidyl-proline 4-dioxygenase activity(GO:0031545) |
| 0.0 | 0.2 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.2 | GO:0005242 | inward rectifier potassium channel activity(GO:0005242) |
| 0.0 | 0.0 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.0 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.0 | GO:0005314 | high-affinity glutamate transmembrane transporter activity(GO:0005314) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.1 | GO:0015168 | glycerol transmembrane transporter activity(GO:0015168) |
| 0.0 | 0.2 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.2 | GO:0004970 | ionotropic glutamate receptor activity(GO:0004970) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0046790 | virion binding(GO:0046790) |
| 0.0 | 0.3 | GO:0005245 | voltage-gated calcium channel activity(GO:0005245) |
| 0.0 | 0.1 | GO:0042285 | UDP-xylosyltransferase activity(GO:0035252) xylosyltransferase activity(GO:0042285) |
| 0.0 | 0.1 | GO:0016813 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in linear amidines(GO:0016813) |
| 0.0 | 0.0 | GO:0033765 | steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) |
| 0.0 | 0.0 | GO:0019958 | C-X-C chemokine binding(GO:0019958) interleukin-8 binding(GO:0019959) |
| 0.0 | 0.0 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.0 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.1 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.0 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0008187 | poly-pyrimidine tract binding(GO:0008187) |
| 0.0 | 0.0 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.0 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.0 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.0 | GO:0019531 | oxalate transmembrane transporter activity(GO:0019531) |
| 0.0 | 0.0 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.0 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.0 | GO:0004673 | protein histidine kinase activity(GO:0004673) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0004738 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.0 | GO:0004829 | threonine-tRNA ligase activity(GO:0004829) |
| 0.0 | 0.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.0 | GO:0047756 | chondroitin 4-sulfotransferase activity(GO:0047756) |
| 0.0 | 0.0 | GO:0000268 | peroxisome targeting sequence binding(GO:0000268) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.0 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.0 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | SIG BCR SIGNALING PATHWAY | Members of the BCR signaling pathway |
| 0.1 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.1 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.0 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.2 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.4 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.1 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 0.1 | PID INTEGRIN A9B1 PATHWAY | Alpha9 beta1 integrin signaling events |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.1 | PID LYSOPHOSPHOLIPID PATHWAY | LPA receptor mediated events |
| 0.0 | 3.5 | NABA CORE MATRISOME | Ensemble of genes encoding core extracellular matrix including ECM glycoproteins, collagens and proteoglycans |
| 0.0 | 0.6 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.3 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.4 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.1 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.5 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.3 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.1 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.2 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.3 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.3 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.1 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.3 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.2 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME LIGAND GATED ION CHANNEL TRANSPORT | Genes involved in Ligand-gated ion channel transport |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.3 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.0 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.0 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME CELL CELL JUNCTION ORGANIZATION | Genes involved in Cell-cell junction organization |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.1 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |