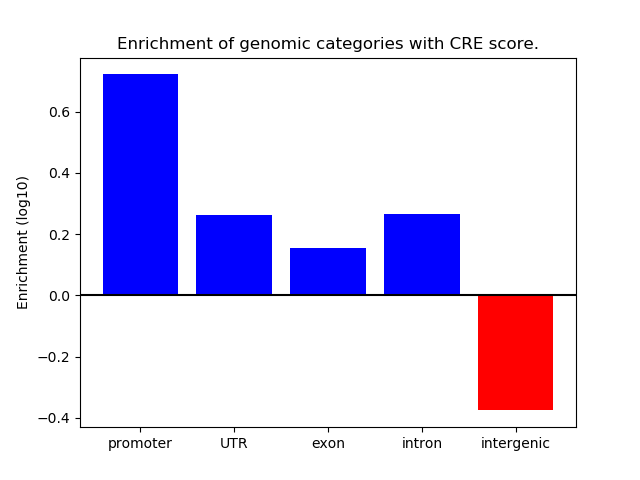
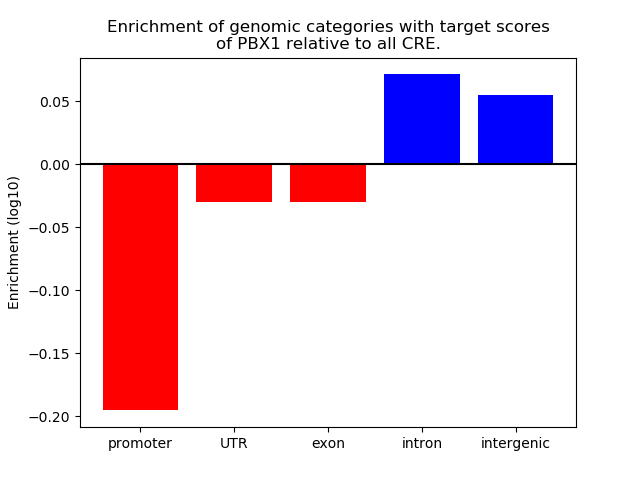
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
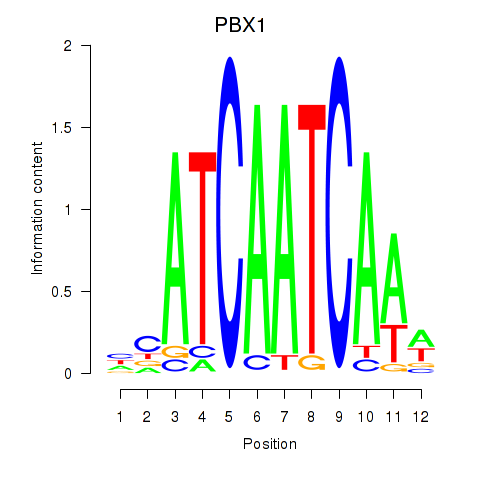
Results for PBX1
Z-value: 0.88
Transcription factors associated with PBX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX1
|
ENSG00000185630.14 | PBX homeobox 1 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr1_164532740_164533333 | PBX1 | 994 | 0.668070 | 0.90 | 1.0e-03 | Click! |
| chr1_164545287_164545502 | PBX1 | 13352 | 0.253667 | 0.84 | 5.1e-03 | Click! |
| chr1_164532131_164532342 | PBX1 | 194 | 0.965561 | 0.75 | 1.9e-02 | Click! |
| chr1_164530790_164530941 | PBX1 | 1177 | 0.609321 | 0.75 | 2.0e-02 | Click! |
| chr1_164545611_164545762 | PBX1 | 13644 | 0.252692 | 0.73 | 2.6e-02 | Click! |
Activity of the PBX1 motif across conditions
Conditions sorted by the z-value of the PBX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr5_54456456_54456818 | 0.39 |
GPX8 |
glutathione peroxidase 8 (putative) |
639 |
0.58 |
| chrX_34673939_34674563 | 0.37 |
TMEM47 |
transmembrane protein 47 |
1154 |
0.68 |
| chr12_85305887_85306488 | 0.36 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
384 |
0.92 |
| chr7_28429293_28429606 | 0.35 |
CREB5 |
cAMP responsive element binding protein 5 |
19557 |
0.28 |
| chr9_134875172_134875430 | 0.35 |
MED27 |
mediator complex subunit 27 |
79952 |
0.1 |
| chr10_33620428_33620906 | 0.33 |
NRP1 |
neuropilin 1 |
2643 |
0.37 |
| chr2_3625481_3625934 | 0.33 |
ENSG00000252531 |
. |
2453 |
0.16 |
| chr5_102202832_102203172 | 0.32 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
1182 |
0.66 |
| chr7_46330968_46331124 | 0.32 |
ENSG00000239004 |
. |
316009 |
0.01 |
| chr6_121756849_121757212 | 0.32 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
192 |
0.94 |
| chr8_19148668_19149118 | 0.31 |
SH2D4A |
SH2 domain containing 4A |
22235 |
0.28 |
| chr10_73725086_73725706 | 0.31 |
CHST3 |
carbohydrate (chondroitin 6) sulfotransferase 3 |
1273 |
0.55 |
| chr6_73263483_73264009 | 0.30 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
67774 |
0.12 |
| chr11_12699031_12699547 | 0.30 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
2239 |
0.43 |
| chr8_126557595_126557859 | 0.30 |
ENSG00000266452 |
. |
100920 |
0.08 |
| chr14_55144332_55144669 | 0.29 |
SAMD4A |
sterile alpha motif domain containing 4A |
77051 |
0.1 |
| chr9_18474095_18474958 | 0.29 |
ADAMTSL1 |
ADAMTS-like 1 |
295 |
0.95 |
| chr18_53214040_53214242 | 0.29 |
TCF4 |
transcription factor 4 |
36141 |
0.18 |
| chr6_165722551_165723097 | 0.28 |
C6orf118 |
chromosome 6 open reading frame 118 |
272 |
0.95 |
| chr7_111002999_111003219 | 0.28 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
29862 |
0.26 |
| chr4_114899439_114899761 | 0.27 |
ARSJ |
arylsulfatase family, member J |
552 |
0.84 |
| chr2_74968036_74968578 | 0.27 |
SEMA4F |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F |
86869 |
0.07 |
| chr15_35598532_35598857 | 0.27 |
ENSG00000265102 |
. |
65871 |
0.14 |
| chr1_164532740_164533333 | 0.26 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
994 |
0.67 |
| chr1_235904828_235905136 | 0.26 |
LYST |
lysosomal trafficking regulator |
10489 |
0.24 |
| chr4_183066487_183066672 | 0.26 |
AC108142.1 |
|
177 |
0.77 |
| chr2_190044545_190044696 | 0.26 |
COL5A2 |
collagen, type V, alpha 2 |
15 |
0.98 |
| chr3_87039220_87039752 | 0.24 |
VGLL3 |
vestigial like 3 (Drosophila) |
366 |
0.93 |
| chr11_122072235_122072431 | 0.24 |
ENSG00000207994 |
. |
49317 |
0.12 |
| chr4_177931083_177931234 | 0.24 |
ENSG00000222859 |
. |
171438 |
0.03 |
| chr21_28215601_28215791 | 0.24 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
163 |
0.97 |
| chr5_42423266_42423460 | 0.24 |
GHR |
growth hormone receptor |
516 |
0.88 |
| chr4_30726767_30727321 | 0.24 |
PCDH7 |
protocadherin 7 |
3067 |
0.38 |
| chr1_51726454_51726959 | 0.24 |
RP11-296A18.6 |
|
5040 |
0.16 |
| chr7_78398482_78399243 | 0.24 |
MAGI2 |
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
1534 |
0.56 |
| chr6_139116835_139117557 | 0.23 |
ECT2L |
epithelial cell transforming sequence 2 oncogene-like |
61 |
0.98 |
| chr5_174162455_174162926 | 0.23 |
MSX2 |
msh homeobox 2 |
11085 |
0.25 |
| chr13_38444595_38444746 | 0.23 |
TRPC4 |
transient receptor potential cation channel, subfamily C, member 4 |
108 |
0.98 |
| chr12_88971091_88971358 | 0.23 |
KITLG |
KIT ligand |
3014 |
0.3 |
| chr3_153840530_153840806 | 0.23 |
ARHGEF26 |
Rho guanine nucleotide exchange factor (GEF) 26 |
1439 |
0.37 |
| chr11_125929052_125929203 | 0.23 |
CDON |
cell adhesion associated, oncogene regulated |
3579 |
0.26 |
| chr12_14537227_14537378 | 0.23 |
ATF7IP |
activating transcription factor 7 interacting protein |
696 |
0.76 |
| chr10_33299780_33299931 | 0.23 |
ITGB1 |
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) |
5135 |
0.26 |
| chr7_54731814_54732170 | 0.23 |
ENSG00000200471 |
. |
43274 |
0.17 |
| chr4_177192718_177192869 | 0.23 |
ASB5 |
ankyrin repeat and SOCS box containing 5 |
2420 |
0.32 |
| chrX_11456470_11456766 | 0.22 |
ARHGAP6 |
Rho GTPase activating protein 6 |
10725 |
0.3 |
| chr6_169652381_169652976 | 0.22 |
THBS2 |
thrombospondin 2 |
432 |
0.89 |
| chr20_14314803_14314963 | 0.22 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
3371 |
0.3 |
| chr13_38172711_38172913 | 0.22 |
POSTN |
periostin, osteoblast specific factor |
51 |
0.99 |
| chr15_72529055_72529849 | 0.22 |
PKM |
pyruvate kinase, muscle |
5288 |
0.17 |
| chrX_33228916_33229133 | 0.22 |
DMD |
dystrophin |
405 |
0.92 |
| chr2_221969079_221969387 | 0.22 |
EPHA4 |
EPH receptor A4 |
398047 |
0.01 |
| chr1_147148877_147149210 | 0.22 |
ACP6 |
acid phosphatase 6, lysophosphatidic |
6425 |
0.22 |
| chr8_37556496_37556710 | 0.22 |
ZNF703 |
zinc finger protein 703 |
3334 |
0.17 |
| chr3_55513844_55514078 | 0.22 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
1263 |
0.55 |
| chr11_35512807_35513203 | 0.22 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
34146 |
0.18 |
| chr5_17021245_17021396 | 0.21 |
BASP1 |
brain abundant, membrane attached signal protein 1 |
44387 |
0.15 |
| chr12_31480010_31480161 | 0.21 |
FAM60A |
family with sequence similarity 60, member A |
93 |
0.96 |
| chr8_119016675_119016955 | 0.21 |
EXT1 |
exostosin glycosyltransferase 1 |
105838 |
0.08 |
| chr1_100112033_100112261 | 0.21 |
PALMD |
palmdelphin |
398 |
0.9 |
| chr1_95009725_95009876 | 0.21 |
F3 |
coagulation factor III (thromboplastin, tissue factor) |
2444 |
0.41 |
| chr4_187646262_187646501 | 0.21 |
FAT1 |
FAT atypical cadherin 1 |
1372 |
0.58 |
| chr3_86992006_86992157 | 0.21 |
VGLL3 |
vestigial like 3 (Drosophila) |
47771 |
0.2 |
| chr1_95388573_95389173 | 0.21 |
CNN3 |
calponin 3, acidic |
2464 |
0.27 |
| chr3_29735259_29735935 | 0.21 |
RBMS3-AS2 |
RBMS3 antisense RNA 2 |
51297 |
0.19 |
| chr8_106743100_106743756 | 0.21 |
RP11-642D21.2 |
|
45435 |
0.16 |
| chr12_54089934_54090280 | 0.21 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
18306 |
0.14 |
| chr3_55514769_55515136 | 0.21 |
WNT5A |
wingless-type MMTV integration site family, member 5A |
272 |
0.94 |
| chr7_112626035_112626431 | 0.21 |
C7orf60 |
chromosome 7 open reading frame 60 |
46262 |
0.17 |
| chr7_80377911_80378104 | 0.21 |
CD36 |
CD36 molecule (thrombospondin receptor) |
75315 |
0.12 |
| chr3_116163075_116163431 | 0.21 |
LSAMP |
limbic system-associated membrane protein |
577 |
0.84 |
| chr1_19947101_19947252 | 0.21 |
NBL1 |
neuroblastoma 1, DAN family BMP antagonist |
19872 |
0.12 |
| chr7_80546807_80546971 | 0.21 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
1610 |
0.56 |
| chr10_89307360_89307659 | 0.21 |
ENSG00000222192 |
. |
15434 |
0.21 |
| chr8_89337580_89337731 | 0.21 |
RP11-586K2.1 |
|
1410 |
0.42 |
| chrX_28605228_28605520 | 0.21 |
IL1RAPL1 |
interleukin 1 receptor accessory protein-like 1 |
142 |
0.98 |
| chr16_80840676_80840857 | 0.20 |
CDYL2 |
chromodomain protein, Y-like 2 |
2540 |
0.35 |
| chr8_38205354_38205751 | 0.20 |
RP11-513D5.2 |
|
12053 |
0.13 |
| chr3_87383057_87383208 | 0.20 |
POU1F1 |
POU class 1 homeobox 1 |
57395 |
0.15 |
| chr20_17553764_17553929 | 0.20 |
DSTN |
destrin (actin depolymerizing factor) |
3119 |
0.21 |
| chr10_36184684_36184984 | 0.20 |
FZD8 |
frizzled family receptor 8 |
254472 |
0.02 |
| chr2_64876893_64877273 | 0.20 |
SERTAD2 |
SERTA domain containing 2 |
3964 |
0.28 |
| chr12_88972313_88972545 | 0.20 |
KITLG |
KIT ligand |
1809 |
0.39 |
| chr16_2177423_2177723 | 0.20 |
ENSG00000200059 |
. |
4279 |
0.06 |
| chr2_235933296_235933447 | 0.20 |
SH3BP4 |
SH3-domain binding protein 4 |
29887 |
0.26 |
| chr6_169653692_169653887 | 0.20 |
THBS2 |
thrombospondin 2 |
350 |
0.92 |
| chr18_25751825_25751976 | 0.20 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
5510 |
0.36 |
| chr3_120168336_120168615 | 0.20 |
FSTL1 |
follistatin-like 1 |
1363 |
0.54 |
| chr9_112813940_112814168 | 0.20 |
AKAP2 |
A kinase (PRKA) anchor protein 2 |
3078 |
0.38 |
| chr9_93058938_93059089 | 0.20 |
ENSG00000263967 |
. |
273196 |
0.02 |
| chr10_65473071_65473288 | 0.20 |
REEP3 |
receptor accessory protein 3 |
192056 |
0.03 |
| chr22_40894900_40895196 | 0.20 |
RP4-591N18.2 |
|
22756 |
0.15 |
| chr22_39711605_39711921 | 0.19 |
ENSG00000209482 |
. |
451 |
0.66 |
| chr17_69629060_69629258 | 0.19 |
ENSG00000222563 |
. |
321850 |
0.01 |
| chr1_95390838_95390989 | 0.19 |
CNN3 |
calponin 3, acidic |
424 |
0.81 |
| chr6_28641788_28642196 | 0.19 |
ENSG00000272278 |
. |
25950 |
0.18 |
| chr13_40176110_40176261 | 0.19 |
LHFP |
lipoma HMGIC fusion partner |
1123 |
0.55 |
| chr18_57362819_57362970 | 0.19 |
RP11-2N1.2 |
|
797 |
0.55 |
| chr12_54391177_54391923 | 0.19 |
HOXC-AS2 |
HOXC cluster antisense RNA 2 |
981 |
0.22 |
| chr7_116167584_116167883 | 0.19 |
CAV1 |
caveolin 1, caveolae protein, 22kDa |
1386 |
0.37 |
| chr2_111206913_111207281 | 0.19 |
RP13-1039J1.3 |
|
278 |
0.91 |
| chr11_27722222_27723091 | 0.19 |
BDNF |
brain-derived neurotrophic factor |
56 |
0.98 |
| chr7_134465998_134466492 | 0.19 |
CALD1 |
caldesmon 1 |
1816 |
0.49 |
| chr18_48571368_48571694 | 0.19 |
SMAD4 |
SMAD family member 4 |
202 |
0.96 |
| chr5_145318270_145318548 | 0.19 |
SH3RF2 |
SH3 domain containing ring finger 2 |
969 |
0.66 |
| chr11_101991103_101991395 | 0.19 |
YAP1 |
Yes-associated protein 1 |
8004 |
0.18 |
| chr21_35899555_35899706 | 0.19 |
RCAN1 |
regulator of calcineurin 1 |
369 |
0.87 |
| chr10_8201743_8201894 | 0.19 |
GATA3 |
GATA binding protein 3 |
105049 |
0.08 |
| chr10_8635845_8635996 | 0.19 |
ENSG00000212505 |
. |
62874 |
0.16 |
| chr2_151339584_151339865 | 0.19 |
RND3 |
Rho family GTPase 3 |
2172 |
0.49 |
| chr6_3156631_3156782 | 0.19 |
TUBB2A |
tubulin, beta 2A class IIa |
1054 |
0.43 |
| chr19_20149056_20149207 | 0.19 |
ZNF682 |
zinc finger protein 682 |
883 |
0.56 |
| chr14_23447971_23448151 | 0.19 |
AJUBA |
ajuba LIM protein |
1115 |
0.25 |
| chr6_44424385_44424803 | 0.19 |
ENSG00000266619 |
. |
21216 |
0.17 |
| chrX_45482024_45482567 | 0.19 |
ENSG00000207870 |
. |
123399 |
0.06 |
| chr12_77457624_77458072 | 0.19 |
E2F7 |
E2F transcription factor 7 |
1482 |
0.56 |
| chr3_149298860_149299373 | 0.18 |
WWTR1 |
WW domain containing transcription regulator 1 |
5077 |
0.23 |
| chr1_20810069_20810220 | 0.18 |
CAMK2N1 |
calcium/calmodulin-dependent protein kinase II inhibitor 1 |
2569 |
0.29 |
| chr15_75942874_75943101 | 0.18 |
SNX33 |
sorting nexin 33 |
890 |
0.37 |
| chr15_40339032_40339183 | 0.18 |
SRP14 |
signal recognition particle 14kDa (homologous Alu RNA binding protein) |
7718 |
0.16 |
| chr1_27425556_27426394 | 0.18 |
SLC9A1 |
solute carrier family 9, subfamily A (NHE1, cation proton antiporter 1), member 1 |
54998 |
0.1 |
| chr8_42063953_42065062 | 0.18 |
PLAT |
plasminogen activator, tissue |
576 |
0.71 |
| chr11_121968787_121969071 | 0.18 |
ENSG00000207971 |
. |
1623 |
0.3 |
| chr13_98826337_98826632 | 0.18 |
RNF113B |
ring finger protein 113B |
3035 |
0.21 |
| chr1_85064609_85064886 | 0.18 |
CTBS |
chitobiase, di-N-acetyl- |
24600 |
0.17 |
| chr11_27739578_27739815 | 0.18 |
BDNF |
brain-derived neurotrophic factor |
1598 |
0.49 |
| chr6_79314382_79314680 | 0.18 |
IRAK1BP1 |
interleukin-1 receptor-associated kinase 1 binding protein 1 |
262658 |
0.02 |
| chr1_209577581_209577868 | 0.18 |
ENSG00000230937 |
. |
27754 |
0.24 |
| chr1_103571993_103572213 | 0.18 |
COL11A1 |
collagen, type XI, alpha 1 |
1631 |
0.56 |
| chr1_172110530_172111247 | 0.18 |
ENSG00000207949 |
. |
2841 |
0.22 |
| chr8_141436376_141436676 | 0.18 |
TRAPPC9 |
trafficking protein particle complex 9 |
24503 |
0.23 |
| chr9_134554105_134554369 | 0.18 |
RAPGEF1 |
Rap guanine nucleotide exchange factor (GEF) 1 |
30992 |
0.18 |
| chr20_14317720_14318261 | 0.18 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
264 |
0.94 |
| chr4_74316638_74316988 | 0.18 |
AFP |
alpha-fetoprotein |
14849 |
0.2 |
| chr7_17411558_17411747 | 0.18 |
ENSG00000199473 |
. |
1298 |
0.57 |
| chr12_90485372_90485572 | 0.18 |
ENSG00000252823 |
. |
337636 |
0.01 |
| chr11_122037044_122037237 | 0.18 |
ENSG00000207994 |
. |
14124 |
0.17 |
| chr17_16286606_16286915 | 0.18 |
RP11-138I1.4 |
|
777 |
0.51 |
| chr9_22683479_22683652 | 0.18 |
DMRTA1 |
DMRT-like family A1 |
236725 |
0.02 |
| chr14_35629027_35629224 | 0.18 |
KIAA0391 |
KIAA0391 |
37160 |
0.15 |
| chr11_6340007_6340271 | 0.18 |
PRKCDBP |
protein kinase C, delta binding protein |
1623 |
0.33 |
| chr5_35130063_35130359 | 0.17 |
PRLR |
prolactin receptor |
235 |
0.95 |
| chr18_18697894_18698383 | 0.17 |
ENSG00000251886 |
. |
2329 |
0.32 |
| chr12_100628376_100628664 | 0.17 |
ENSG00000206790 |
. |
23048 |
0.11 |
| chr2_224477610_224477904 | 0.17 |
SCG2 |
secretogranin II |
10536 |
0.31 |
| chr4_177712658_177713139 | 0.17 |
VEGFC |
vascular endothelial growth factor C |
983 |
0.71 |
| chr16_65152808_65153250 | 0.17 |
CDH11 |
cadherin 11, type 2, OB-cadherin (osteoblast) |
2804 |
0.44 |
| chr3_159847220_159847374 | 0.17 |
IL12A-AS1 |
IL12A antisense RNA 1 |
77582 |
0.09 |
| chr8_20126437_20126669 | 0.17 |
LZTS1 |
leucine zipper, putative tumor suppressor 1 |
13750 |
0.19 |
| chr1_64096235_64096386 | 0.17 |
PGM1 |
phosphoglucomutase 1 |
7546 |
0.2 |
| chr3_171814831_171814982 | 0.17 |
FNDC3B |
fibronectin type III domain containing 3B |
29858 |
0.23 |
| chr4_110910630_110910781 | 0.17 |
ENSG00000207260 |
. |
2776 |
0.34 |
| chr15_96878987_96879612 | 0.17 |
ENSG00000222651 |
. |
2809 |
0.21 |
| chr4_144303651_144303881 | 0.17 |
GAB1 |
GRB2-associated binding protein 1 |
668 |
0.76 |
| chr22_46462951_46463814 | 0.17 |
RP6-109B7.4 |
|
2389 |
0.16 |
| chr6_52532294_52532590 | 0.17 |
RP1-152L7.5 |
|
2370 |
0.29 |
| chr2_30888986_30889287 | 0.17 |
CAPN13 |
calpain 13 |
141175 |
0.05 |
| chr3_99593001_99593152 | 0.17 |
FILIP1L |
filamin A interacting protein 1-like |
1872 |
0.43 |
| chr12_119617594_119617745 | 0.17 |
HSPB8 |
heat shock 22kDa protein 8 |
274 |
0.89 |
| chr8_89338361_89338512 | 0.17 |
RP11-586K2.1 |
|
629 |
0.66 |
| chr12_13025379_13025989 | 0.17 |
GPRC5A |
G protein-coupled receptor, family C, group 5, member A |
18032 |
0.14 |
| chr8_119120675_119121215 | 0.17 |
EXT1 |
exostosin glycosyltransferase 1 |
1708 |
0.54 |
| chrX_16840604_16840755 | 0.17 |
RBBP7 |
retinoblastoma binding protein 7 |
29509 |
0.14 |
| chr1_32168571_32169014 | 0.17 |
COL16A1 |
collagen, type XVI, alpha 1 |
976 |
0.48 |
| chr7_41737032_41737183 | 0.17 |
INHBA |
inhibin, beta A |
3100 |
0.25 |
| chr14_53416235_53416562 | 0.17 |
FERMT2 |
fermitin family member 2 |
888 |
0.69 |
| chr13_100635534_100635685 | 0.17 |
ZIC2 |
Zic family member 2 |
1583 |
0.38 |
| chr5_9047596_9047771 | 0.17 |
ENSG00000266415 |
. |
6324 |
0.29 |
| chr11_93865075_93865400 | 0.17 |
PANX1 |
pannexin 1 |
3142 |
0.34 |
| chr3_61549164_61549336 | 0.17 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
1665 |
0.55 |
| chr17_36623721_36623872 | 0.17 |
ARHGAP23 |
Rho GTPase activating protein 23 |
4384 |
0.19 |
| chr13_50650853_50651004 | 0.17 |
DLEU1 |
deleted in lymphocytic leukemia 1 (non-protein coding) |
5379 |
0.18 |
| chr1_87244034_87244185 | 0.17 |
SH3GLB1 |
SH3-domain GRB2-like endophilin B1 |
73531 |
0.09 |
| chr18_74798828_74799607 | 0.17 |
MBP |
myelin basic protein |
18000 |
0.27 |
| chr15_81614829_81615120 | 0.16 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
1492 |
0.33 |
| chr19_54538995_54539193 | 0.16 |
VSTM1 |
V-set and transmembrane domain containing 1 |
22583 |
0.08 |
| chr12_78334342_78334902 | 0.16 |
NAV3 |
neuron navigator 3 |
25434 |
0.27 |
| chr3_156409477_156409860 | 0.16 |
TIPARP |
TCDD-inducible poly(ADP-ribose) polymerase |
10713 |
0.23 |
| chr10_61385710_61385861 | 0.16 |
SLC16A9 |
solute carrier family 16, member 9 |
84052 |
0.11 |
| chr6_121757423_121758012 | 0.16 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
879 |
0.59 |
| chr21_43085116_43085274 | 0.16 |
LINC00112 |
long intergenic non-protein coding RNA 112 |
51401 |
0.12 |
| chr22_37828954_37829105 | 0.16 |
ELFN2 |
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
5524 |
0.16 |
| chr1_183132842_183132993 | 0.16 |
LAMC2 |
laminin, gamma 2 |
22456 |
0.18 |
| chr14_50453745_50453978 | 0.16 |
C14orf182 |
chromosome 14 open reading frame 182 |
20377 |
0.16 |
| chr3_65941462_65941807 | 0.16 |
MAGI1-IT1 |
MAGI1 intronic transcript 1 (non-protein coding) |
1401 |
0.43 |
| chr19_18655304_18655455 | 0.16 |
FKBP8 |
FK506 binding protein 8, 38kDa |
492 |
0.6 |
| chr22_31033608_31033759 | 0.16 |
SLC35E4 |
solute carrier family 35, member E4 |
1173 |
0.36 |
| chr1_154597059_154597346 | 0.16 |
ADAR |
adenosine deaminase, RNA-specific |
3238 |
0.19 |
| chr22_46480425_46480827 | 0.16 |
FLJ27365 |
hsa-mir-4763 |
1257 |
0.28 |
| chr4_184880881_184881125 | 0.16 |
STOX2 |
storkhead box 2 |
409 |
0.89 |
| chr20_30292538_30292716 | 0.16 |
AL160175.1 |
|
16374 |
0.12 |
| chr7_137402579_137403024 | 0.16 |
DGKI |
diacylglycerol kinase, iota |
128481 |
0.05 |
| chr3_98619314_98619465 | 0.16 |
DCBLD2 |
discoidin, CUB and LCCL domain containing 2 |
626 |
0.66 |
| chr17_750320_750517 | 0.16 |
NXN |
nucleoredoxin |
16919 |
0.14 |
| chr7_27142394_27142575 | 0.16 |
HOXA2 |
homeobox A2 |
54 |
0.93 |
| chrX_114467670_114468385 | 0.16 |
LRCH2 |
leucine-rich repeats and calponin homology (CH) domain containing 2 |
601 |
0.77 |
| chr9_128728923_128729074 | 0.16 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
101448 |
0.08 |
| chr17_79372410_79373082 | 0.16 |
RP11-1055B8.7 |
BAH and coiled-coil domain-containing protein 1 |
794 |
0.45 |
| chr6_35337873_35338102 | 0.16 |
PPARD |
peroxisome proliferator-activated receptor delta |
27585 |
0.16 |
| chr1_94701176_94701460 | 0.16 |
ARHGAP29 |
Rho GTPase activating protein 29 |
1803 |
0.43 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0015803 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.1 | 0.4 | GO:0051964 | negative regulation of synapse assembly(GO:0051964) |
| 0.1 | 0.5 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.4 | GO:0051503 | adenine nucleotide transport(GO:0051503) |
| 0.1 | 0.2 | GO:0046449 | allantoin metabolic process(GO:0000255) creatinine metabolic process(GO:0046449) cellular lactam metabolic process(GO:0072338) |
| 0.1 | 0.3 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.2 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.2 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.1 | 0.2 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.2 | GO:0033024 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.1 | 0.3 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.1 | 0.2 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.1 | 0.1 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.1 | GO:0048343 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.1 | 0.2 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.1 | 0.2 | GO:0002551 | mast cell chemotaxis(GO:0002551) regulation of mast cell chemotaxis(GO:0060753) positive regulation of mast cell chemotaxis(GO:0060754) mast cell migration(GO:0097531) |
| 0.1 | 0.2 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.1 | 0.1 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.0 | 0.1 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.1 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.3 | GO:0032906 | transforming growth factor beta2 production(GO:0032906) regulation of transforming growth factor beta2 production(GO:0032909) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.3 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.2 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:1903054 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.5 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.1 | GO:0010755 | regulation of plasminogen activation(GO:0010755) negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.2 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0070208 | protein heterotrimerization(GO:0070208) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.0 | GO:0036336 | dendritic cell chemotaxis(GO:0002407) dendritic cell migration(GO:0036336) |
| 0.0 | 0.1 | GO:0006531 | aspartate metabolic process(GO:0006531) aspartate catabolic process(GO:0006533) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.1 | GO:0072677 | eosinophil chemotaxis(GO:0048245) eosinophil migration(GO:0072677) |
| 0.0 | 0.2 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.0 | 0.1 | GO:0060841 | venous blood vessel development(GO:0060841) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:1900115 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.2 | GO:0010799 | regulation of peptidyl-threonine phosphorylation(GO:0010799) |
| 0.0 | 0.1 | GO:0040037 | negative regulation of fibroblast growth factor receptor signaling pathway(GO:0040037) |
| 0.0 | 0.1 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.1 | GO:0032836 | glomerular basement membrane development(GO:0032836) |
| 0.0 | 0.2 | GO:0007185 | transmembrane receptor protein tyrosine phosphatase signaling pathway(GO:0007185) |
| 0.0 | 0.1 | GO:0042532 | negative regulation of tyrosine phosphorylation of STAT protein(GO:0042532) |
| 0.0 | 0.1 | GO:0000492 | small nucleolar ribonucleoprotein complex assembly(GO:0000491) box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.1 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.0 | GO:0090206 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0008049 | male courtship behavior(GO:0008049) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.0 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.5 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.2 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.0 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.0 | GO:0010996 | response to auditory stimulus(GO:0010996) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.3 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.1 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.0 | 0.0 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.1 | GO:0032730 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.1 | GO:0007100 | mitotic centrosome separation(GO:0007100) |
| 0.0 | 0.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.3 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.2 | GO:0051044 | positive regulation of membrane protein ectodomain proteolysis(GO:0051044) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.0 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.0 | 0.1 | GO:0060390 | regulation of SMAD protein import into nucleus(GO:0060390) |
| 0.0 | 0.1 | GO:0072386 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.0 | GO:0022038 | corpus callosum development(GO:0022038) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.0 | GO:0055089 | fatty acid homeostasis(GO:0055089) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.0 | GO:0019511 | peptidyl-proline hydroxylation to 4-hydroxy-L-proline(GO:0018401) 4-hydroxyproline metabolic process(GO:0019471) peptidyl-proline hydroxylation(GO:0019511) |
| 0.0 | 0.1 | GO:0014912 | negative regulation of smooth muscle cell migration(GO:0014912) |
| 0.0 | 0.1 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.1 | GO:0008228 | opsonization(GO:0008228) |
| 0.0 | 0.1 | GO:0043584 | nose development(GO:0043584) |
| 0.0 | 0.1 | GO:0048844 | artery morphogenesis(GO:0048844) |
| 0.0 | 0.0 | GO:0071462 | cellular response to water stimulus(GO:0071462) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.0 | GO:1903519 | negative regulation of immature T cell proliferation(GO:0033087) negative regulation of immature T cell proliferation in thymus(GO:0033088) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.0 | 0.0 | GO:0060206 | estrous cycle phase(GO:0060206) |
| 0.0 | 0.0 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.0 | 0.0 | GO:1903332 | regulation of protein folding(GO:1903332) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.1 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.0 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.0 | GO:0090273 | regulation of somatostatin secretion(GO:0090273) |
| 0.0 | 0.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.0 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.0 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.0 | GO:0006573 | valine metabolic process(GO:0006573) |
| 0.0 | 0.0 | GO:0042109 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.0 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0043032 | positive regulation of macrophage activation(GO:0043032) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.0 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.0 | GO:0071379 | cellular response to prostaglandin stimulus(GO:0071379) |
| 0.0 | 0.0 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.0 | GO:0014850 | response to muscle activity(GO:0014850) |
| 0.0 | 0.0 | GO:0042482 | positive regulation of odontogenesis(GO:0042482) |
| 0.0 | 0.0 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.2 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.0 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.0 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.0 | GO:0051590 | positive regulation of neurotransmitter transport(GO:0051590) |
| 0.0 | 0.0 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0032230 | positive regulation of synaptic transmission, GABAergic(GO:0032230) |
| 0.0 | 0.1 | GO:0097061 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.1 | GO:0007263 | nitric oxide mediated signal transduction(GO:0007263) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
| 0.0 | 0.1 | GO:0002069 | columnar/cuboidal epithelial cell maturation(GO:0002069) |
| 0.0 | 0.0 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.0 | GO:0032148 | activation of protein kinase B activity(GO:0032148) |
| 0.0 | 0.2 | GO:0051353 | positive regulation of oxidoreductase activity(GO:0051353) |
| 0.0 | 0.0 | GO:0015825 | L-serine transport(GO:0015825) |
| 0.0 | 0.0 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0008088 | axo-dendritic transport(GO:0008088) |
| 0.0 | 0.0 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0045885 | obsolete positive regulation of survival gene product expression(GO:0045885) |
| 0.0 | 0.1 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0030033 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.0 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.0 | GO:0035413 | positive regulation of catenin import into nucleus(GO:0035413) |
| 0.0 | 0.0 | GO:0051580 | regulation of neurotransmitter uptake(GO:0051580) regulation of dopamine uptake involved in synaptic transmission(GO:0051584) regulation of catecholamine uptake involved in synaptic transmission(GO:0051940) |
| 0.0 | 0.0 | GO:2000189 | regulation of cholesterol homeostasis(GO:2000188) positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.6 | GO:0030198 | extracellular matrix organization(GO:0030198) extracellular structure organization(GO:0043062) |
| 0.0 | 0.1 | GO:0010452 | histone H3-K36 methylation(GO:0010452) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.0 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.0 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.4 | GO:0007272 | ensheathment of neurons(GO:0007272) axon ensheathment(GO:0008366) |
| 0.0 | 0.0 | GO:0046607 | positive regulation of centrosome cycle(GO:0046607) |
| 0.0 | 0.0 | GO:0042268 | regulation of cytolysis(GO:0042268) |
| 0.0 | 0.0 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.1 | GO:0051382 | kinetochore assembly(GO:0051382) |
| 0.0 | 0.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0046113 | nucleobase catabolic process(GO:0046113) |
| 0.0 | 0.1 | GO:0060037 | pharyngeal system development(GO:0060037) |
| 0.0 | 0.0 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.0 | GO:0034384 | high-density lipoprotein particle clearance(GO:0034384) |
| 0.0 | 0.0 | GO:0032455 | nerve growth factor processing(GO:0032455) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.1 | 0.6 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.4 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.2 | GO:0032444 | activin responsive factor complex(GO:0032444) |
| 0.0 | 0.2 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.3 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.3 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.3 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 1.1 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.0 | GO:0014704 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0048500 | signal recognition particle, endoplasmic reticulum targeting(GO:0005786) signal recognition particle(GO:0048500) |
| 0.0 | 0.3 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.2 | GO:0005581 | collagen trimer(GO:0005581) |
| 0.0 | 0.0 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0016585 | obsolete chromatin remodeling complex(GO:0016585) |
| 0.0 | 0.6 | GO:0043197 | dendritic spine(GO:0043197) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.0 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.1 | GO:0070761 | pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0030686 | 90S preribosome(GO:0030686) |
| 0.0 | 0.6 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.0 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 1.0 | GO:0030016 | myofibril(GO:0030016) |
| 0.0 | 0.2 | GO:0032154 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.3 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.2 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.2 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.1 | 0.4 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.0 | 0.2 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.5 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.3 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.7 | GO:0005546 | phosphatidylinositol-4,5-bisphosphate binding(GO:0005546) |
| 0.0 | 0.2 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.2 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.3 | GO:0005072 | transforming growth factor beta receptor, cytoplasmic mediator activity(GO:0005072) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.4 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.3 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.3 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.4 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 0.2 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0034190 | apolipoprotein receptor binding(GO:0034190) |
| 0.0 | 0.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.1 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.1 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.1 | GO:0043422 | protein kinase B binding(GO:0043422) |
| 0.0 | 0.1 | GO:0035198 | miRNA binding(GO:0035198) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0034584 | piRNA binding(GO:0034584) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.2 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0030297 | transmembrane receptor protein tyrosine kinase activator activity(GO:0030297) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.0 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.2 | GO:0001540 | beta-amyloid binding(GO:0001540) |
| 0.0 | 0.1 | GO:0001012 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.0 | GO:0004946 | bombesin receptor activity(GO:0004946) |
| 0.0 | 1.1 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 1.3 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0015194 | L-serine transmembrane transporter activity(GO:0015194) serine transmembrane transporter activity(GO:0022889) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.0 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0050694 | galactosylceramide sulfotransferase activity(GO:0001733) galactose 3-O-sulfotransferase activity(GO:0050694) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.0 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.0 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0043395 | proteoglycan binding(GO:0043394) heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.0 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.0 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0045502 | dynein binding(GO:0045502) |
| 0.0 | 0.0 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.1 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.4 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 1.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.2 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 0.1 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.7 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.0 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.4 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.2 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.1 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.4 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.2 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.4 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.0 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.4 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.2 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.2 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.0 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME REGULATION OF WATER BALANCE BY RENAL AQUAPORINS | Genes involved in Regulation of Water Balance by Renal Aquaporins |
| 0.0 | 0.4 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.1 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.0 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF MITOTIC PROTEINS | Genes involved in APC/C:Cdc20 mediated degradation of mitotic proteins |
| 0.0 | 0.1 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.1 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.2 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.2 | REACTOME ENOS ACTIVATION AND REGULATION | Genes involved in eNOS activation and regulation |
| 0.0 | 0.3 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME MUSCLE CONTRACTION | Genes involved in Muscle contraction |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME GABA B RECEPTOR ACTIVATION | Genes involved in GABA B receptor activation |
| 0.0 | 0.1 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |