Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
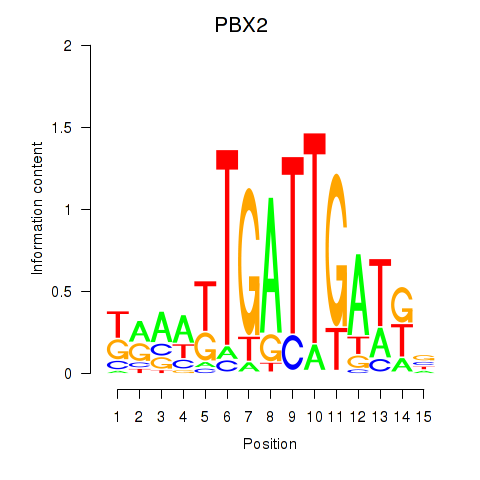
Results for PBX2
Z-value: 0.59
Transcription factors associated with PBX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX2
|
ENSG00000204304.7 | PBX homeobox 2 |
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr6_32158116_32158500 | PBX2 | 345 | 0.703432 | 0.75 | 2.1e-02 | Click! |
| chr6_32156796_32157600 | PBX2 | 765 | 0.383508 | 0.69 | 3.9e-02 | Click! |
| chr6_32158925_32159076 | PBX2 | 1037 | 0.259074 | 0.60 | 8.5e-02 | Click! |
| chr6_32156474_32156775 | PBX2 | 1339 | 0.204998 | 0.57 | 1.1e-01 | Click! |
| chr6_32158563_32158714 | PBX2 | 675 | 0.425087 | 0.46 | 2.1e-01 | Click! |
Activity of the PBX2 motif across conditions
Conditions sorted by the z-value of the PBX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
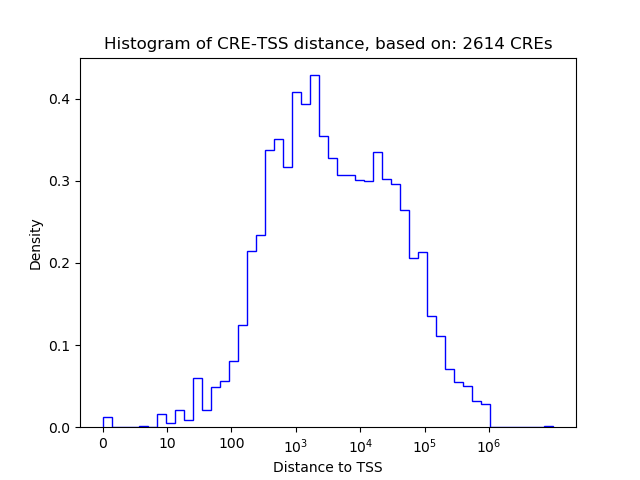
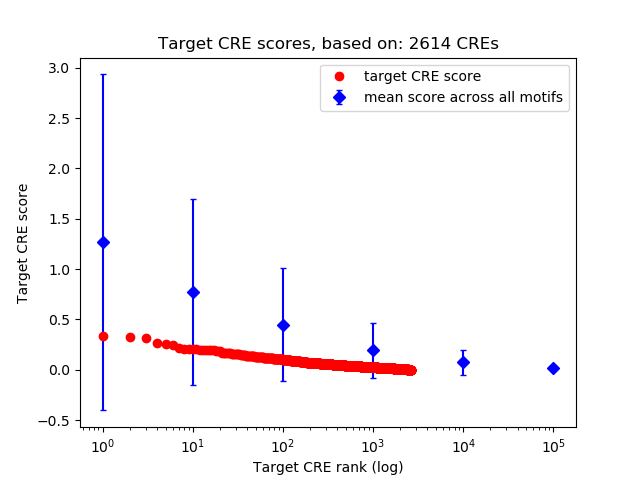
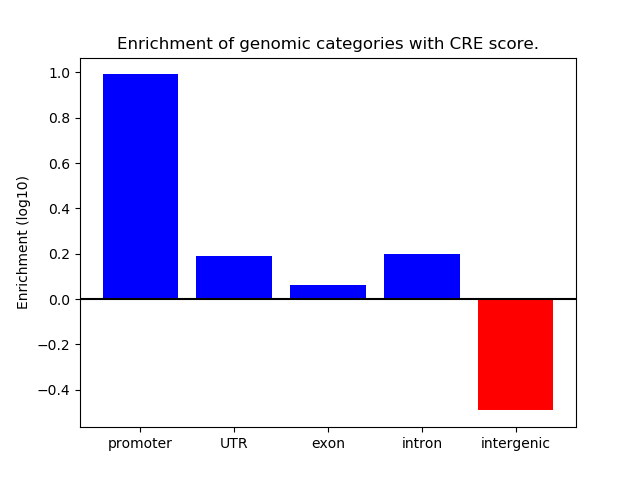
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr2_43448588_43448818 | 0.33 |
ZFP36L2 |
ZFP36 ring finger protein-like 2 |
5045 |
0.26 |
| chr19_1363575_1363781 | 0.33 |
MUM1 |
melanoma associated antigen (mutated) 1 |
7315 |
0.09 |
| chr8_122738565_122738716 | 0.31 |
HAS2-AS1 |
HAS2 antisense RNA 1 |
84964 |
0.1 |
| chr11_67979557_67979856 | 0.27 |
SUV420H1 |
suppressor of variegation 4-20 homolog 1 (Drosophila) |
515 |
0.8 |
| chr4_73711574_73711725 | 0.26 |
ENSG00000252955 |
. |
119246 |
0.06 |
| chr1_9713633_9713784 | 0.25 |
C1orf200 |
chromosome 1 open reading frame 200 |
936 |
0.49 |
| chr3_105136664_105136859 | 0.21 |
ALCAM |
activated leukocyte cell adhesion molecule |
50574 |
0.2 |
| chr12_47607778_47608233 | 0.21 |
PCED1B |
PC-esterase domain containing 1B |
2047 |
0.36 |
| chr13_75899167_75899318 | 0.21 |
TBC1D4 |
TBC1 domain family, member 4 |
16425 |
0.24 |
| chr6_6396318_6396469 | 0.20 |
F13A1 |
coagulation factor XIII, A1 polypeptide |
75147 |
0.11 |
| chr9_273424_273720 | 0.20 |
DOCK8 |
dedicator of cytokinesis 8 |
502 |
0.5 |
| chr10_100227423_100227662 | 0.20 |
HPS1 |
Hermansky-Pudlak syndrome 1 |
20859 |
0.19 |
| chr10_7227326_7227477 | 0.20 |
SFMBT2 |
Scm-like with four mbt domains 2 |
223306 |
0.02 |
| chr13_65531787_65531938 | 0.20 |
ENSG00000221685 |
. |
906443 |
0.0 |
| chr19_1021420_1021571 | 0.20 |
ENSG00000207357 |
. |
26 |
0.83 |
| chr16_11677800_11677980 | 0.20 |
LITAF |
lipopolysaccharide-induced TNF factor |
2339 |
0.3 |
| chr3_58292298_58292509 | 0.19 |
RPP14 |
ribonuclease P/MRP 14kDa subunit |
366 |
0.88 |
| chr16_81481663_81481945 | 0.19 |
CMIP |
c-Maf inducing protein |
3029 |
0.33 |
| chr1_27126960_27127111 | 0.19 |
PIGV |
phosphatidylinositol glycan anchor biosynthesis, class V |
11616 |
0.11 |
| chr8_133771865_133772108 | 0.19 |
TMEM71 |
transmembrane protein 71 |
808 |
0.63 |
| chr2_162102268_162102419 | 0.17 |
AC009299.2 |
|
5330 |
0.22 |
| chr1_161195567_161195718 | 0.17 |
TOMM40L |
translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
151 |
0.85 |
| chr11_100896662_100896813 | 0.17 |
TMEM133 |
transmembrane protein 133 |
33926 |
0.2 |
| chr18_43780070_43780221 | 0.17 |
C18orf25 |
chromosome 18 open reading frame 25 |
26145 |
0.19 |
| chr5_169074909_169075202 | 0.17 |
DOCK2 |
dedicator of cytokinesis 2 |
10804 |
0.25 |
| chr1_101360811_101360994 | 0.17 |
RP4-549L20.3 |
|
418 |
0.45 |
| chr1_42419485_42419636 | 0.16 |
HIVEP3 |
human immunodeficiency virus type I enhancer binding protein 3 |
35182 |
0.22 |
| chr3_64017563_64017714 | 0.16 |
PSMD6 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
7980 |
0.12 |
| chr19_48760988_48761397 | 0.16 |
CARD8 |
caspase recruitment domain family, member 8 |
1989 |
0.2 |
| chr20_62270981_62271132 | 0.16 |
CTD-3184A7.4 |
|
12453 |
0.09 |
| chr9_107687239_107687390 | 0.16 |
RP11-217B7.2 |
|
2520 |
0.3 |
| chr14_77463876_77464027 | 0.15 |
ENSG00000266553 |
. |
29843 |
0.15 |
| chr8_131144625_131144776 | 0.15 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
13932 |
0.2 |
| chr1_88526600_88526751 | 0.15 |
ENSG00000239504 |
. |
417122 |
0.01 |
| chr13_48612227_48612453 | 0.15 |
SUCLA2 |
succinate-CoA ligase, ADP-forming, beta subunit |
215 |
0.81 |
| chr22_43659737_43659896 | 0.15 |
SCUBE1 |
signal peptide, CUB domain, EGF-like 1 |
1011 |
0.5 |
| chr11_104914640_104914826 | 0.15 |
CARD16 |
caspase recruitment domain family, member 16 |
1301 |
0.41 |
| chr9_117129321_117129472 | 0.15 |
AKNA |
AT-hook transcription factor |
9848 |
0.2 |
| chr18_61148002_61148153 | 0.14 |
SERPINB5 |
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
3717 |
0.21 |
| chr11_17300725_17300876 | 0.14 |
NUCB2 |
nucleobindin 2 |
2230 |
0.33 |
| chr1_198613830_198613981 | 0.14 |
PTPRC |
protein tyrosine phosphatase, receptor type, C |
5613 |
0.26 |
| chr14_101158298_101158449 | 0.14 |
DLK1 |
delta-like 1 homolog (Drosophila) |
33669 |
0.09 |
| chr6_122225746_122225897 | 0.14 |
ENSG00000199932 |
. |
306973 |
0.01 |
| chr13_67288038_67288189 | 0.14 |
ENSG00000238500 |
. |
83205 |
0.1 |
| chr1_14028826_14029045 | 0.13 |
PRDM2 |
PR domain containing 2, with ZNF domain |
2200 |
0.32 |
| chr5_179044969_179045120 | 0.13 |
HNRNPH1 |
heterogeneous nuclear ribonucleoprotein H1 (H) |
0 |
0.96 |
| chr18_2573280_2573731 | 0.13 |
NDC80 |
NDC80 kinetochore complex component |
1941 |
0.23 |
| chr7_7196815_7197697 | 0.13 |
C1GALT1 |
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
691 |
0.7 |
| chr18_60822617_60822842 | 0.13 |
RP11-299P2.1 |
|
4176 |
0.28 |
| chr19_44577362_44577513 | 0.13 |
ZNF284 |
zinc finger protein 284 |
1140 |
0.28 |
| chr17_8191755_8191938 | 0.13 |
RANGRF |
RAN guanine nucleotide release factor |
31 |
0.95 |
| chr10_89624184_89624835 | 0.13 |
KLLN |
killin, p53-regulated DNA replication inhibitor |
1315 |
0.3 |
| chr14_52000304_52000455 | 0.13 |
ENSG00000252400 |
. |
10797 |
0.23 |
| chr6_100679332_100679483 | 0.13 |
RP1-121G13.2 |
|
195587 |
0.03 |
| chr17_64297348_64297511 | 0.13 |
PRKCA |
protein kinase C, alpha |
1515 |
0.42 |
| chr15_77288409_77288609 | 0.13 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
594 |
0.75 |
| chr9_88357119_88357540 | 0.12 |
AGTPBP1 |
ATP/GTP binding protein 1 |
385 |
0.93 |
| chr21_15917471_15917848 | 0.12 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
1003 |
0.63 |
| chr2_174518145_174518296 | 0.12 |
CDCA7 |
cell division cycle associated 7 |
298620 |
0.01 |
| chr8_56831921_56832112 | 0.12 |
ENSG00000216204 |
. |
10496 |
0.14 |
| chr1_176965552_176965703 | 0.12 |
ENSG00000202609 |
. |
32954 |
0.24 |
| chr17_71641526_71641788 | 0.12 |
SDK2 |
sidekick cell adhesion molecule 2 |
1429 |
0.46 |
| chr3_147337841_147337992 | 0.12 |
ZIC1 |
Zic family member 1 |
210745 |
0.02 |
| chr20_47441910_47442200 | 0.12 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
2365 |
0.4 |
| chr8_82597903_82598285 | 0.12 |
IMPA1 |
inositol(myo)-1(or 4)-monophosphatase 1 |
27 |
0.97 |
| chr1_161035238_161035389 | 0.12 |
AL591806.1 |
Uncharacterized protein |
342 |
0.73 |
| chr13_107220642_107220857 | 0.12 |
ARGLU1 |
arginine and glutamate rich 1 |
237 |
0.95 |
| chr1_90223723_90223893 | 0.12 |
ENSG00000239176 |
. |
10069 |
0.21 |
| chrX_119737518_119737925 | 0.12 |
MCTS1 |
malignant T cell amplified sequence 1 |
114 |
0.97 |
| chr12_65927555_65927706 | 0.12 |
MSRB3 |
methionine sulfoxide reductase B3 |
206975 |
0.02 |
| chr13_106828579_106828730 | 0.12 |
ENSG00000222682 |
. |
20815 |
0.25 |
| chr15_74834439_74834753 | 0.12 |
ARID3B |
AT rich interactive domain 3B (BRIGHT-like) |
1078 |
0.48 |
| chr4_47035027_47035178 | 0.11 |
GABRB1 |
gamma-aminobutyric acid (GABA) A receptor, beta 1 |
1452 |
0.52 |
| chr9_80644809_80645465 | 0.11 |
GNAQ |
guanine nucleotide binding protein (G protein), q polypeptide |
383 |
0.92 |
| chr2_43242041_43242199 | 0.11 |
ENSG00000207087 |
. |
76512 |
0.11 |
| chr11_3010488_3011464 | 0.11 |
AC131971.1 |
HCG1782999; PRO0943; Uncharacterized protein |
117 |
0.83 |
| chr3_46142263_46142414 | 0.11 |
CCR3 |
chemokine (C-C motif) receptor 3 |
62758 |
0.1 |
| chr2_71677518_71677669 | 0.11 |
DYSF |
dysferlin |
3259 |
0.3 |
| chr3_112693608_112694254 | 0.11 |
CD200R1 |
CD200 receptor 1 |
6 |
0.97 |
| chr12_40077019_40077550 | 0.11 |
C12orf40 |
chromosome 12 open reading frame 40 |
57299 |
0.13 |
| chr8_124186554_124186705 | 0.11 |
FAM83A |
family with sequence similarity 83, member A |
4658 |
0.13 |
| chr14_98668445_98668596 | 0.11 |
ENSG00000222066 |
. |
129567 |
0.06 |
| chr12_69419070_69419221 | 0.11 |
CPM |
carboxypeptidase M |
62096 |
0.11 |
| chr17_7744594_7744818 | 0.11 |
KDM6B |
lysine (K)-specific demethylase 6B |
1426 |
0.25 |
| chr19_8645059_8645210 | 0.11 |
AC130469.2 |
|
910 |
0.44 |
| chr2_163173016_163173250 | 0.11 |
IFIH1 |
interferon induced with helicase C domain 1 |
2061 |
0.31 |
| chr8_2590819_2591169 | 0.11 |
MYOM2 |
myomesin 2 |
597810 |
0.0 |
| chr12_96427792_96427943 | 0.11 |
LTA4H |
leukotriene A4 hydrolase |
1572 |
0.33 |
| chr4_159689303_159689454 | 0.11 |
FNIP2 |
folliculin interacting protein 2 |
912 |
0.58 |
| chr11_102192518_102192994 | 0.11 |
BIRC3 |
baculoviral IAP repeat containing 3 |
3196 |
0.22 |
| chr16_85096756_85096907 | 0.10 |
KIAA0513 |
KIAA0513 |
13 |
0.98 |
| chr6_11094574_11095155 | 0.10 |
SMIM13 |
small integral membrane protein 13 |
598 |
0.69 |
| chr11_30606067_30606222 | 0.10 |
RP5-1024C24.1 |
|
434 |
0.82 |
| chr6_6604888_6605039 | 0.10 |
LY86 |
lymphocyte antigen 86 |
16061 |
0.23 |
| chr1_116197547_116197911 | 0.10 |
VANGL1 |
VANGL planar cell polarity protein 1 |
3706 |
0.26 |
| chr7_69061838_69061989 | 0.10 |
AUTS2 |
autism susceptibility candidate 2 |
1992 |
0.44 |
| chr18_60984369_60984520 | 0.10 |
BCL2 |
B-cell CLL/lymphoma 2 |
1601 |
0.34 |
| chr8_124049779_124050367 | 0.10 |
TBC1D31 |
TBC1 domain family, member 31 |
4135 |
0.17 |
| chr2_225876272_225876423 | 0.10 |
ENSG00000263828 |
. |
1090 |
0.63 |
| chr13_114439745_114439896 | 0.10 |
TMEM255B |
transmembrane protein 255B |
22396 |
0.21 |
| chr6_24934559_24934719 | 0.10 |
FAM65B |
family with sequence similarity 65, member B |
1549 |
0.46 |
| chr16_23159267_23159418 | 0.10 |
USP31 |
ubiquitin specific peptidase 31 |
1249 |
0.52 |
| chr17_28087229_28087445 | 0.10 |
RP11-82O19.1 |
|
784 |
0.6 |
| chr1_8482394_8483668 | 0.10 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
695 |
0.63 |
| chr2_204732928_204733209 | 0.10 |
CTLA4 |
cytotoxic T-lymphocyte-associated protein 4 |
402 |
0.9 |
| chr8_21763339_21763611 | 0.10 |
DOK2 |
docking protein 2, 56kDa |
7699 |
0.2 |
| chr18_57568186_57568738 | 0.10 |
PMAIP1 |
phorbol-12-myristate-13-acetate-induced protein 1 |
1214 |
0.58 |
| chr1_89591012_89591781 | 0.10 |
GBP2 |
guanylate binding protein 2, interferon-inducible |
401 |
0.85 |
| chr7_72705892_72706043 | 0.10 |
GTF2IRD2P1 |
GTF2I repeat domain containing 2 pseudogene 1 |
11824 |
0.13 |
| chr15_71006763_71006990 | 0.10 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
11262 |
0.26 |
| chr6_5997225_5997376 | 0.10 |
NRN1 |
neuritin 1 |
9900 |
0.25 |
| chr2_137926013_137926164 | 0.10 |
THSD7B |
thrombospondin, type I, domain containing 7B |
111814 |
0.08 |
| chr7_75057272_75057423 | 0.10 |
NSUN5P1 |
NOP2/Sun domain family, member 5 pseudogene 1 |
12165 |
0.11 |
| chr17_79361447_79361695 | 0.09 |
RP11-1055B8.6 |
Uncharacterized protein |
7704 |
0.12 |
| chr13_41588793_41589349 | 0.09 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
4379 |
0.23 |
| chr2_27255514_27255731 | 0.09 |
TMEM214 |
transmembrane protein 214 |
200 |
0.85 |
| chr9_27454069_27454220 | 0.09 |
RP11-298E2.2 |
|
62412 |
0.11 |
| chr8_16870402_16870553 | 0.09 |
FGF20 |
fibroblast growth factor 20 |
10787 |
0.23 |
| chrX_77361758_77362293 | 0.09 |
PGK1 |
phosphoglycerate kinase 1 |
166 |
0.97 |
| chr3_71834842_71834993 | 0.09 |
PROK2 |
prokineticin 2 |
560 |
0.77 |
| chr20_35830256_35830797 | 0.09 |
MROH8 |
maestro heat-like repeat family member 8 |
22535 |
0.17 |
| chr2_106367063_106367214 | 0.09 |
NCK2 |
NCK adaptor protein 2 |
4950 |
0.33 |
| chr20_52295216_52295367 | 0.09 |
ENSG00000238468 |
. |
9994 |
0.25 |
| chr7_16794353_16794513 | 0.09 |
TSPAN13 |
tetraspanin 13 |
1273 |
0.44 |
| chr1_207093221_207093431 | 0.09 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
1886 |
0.27 |
| chr3_151914442_151914993 | 0.09 |
MBNL1 |
muscleblind-like splicing regulator 1 |
71112 |
0.11 |
| chr4_154389972_154390290 | 0.09 |
KIAA0922 |
KIAA0922 |
2630 |
0.35 |
| chrX_70585572_70585765 | 0.09 |
TAF1 |
TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa |
446 |
0.84 |
| chr13_100630096_100630247 | 0.09 |
ZIC2 |
Zic family member 2 |
3855 |
0.22 |
| chr16_89034372_89034523 | 0.09 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
8954 |
0.15 |
| chr2_5727897_5728048 | 0.09 |
AC108025.2 |
|
103206 |
0.07 |
| chrX_3189331_3189985 | 0.09 |
CXorf28 |
chromosome X open reading frame 28 |
203 |
0.96 |
| chr14_55137368_55137519 | 0.09 |
SAMD4A |
sterile alpha motif domain containing 4A |
84108 |
0.09 |
| chr13_46038649_46038966 | 0.09 |
COG3 |
component of oligomeric golgi complex 3 |
253 |
0.93 |
| chr13_37575523_37575689 | 0.09 |
EXOSC8 |
exosome component 8 |
928 |
0.49 |
| chr3_188152189_188152340 | 0.09 |
LPP-AS1 |
LPP antisense RNA 1 |
134190 |
0.05 |
| chr4_40209794_40209945 | 0.09 |
RHOH |
ras homolog family member H |
7905 |
0.22 |
| chr20_23400882_23401206 | 0.09 |
NAPB |
N-ethylmaleimide-sensitive factor attachment protein, beta |
1054 |
0.39 |
| chr1_247123081_247123305 | 0.09 |
AHCTF1 |
AT hook containing transcription factor 1 |
27913 |
0.17 |
| chr20_61408872_61409023 | 0.09 |
LINC00659 |
long intergenic non-protein coding RNA 659 |
2183 |
0.2 |
| chr1_40848541_40848692 | 0.09 |
SMAP2 |
small ArfGAP2 |
8296 |
0.18 |
| chr15_93445330_93445481 | 0.09 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
1840 |
0.31 |
| chr13_100622367_100622591 | 0.09 |
ZIC5 |
Zic family member 5 |
1684 |
0.37 |
| chr6_77599653_77599804 | 0.09 |
ENSG00000272445 |
. |
332257 |
0.01 |
| chr5_95155170_95155321 | 0.09 |
GLRX |
glutaredoxin (thioltransferase) |
3170 |
0.2 |
| chr6_152505186_152505339 | 0.09 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
15763 |
0.29 |
| chr6_31514651_31514955 | 0.09 |
NFKBIL1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor-like 1 |
156 |
0.49 |
| chr1_145396978_145397482 | 0.08 |
ENSG00000201558 |
. |
14473 |
0.13 |
| chr3_89033455_89033606 | 0.08 |
EPHA3 |
EPH receptor A3 |
123144 |
0.07 |
| chr5_110560941_110561277 | 0.08 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
1325 |
0.51 |
| chr2_231082753_231082904 | 0.08 |
SP110 |
SP110 nuclear body protein |
1789 |
0.35 |
| chr4_84036093_84036262 | 0.08 |
PLAC8 |
placenta-specific 8 |
222 |
0.95 |
| chrX_14515159_14515412 | 0.08 |
GLRA2 |
glycine receptor, alpha 2 |
32135 |
0.25 |
| chr9_111063457_111063608 | 0.08 |
ENSG00000222512 |
. |
57677 |
0.17 |
| chr11_66883336_66883487 | 0.08 |
KDM2A |
lysine (K)-specific demethylase 2A |
3329 |
0.22 |
| chr10_22630594_22630745 | 0.08 |
SPAG6 |
sperm associated antigen 6 |
3730 |
0.19 |
| chr3_112217855_112218341 | 0.08 |
BTLA |
B and T lymphocyte associated |
107 |
0.98 |
| chr4_108910417_108910832 | 0.08 |
HADH |
hydroxyacyl-CoA dehydrogenase |
246 |
0.92 |
| chr17_72463483_72463716 | 0.08 |
CD300A |
CD300a molecule |
598 |
0.68 |
| chr19_18668411_18668562 | 0.08 |
KXD1 |
KxDL motif containing 1 |
86 |
0.92 |
| chr12_67662728_67662890 | 0.08 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
252 |
0.96 |
| chr6_138192975_138193775 | 0.08 |
RP11-356I2.4 |
|
4005 |
0.25 |
| chr2_86116312_86117036 | 0.08 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
537 |
0.8 |
| chr1_153643822_153644080 | 0.08 |
ILF2 |
interleukin enhancer binding factor 2 |
427 |
0.65 |
| chr8_31158022_31158173 | 0.08 |
RP11-363L24.3 |
|
123414 |
0.06 |
| chr17_27919309_27919602 | 0.08 |
RP11-68I3.7 |
|
196 |
0.81 |
| chr2_192014463_192014614 | 0.08 |
STAT4 |
signal transducer and activator of transcription 4 |
1159 |
0.54 |
| chr3_16550538_16550689 | 0.08 |
RFTN1 |
raftlin, lipid raft linker 1 |
3798 |
0.32 |
| chr3_101442973_101443124 | 0.08 |
CEP97 |
centrosomal protein 97kDa |
279 |
0.88 |
| chr6_119027020_119027704 | 0.08 |
CEP85L |
centrosomal protein 85kDa-like |
3869 |
0.35 |
| chr11_61780107_61780258 | 0.08 |
AP003733.1 |
Uncharacterized protein; cDNA FLJ36460 fis, clone THYMU2014801 |
44729 |
0.1 |
| chr4_109062684_109062835 | 0.08 |
LEF1 |
lymphoid enhancer-binding factor 1 |
24698 |
0.2 |
| chr2_68963544_68963853 | 0.08 |
ARHGAP25 |
Rho GTPase activating protein 25 |
1684 |
0.46 |
| chr17_38177630_38177781 | 0.07 |
MED24 |
mediator complex subunit 24 |
1708 |
0.23 |
| chr4_143321559_143321710 | 0.07 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
30778 |
0.27 |
| chr14_78081630_78082591 | 0.07 |
SPTLC2 |
serine palmitoyltransferase, long chain base subunit 2 |
1006 |
0.56 |
| chr2_144006550_144006740 | 0.07 |
RP11-190J23.1 |
|
76904 |
0.11 |
| chr9_132402475_132402626 | 0.07 |
RP11-483H20.4 |
|
205 |
0.89 |
| chr1_24882080_24882472 | 0.07 |
NCMAP |
noncompact myelin associated protein |
326 |
0.88 |
| chr8_116601507_116601658 | 0.07 |
TRPS1 |
trichorhinophalangeal syndrome I |
30715 |
0.24 |
| chr5_90677845_90678028 | 0.07 |
ARRDC3 |
arrestin domain containing 3 |
1240 |
0.57 |
| chr3_107806842_107807168 | 0.07 |
CD47 |
CD47 molecule |
2856 |
0.4 |
| chr4_47913848_47913999 | 0.07 |
NFXL1 |
nuclear transcription factor, X-box binding-like 1 |
2633 |
0.29 |
| chr19_24155336_24155487 | 0.07 |
CTB-92J24.3 |
|
14679 |
0.18 |
| chr8_8570882_8571033 | 0.07 |
CLDN23 |
claudin 23 |
11509 |
0.26 |
| chr7_104643472_104643623 | 0.07 |
LINC01004 |
long intergenic non-protein coding RNA 1004 |
9924 |
0.16 |
| chr10_15211985_15212371 | 0.07 |
NMT2 |
N-myristoyltransferase 2 |
1486 |
0.45 |
| chr9_139434770_139435052 | 0.07 |
RP11-413M3.4 |
|
2422 |
0.15 |
| chr12_9903729_9904030 | 0.07 |
CD69 |
CD69 molecule |
9618 |
0.15 |
| chr10_7451836_7452578 | 0.07 |
SFMBT2 |
Scm-like with four mbt domains 2 |
904 |
0.71 |
| chr2_11893696_11894011 | 0.07 |
LPIN1 |
lipin 1 |
7089 |
0.15 |
| chr3_177321275_177321426 | 0.07 |
ENSG00000200288 |
. |
20847 |
0.27 |
| chr18_27836555_27836706 | 0.07 |
ENSG00000251702 |
. |
727442 |
0.0 |
| chr4_153600093_153601094 | 0.07 |
TMEM154 |
transmembrane protein 154 |
571 |
0.78 |
| chr4_152588470_152588621 | 0.07 |
RP11-164P12.4 |
|
169 |
0.96 |
| chr1_149969239_149969390 | 0.07 |
OTUD7B |
OTU domain containing 7B |
13310 |
0.14 |
| chr17_46646140_46646364 | 0.07 |
HOXB3 |
homeobox B3 |
5133 |
0.08 |
| chr4_37075132_37075283 | 0.07 |
ENSG00000264319 |
. |
168406 |
0.04 |
| chr2_208395832_208396657 | 0.07 |
CREB1 |
cAMP responsive element binding protein 1 |
1416 |
0.37 |
| chr3_187637243_187637630 | 0.07 |
BCL6 |
B-cell CLL/lymphoma 6 |
173921 |
0.03 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.2 | GO:0050932 | regulation of melanocyte differentiation(GO:0045634) regulation of pigment cell differentiation(GO:0050932) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.1 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.2 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.2 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0031125 | rRNA 3'-end processing(GO:0031125) |
| 0.0 | 0.1 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.0 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.0 | GO:0046101 | hypoxanthine biosynthetic process(GO:0046101) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.0 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.0 | GO:0003352 | regulation of cilium movement(GO:0003352) regulation of cilium beat frequency(GO:0003356) cilium movement involved in cell motility(GO:0060294) regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.0 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.0 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.0 | GO:0010447 | response to acidic pH(GO:0010447) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.0 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0072577 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.0 | GO:0032488 | Cdc42 protein signal transduction(GO:0032488) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.0 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.0 | 0.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0000780 | condensed nuclear chromosome, centromeric region(GO:0000780) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.0 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.2 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0004526 | ribonuclease P activity(GO:0004526) |
| 0.0 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.0 | GO:0016314 | phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.1 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.0 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.0 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.0 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.0 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |