Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
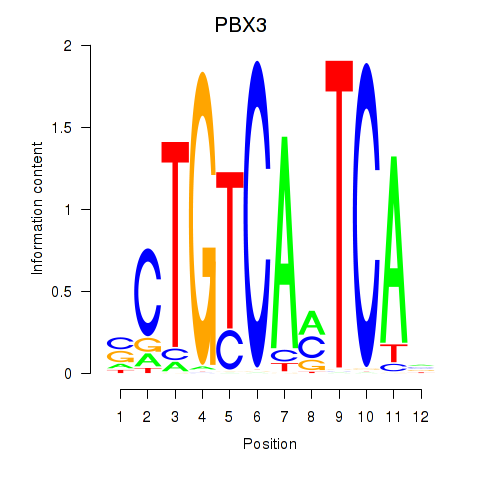
Results for PBX3
Z-value: 1.16
Transcription factors associated with PBX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PBX3
|
ENSG00000167081.12 | PBX homeobox 3 |
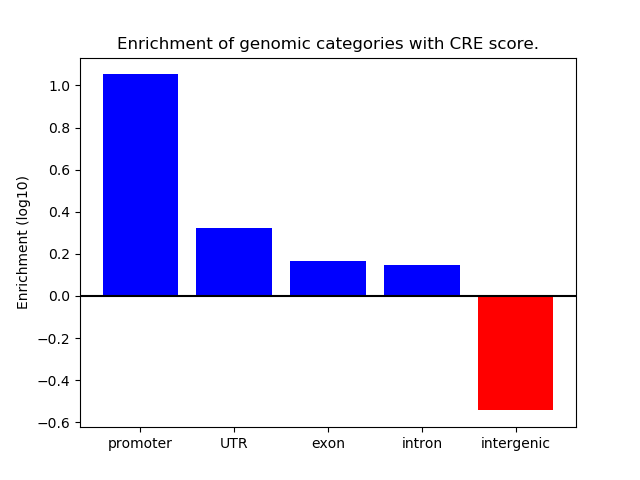
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr9_128514663_128514814 | PBX3 | 4260 | 0.307116 | -0.93 | 2.8e-04 | Click! |
| chr9_128664438_128664589 | PBX3 | 36963 | 0.227213 | -0.92 | 5.2e-04 | Click! |
| chr9_128524004_128524155 | PBX3 | 13601 | 0.262021 | -0.91 | 7.2e-04 | Click! |
| chr9_128516456_128516607 | PBX3 | 6053 | 0.285353 | -0.88 | 1.9e-03 | Click! |
| chr9_128512019_128512257 | PBX3 | 1660 | 0.479975 | -0.87 | 2.6e-03 | Click! |
Activity of the PBX3 motif across conditions
Conditions sorted by the z-value of the PBX3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
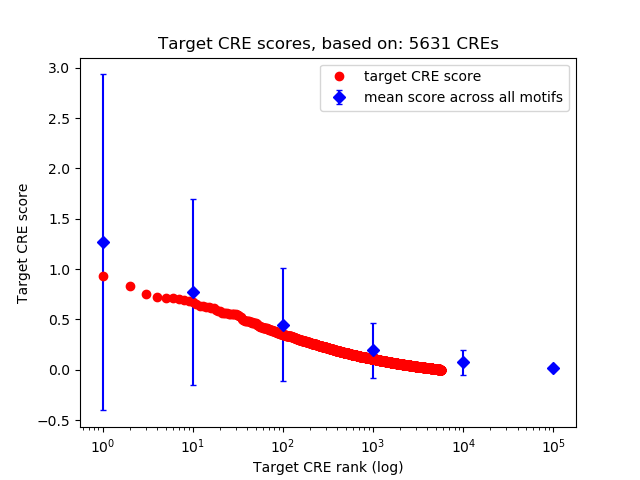
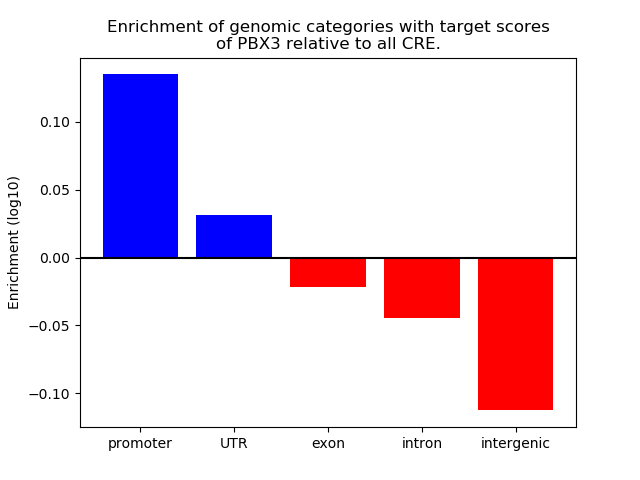
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_46612034_46612360 | 0.94 |
SLC38A1 |
solute carrier family 38, member 1 |
49287 |
0.18 |
| chr22_40298890_40299260 | 0.83 |
GRAP2 |
GRB2-related adaptor protein 2 |
1962 |
0.3 |
| chr7_150148386_150148563 | 0.76 |
GIMAP8 |
GTPase, IMAP family member 8 |
756 |
0.63 |
| chrX_135731409_135731662 | 0.72 |
CD40LG |
CD40 ligand |
1149 |
0.47 |
| chr13_24827167_24827318 | 0.72 |
SPATA13-AS1 |
SPATA13 antisense RNA 1 |
1335 |
0.33 |
| chr9_130539941_130540132 | 0.71 |
SH2D3C |
SH2 domain containing 3C |
984 |
0.33 |
| chr1_154927193_154927621 | 0.70 |
PBXIP1 |
pre-B-cell leukemia homeobox interacting protein 1 |
1173 |
0.25 |
| chr5_35859258_35859647 | 0.69 |
IL7R |
interleukin 7 receptor |
2458 |
0.3 |
| chr12_55362139_55362302 | 0.68 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
5118 |
0.26 |
| chr14_91866311_91866542 | 0.68 |
CCDC88C |
coiled-coil domain containing 88C |
17264 |
0.22 |
| chr6_149805654_149805958 | 0.65 |
ZC3H12D |
zinc finger CCCH-type containing 12D |
320 |
0.87 |
| chr1_156785806_156786457 | 0.64 |
SH2D2A |
SH2 domain containing 2A |
495 |
0.51 |
| chr19_39825870_39826313 | 0.64 |
GMFG |
glia maturation factor, gamma |
554 |
0.58 |
| chr16_21519015_21519166 | 0.62 |
ENSG00000265462 |
. |
1634 |
0.33 |
| chr20_52199257_52199767 | 0.62 |
ZNF217 |
zinc finger protein 217 |
124 |
0.96 |
| chr17_75663137_75663644 | 0.62 |
SEPT9 |
septin 9 |
185395 |
0.03 |
| chr14_22969065_22969423 | 0.61 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
13073 |
0.1 |
| chr1_239883177_239883989 | 0.60 |
CHRM3 |
cholinergic receptor, muscarinic 3 |
740 |
0.59 |
| chr21_15917916_15918619 | 0.59 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
395 |
0.89 |
| chr17_76128814_76128979 | 0.58 |
TMC6 |
transmembrane channel-like 6 |
408 |
0.73 |
| chr14_22974481_22974956 | 0.57 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
18547 |
0.09 |
| chr10_129861850_129862130 | 0.56 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
16156 |
0.26 |
| chr5_177236761_177236962 | 0.56 |
FAM153A |
family with sequence similarity 153, member A |
26462 |
0.14 |
| chr6_159464766_159465080 | 0.56 |
TAGAP |
T-cell activation RhoGTPase activating protein |
1127 |
0.51 |
| chr13_99910339_99910626 | 0.56 |
GPR18 |
G protein-coupled receptor 18 |
146 |
0.96 |
| chr15_55547653_55548107 | 0.56 |
RAB27A |
RAB27A, member RAS oncogene family |
6647 |
0.22 |
| chr18_13383462_13383613 | 0.56 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
984 |
0.5 |
| chr11_102192518_102192994 | 0.55 |
BIRC3 |
baculoviral IAP repeat containing 3 |
3196 |
0.22 |
| chr5_175461188_175461339 | 0.55 |
THOC3 |
THO complex 3 |
420 |
0.79 |
| chr2_204801338_204802214 | 0.55 |
ICOS |
inducible T-cell co-stimulator |
273 |
0.95 |
| chr11_46354220_46354463 | 0.55 |
DGKZ |
diacylglycerol kinase, zeta |
114 |
0.96 |
| chr22_50631230_50631436 | 0.54 |
TRABD |
TraB domain containing |
110 |
0.82 |
| chr16_81748402_81748687 | 0.54 |
PLCG2 |
phospholipase C, gamma 2 (phosphatidylinositol-specific) |
24158 |
0.24 |
| chr2_235362717_235362976 | 0.52 |
ARL4C |
ADP-ribosylation factor-like 4C |
42398 |
0.22 |
| chr6_35279100_35279566 | 0.50 |
DEF6 |
differentially expressed in FDCP 6 homolog (mouse) |
1818 |
0.34 |
| chr22_40781561_40781851 | 0.50 |
RP5-1042K10.10 |
|
2515 |
0.24 |
| chr14_22471633_22471925 | 0.49 |
ENSG00000238634 |
. |
139108 |
0.04 |
| chr11_65379543_65379824 | 0.49 |
MAP3K11 |
mitogen-activated protein kinase kinase kinase 11 |
911 |
0.31 |
| chr9_36142717_36143534 | 0.48 |
GLIPR2 |
GLI pathogenesis-related 2 |
6383 |
0.2 |
| chr1_117342787_117343202 | 0.48 |
CD2 |
CD2 molecule |
45905 |
0.14 |
| chr2_182174449_182174635 | 0.48 |
ENSG00000266705 |
. |
4163 |
0.36 |
| chr11_121329971_121330303 | 0.48 |
RP11-730K11.1 |
|
6415 |
0.25 |
| chr19_16479476_16479627 | 0.48 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
6787 |
0.16 |
| chr19_18414393_18414628 | 0.47 |
LSM4 |
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
12559 |
0.09 |
| chr5_35859676_35859963 | 0.47 |
IL7R |
interleukin 7 receptor |
2825 |
0.28 |
| chr1_169665907_169666062 | 0.47 |
SELL |
selectin L |
14855 |
0.19 |
| chr11_118213949_118214100 | 0.47 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
565 |
0.54 |
| chr2_206809495_206809646 | 0.47 |
ENSG00000201875 |
. |
44216 |
0.13 |
| chr14_91707602_91707801 | 0.46 |
CTD-2547L24.3 |
HCG1816139; Uncharacterized protein |
1402 |
0.37 |
| chrX_44203680_44203969 | 0.46 |
EFHC2 |
EF-hand domain (C-terminal) containing 2 |
906 |
0.71 |
| chr11_2323777_2324127 | 0.45 |
TSPAN32 |
tetraspanin 32 |
1 |
0.92 |
| chr17_28033493_28033909 | 0.44 |
RP11-82O19.1 |
|
54420 |
0.07 |
| chr1_21620200_21620457 | 0.44 |
RP5-1071N3.1 |
|
545 |
0.47 |
| chr13_31311093_31311268 | 0.44 |
ALOX5AP |
arachidonate 5-lipoxygenase-activating protein |
1535 |
0.52 |
| chr7_5572395_5572659 | 0.44 |
ACTB |
actin, beta |
2187 |
0.2 |
| chr5_100116307_100116504 | 0.43 |
ENSG00000221263 |
. |
35864 |
0.2 |
| chr6_119131761_119131953 | 0.43 |
MCM9 |
minichromosome maintenance complex component 9 |
18559 |
0.24 |
| chr14_22895521_22895672 | 0.43 |
ENSG00000251002 |
. |
6123 |
0.13 |
| chr21_46337373_46337524 | 0.43 |
ITGB2 |
integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) |
3250 |
0.13 |
| chr11_64216918_64217087 | 0.42 |
AP003774.4 |
HCG1652096, isoform CRA_a; Uncharacterized protein; cDNA FLJ37045 fis, clone BRACE2012185 |
456 |
0.79 |
| chr10_8446299_8446474 | 0.42 |
ENSG00000212505 |
. |
252408 |
0.02 |
| chr17_4389201_4389708 | 0.41 |
RP13-580F15.2 |
|
194 |
0.92 |
| chr4_40203939_40204397 | 0.41 |
RHOH |
ras homolog family member H |
2204 |
0.34 |
| chr16_11677800_11677980 | 0.41 |
LITAF |
lipopolysaccharide-induced TNF factor |
2339 |
0.3 |
| chr16_30486681_30486832 | 0.41 |
ITGAL |
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
1434 |
0.16 |
| chr1_207088744_207089040 | 0.41 |
FAIM3 |
Fas apoptotic inhibitory molecule 3 |
6320 |
0.15 |
| chr1_19804030_19804234 | 0.41 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
6447 |
0.18 |
| chr12_55375935_55376187 | 0.41 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
439 |
0.86 |
| chr14_91834289_91834440 | 0.41 |
ENSG00000265856 |
. |
34307 |
0.17 |
| chr16_68113033_68113320 | 0.41 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
6071 |
0.11 |
| chr13_99960008_99960188 | 0.40 |
GPR183 |
G protein-coupled receptor 183 |
439 |
0.84 |
| chr1_145454503_145454819 | 0.40 |
TXNIP |
thioredoxin interacting protein |
15355 |
0.1 |
| chr2_69867626_69867777 | 0.39 |
AAK1 |
AP2 associated kinase 1 |
3085 |
0.28 |
| chr11_118094529_118095128 | 0.39 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
26 |
0.97 |
| chr10_70849708_70849969 | 0.39 |
SRGN |
serglycin |
1964 |
0.33 |
| chr1_19803053_19803337 | 0.39 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
7384 |
0.17 |
| chr4_90227728_90227903 | 0.39 |
GPRIN3 |
GPRIN family member 3 |
1346 |
0.6 |
| chr17_143001_143152 | 0.39 |
RP11-1260E13.1 |
|
28107 |
0.13 |
| chr5_39200999_39201150 | 0.38 |
FYB |
FYN binding protein |
2055 |
0.44 |
| chr14_22865587_22865738 | 0.38 |
ENSG00000251002 |
. |
36057 |
0.11 |
| chr1_111742376_111742661 | 0.38 |
DENND2D |
DENN/MADD domain containing 2D |
793 |
0.38 |
| chr16_85936780_85936931 | 0.38 |
IRF8 |
interferon regulatory factor 8 |
411 |
0.88 |
| chr10_5336313_5336469 | 0.38 |
AKR1C7P |
aldo-keto reductase family 1, member C7, pseudogene |
5958 |
0.22 |
| chr17_56421423_56421650 | 0.38 |
BZRAP1-AS1 |
BZRAP1 antisense RNA 1 |
6817 |
0.11 |
| chr14_22964891_22965136 | 0.37 |
TRAJ51 |
T cell receptor alpha joining 51 (pseudogene) |
8842 |
0.1 |
| chr7_99819175_99819326 | 0.37 |
ENSG00000222482 |
. |
1507 |
0.2 |
| chr5_156698426_156698828 | 0.37 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
2265 |
0.23 |
| chr5_39203555_39203988 | 0.37 |
FYB |
FYN binding protein |
642 |
0.81 |
| chr22_39547258_39547409 | 0.37 |
CBX7 |
chromobox homolog 7 |
1116 |
0.46 |
| chr3_185826921_185827162 | 0.36 |
ETV5 |
ets variant 5 |
140 |
0.97 |
| chr21_44166870_44167151 | 0.36 |
AP001627.1 |
|
5142 |
0.26 |
| chr4_2799841_2800188 | 0.36 |
SH3BP2 |
SH3-domain binding protein 2 |
277 |
0.92 |
| chr11_35194712_35194909 | 0.36 |
CD44 |
CD44 molecule (Indian blood group) |
3308 |
0.21 |
| chrX_48793999_48794379 | 0.36 |
PIM2 |
pim-2 oncogene |
17888 |
0.08 |
| chr19_14477270_14477421 | 0.36 |
CD97 |
CD97 molecule |
14623 |
0.15 |
| chr5_66461571_66461722 | 0.36 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
23384 |
0.22 |
| chr10_82220206_82220414 | 0.35 |
TSPAN14 |
tetraspanin 14 |
1252 |
0.5 |
| chr1_1136100_1136251 | 0.35 |
TNFRSF18 |
tumor necrosis factor receptor superfamily, member 18 |
4885 |
0.08 |
| chr2_96986640_96986853 | 0.35 |
AC021188.4 |
|
723 |
0.57 |
| chr2_197032904_197033284 | 0.35 |
STK17B |
serine/threonine kinase 17b |
2630 |
0.28 |
| chr18_9107105_9107411 | 0.35 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
3178 |
0.21 |
| chr22_22089210_22089361 | 0.34 |
YPEL1 |
yippee-like 1 (Drosophila) |
759 |
0.55 |
| chr19_22605163_22605314 | 0.34 |
ZNF98 |
zinc finger protein 98 |
79 |
0.98 |
| chr15_63798708_63798859 | 0.34 |
USP3 |
ubiquitin specific peptidase 3 |
1928 |
0.4 |
| chr2_240302618_240303103 | 0.34 |
HDAC4 |
histone deacetylase 4 |
19783 |
0.17 |
| chr14_99710276_99710624 | 0.34 |
AL109767.1 |
|
18835 |
0.2 |
| chr19_12512149_12512361 | 0.34 |
ZNF799 |
zinc finger protein 799 |
170 |
0.92 |
| chr3_30670169_30670603 | 0.34 |
TGFBR2 |
transforming growth factor, beta receptor II (70/80kDa) |
22293 |
0.26 |
| chr16_27413508_27414237 | 0.34 |
IL21R |
interleukin 21 receptor |
377 |
0.88 |
| chrX_71321072_71321371 | 0.34 |
RGAG4 |
retrotransposon gag domain containing 4 |
30457 |
0.14 |
| chr5_137416910_137417200 | 0.34 |
WNT8A |
wingless-type MMTV integration site family, member 8A |
2526 |
0.17 |
| chr17_33541971_33542122 | 0.34 |
SLC35G3 |
solute carrier family 35, member G3 |
20634 |
0.11 |
| chr2_74432517_74432668 | 0.34 |
MTHFD2 |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
6849 |
0.14 |
| chr14_106950202_106950486 | 0.34 |
IGHVII-44-2 |
immunoglobulin heavy variable (II)-44-2 (pseudogene) |
53 |
0.89 |
| chr8_91018652_91018803 | 0.34 |
DECR1 |
2,4-dienoyl CoA reductase 1, mitochondrial |
4982 |
0.24 |
| chr13_99856162_99856313 | 0.34 |
ENSG00000201793 |
. |
1725 |
0.34 |
| chr1_204941075_204941226 | 0.33 |
NFASC |
neurofascin |
9834 |
0.21 |
| chr12_66662591_66662742 | 0.33 |
RP11-335I12.2 |
|
11452 |
0.15 |
| chr2_27485345_27486489 | 0.33 |
SLC30A3 |
solute carrier family 30 (zinc transporter), member 3 |
178 |
0.89 |
| chr9_82185992_82186420 | 0.33 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
482 |
0.9 |
| chr14_75715883_75716034 | 0.33 |
RP11-293M10.2 |
|
10156 |
0.15 |
| chr7_50345819_50346403 | 0.33 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
1733 |
0.5 |
| chr16_23846602_23847146 | 0.33 |
PRKCB |
protein kinase C, beta |
448 |
0.87 |
| chr1_111423669_111423843 | 0.33 |
CD53 |
CD53 molecule |
7980 |
0.2 |
| chr17_43487096_43487739 | 0.33 |
ARHGAP27 |
Rho GTPase activating protein 27 |
335 |
0.82 |
| chr7_21467616_21467934 | 0.33 |
SP4 |
Sp4 transcription factor |
114 |
0.97 |
| chr6_126153129_126153280 | 0.33 |
NCOA7-AS1 |
NCOA7 antisense RNA 1 |
13200 |
0.18 |
| chr16_84582402_84582610 | 0.33 |
TLDC1 |
TBC/LysM-associated domain containing 1 |
5133 |
0.21 |
| chr1_168390803_168391087 | 0.32 |
ENSG00000207974 |
. |
46183 |
0.16 |
| chr12_48551760_48551911 | 0.32 |
ASB8 |
ankyrin repeat and SOCS box containing 8 |
457 |
0.75 |
| chr19_6926549_6926700 | 0.32 |
RP11-1137G4.3 |
|
26598 |
0.12 |
| chr16_3116517_3116777 | 0.32 |
IL32 |
interleukin 32 |
834 |
0.3 |
| chr12_57441357_57441508 | 0.32 |
MYO1A |
myosin IA |
182 |
0.9 |
| chr22_17567910_17568443 | 0.32 |
IL17RA |
interleukin 17 receptor A |
2327 |
0.26 |
| chr2_128395227_128395389 | 0.32 |
LIMS2 |
LIM and senescent cell antigen-like domains 2 |
4398 |
0.16 |
| chr6_166954866_166955076 | 0.32 |
RPS6KA2 |
ribosomal protein S6 kinase, 90kDa, polypeptide 2 |
967 |
0.56 |
| chr4_6928559_6928859 | 0.31 |
TBC1D14 |
TBC1 domain family, member 14 |
16734 |
0.16 |
| chr1_26158794_26158945 | 0.31 |
RP1-317E23.7 |
|
244 |
0.84 |
| chr1_84902979_84903163 | 0.31 |
DNASE2B |
deoxyribonuclease II beta |
29158 |
0.16 |
| chr6_159074554_159074705 | 0.31 |
SYTL3 |
synaptotagmin-like 3 |
3583 |
0.22 |
| chr17_45297902_45298194 | 0.31 |
ENSG00000252088 |
. |
40 |
0.92 |
| chr11_128566078_128566229 | 0.31 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
235 |
0.91 |
| chr2_198651086_198651566 | 0.31 |
BOLL |
boule-like RNA-binding protein |
28 |
0.98 |
| chr9_117152254_117152405 | 0.31 |
AKNA |
AT-hook transcription factor |
1968 |
0.35 |
| chr14_100533458_100533776 | 0.31 |
EVL |
Enah/Vasp-like |
843 |
0.55 |
| chr15_74834439_74834753 | 0.31 |
ARID3B |
AT rich interactive domain 3B (BRIGHT-like) |
1078 |
0.48 |
| chr16_68403596_68403824 | 0.30 |
SMPD3 |
sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) |
1198 |
0.37 |
| chr17_75606628_75606867 | 0.30 |
SEPT9 |
septin 9 |
128752 |
0.05 |
| chr19_42703347_42703984 | 0.30 |
ENSG00000265122 |
. |
6435 |
0.1 |
| chr19_3036283_3036702 | 0.30 |
TLE2 |
transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) |
6880 |
0.13 |
| chr17_43305439_43305590 | 0.30 |
CTD-2020K17.1 |
|
5925 |
0.1 |
| chr4_184642687_184643147 | 0.30 |
ENSG00000251739 |
. |
15833 |
0.17 |
| chr18_24289152_24289303 | 0.30 |
ENSG00000265369 |
. |
19734 |
0.21 |
| chr7_50351765_50352753 | 0.30 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
3941 |
0.34 |
| chrX_64917369_64917808 | 0.30 |
MSN |
moesin |
30051 |
0.25 |
| chr6_135450306_135450457 | 0.30 |
HBS1L |
HBS1-like (S. cerevisiae) |
26187 |
0.17 |
| chr5_39207439_39207590 | 0.30 |
FYB |
FYN binding protein |
4385 |
0.32 |
| chr9_95945997_95946357 | 0.30 |
WNK2 |
WNK lysine deficient protein kinase 2 |
1021 |
0.58 |
| chr10_126182799_126183006 | 0.29 |
LHPP |
phospholysine phosphohistidine inorganic pyrophosphate phosphatase |
32490 |
0.16 |
| chr2_106572142_106572293 | 0.29 |
AC009505.2 |
|
98584 |
0.07 |
| chr17_43302789_43303077 | 0.29 |
CTD-2020K17.1 |
|
3344 |
0.12 |
| chr17_17654361_17654512 | 0.29 |
RAI1-AS1 |
RAI1 antisense RNA 1 |
19699 |
0.13 |
| chrX_19815653_19816006 | 0.29 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
2040 |
0.46 |
| chr1_156933589_156933740 | 0.29 |
ARHGEF11 |
Rho guanine nucleotide exchange factor (GEF) 11 |
14858 |
0.14 |
| chr1_231113171_231113322 | 0.29 |
TTC13 |
tetratricopeptide repeat domain 13 |
1296 |
0.3 |
| chr14_20811572_20811723 | 0.29 |
RPPH1 |
ribonuclease P RNA component H1 |
81 |
0.5 |
| chr6_2222017_2222664 | 0.29 |
GMDS |
GDP-mannose 4,6-dehydratase |
23586 |
0.28 |
| chr16_1585013_1585440 | 0.29 |
TMEM204 |
transmembrane protein 204 |
1652 |
0.19 |
| chrX_153267742_153268222 | 0.29 |
IRAK1 |
interleukin-1 receptor-associated kinase 1 |
11715 |
0.09 |
| chr1_120642387_120642538 | 0.29 |
ENSG00000207149 |
. |
27100 |
0.19 |
| chr21_45579276_45579734 | 0.28 |
AP001055.1 |
|
14075 |
0.13 |
| chr6_111180428_111180655 | 0.28 |
ENSG00000199360 |
. |
2921 |
0.21 |
| chr19_22193781_22193932 | 0.28 |
ZNF208 |
zinc finger protein 208 |
105 |
0.98 |
| chr2_98279402_98280293 | 0.28 |
ACTR1B |
ARP1 actin-related protein 1 homolog B, centractin beta (yeast) |
723 |
0.38 |
| chr1_89664664_89664873 | 0.28 |
GBP4 |
guanylate binding protein 4 |
153 |
0.96 |
| chr22_42831151_42831302 | 0.28 |
NFAM1 |
NFAT activating protein with ITAM motif 1 |
2825 |
0.27 |
| chr6_160241186_160241397 | 0.28 |
PNLDC1 |
poly(A)-specific ribonuclease (PARN)-like domain containing 1 |
19985 |
0.12 |
| chr8_24151584_24151953 | 0.28 |
ADAM28 |
ADAM metallopeptidase domain 28 |
144 |
0.97 |
| chr2_45168726_45169073 | 0.28 |
SIX3 |
SIX homeobox 3 |
3 |
0.61 |
| chr1_207997626_207997777 | 0.28 |
ENSG00000203709 |
. |
21833 |
0.22 |
| chr17_72459271_72459541 | 0.28 |
CD300A |
CD300a molecule |
3149 |
0.19 |
| chr1_160614457_160614608 | 0.28 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
2279 |
0.26 |
| chr16_29611589_29612053 | 0.28 |
ENSG00000266758 |
. |
1235 |
0.4 |
| chr6_90120799_90121205 | 0.28 |
RRAGD |
Ras-related GTP binding D |
787 |
0.65 |
| chr16_83760457_83760608 | 0.28 |
RP11-298D21.2 |
|
550 |
0.74 |
| chr16_89043094_89043288 | 0.28 |
CBFA2T3 |
core-binding factor, runt domain, alpha subunit 2; translocated to, 3 |
210 |
0.92 |
| chr2_68966290_68966892 | 0.28 |
ARHGAP25 |
Rho GTPase activating protein 25 |
4577 |
0.29 |
| chr4_186706311_186706462 | 0.28 |
SORBS2 |
sorbin and SH3 domain containing 2 |
9320 |
0.25 |
| chr8_121744015_121744417 | 0.28 |
RP11-713M15.1 |
|
29277 |
0.22 |
| chr11_128595669_128596120 | 0.28 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
29976 |
0.16 |
| chr9_66109751_66110085 | 0.28 |
ENSG00000202474 |
. |
348344 |
0.01 |
| chr15_45005178_45005397 | 0.28 |
B2M |
beta-2-microglobulin |
1572 |
0.31 |
| chr2_197034753_197035766 | 0.27 |
STK17B |
serine/threonine kinase 17b |
465 |
0.83 |
| chr12_9948574_9948725 | 0.27 |
KLRF1 |
killer cell lectin-like receptor subfamily F, member 1 |
31428 |
0.12 |
| chr12_6570039_6570337 | 0.27 |
TAPBPL |
TAP binding protein-like |
8938 |
0.09 |
| chr19_21182444_21182737 | 0.27 |
ZNF430 |
zinc finger protein 430 |
20836 |
0.19 |
| chr10_26737911_26738372 | 0.27 |
APBB1IP |
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
10787 |
0.27 |
| chr10_81947502_81948014 | 0.27 |
ANXA11 |
annexin A11 |
14981 |
0.2 |
| chr7_142180665_142180816 | 0.27 |
PRSS3P3 |
protease, serine, 3 pseudogene 3 |
191131 |
0.02 |
| chr4_99124752_99124919 | 0.27 |
ENSG00000200658 |
. |
29900 |
0.18 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035405 | histone-threonine phosphorylation(GO:0035405) |
| 0.1 | 0.5 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.1 | 0.2 | GO:0002540 | arachidonic acid metabolite production involved in inflammatory response(GO:0002538) leukotriene production involved in inflammatory response(GO:0002540) |
| 0.1 | 0.4 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.1 | 0.4 | GO:1904035 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.1 | 0.4 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.1 | 0.3 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.1 | 0.2 | GO:0045074 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.1 | 0.2 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.1 | 0.2 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.1 | 0.5 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.1 | 0.2 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.1 | 0.2 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.1 | 0.2 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.1 | 0.5 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.1 | 0.8 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.1 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.3 | GO:0034616 | response to laminar fluid shear stress(GO:0034616) |
| 0.1 | 0.2 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.1 | 0.3 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.1 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.4 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.1 | 0.2 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.1 | 0.2 | GO:0022614 | membrane to membrane docking(GO:0022614) |
| 0.1 | 0.2 | GO:0032543 | mitochondrial translation(GO:0032543) |
| 0.0 | 0.1 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.2 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0014874 | response to stimulus involved in regulation of muscle adaptation(GO:0014874) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0045759 | negative regulation of action potential(GO:0045759) |
| 0.0 | 0.2 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.0 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.2 | GO:0006356 | regulation of transcription from RNA polymerase I promoter(GO:0006356) |
| 0.0 | 0.1 | GO:0048143 | astrocyte activation(GO:0048143) |
| 0.0 | 0.1 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.2 | GO:0032528 | microvillus assembly(GO:0030033) microvillus organization(GO:0032528) |
| 0.0 | 0.0 | GO:0002691 | regulation of cellular extravasation(GO:0002691) |
| 0.0 | 0.4 | GO:0030851 | granulocyte differentiation(GO:0030851) |
| 0.0 | 0.1 | GO:0002282 | microglial cell activation involved in immune response(GO:0002282) |
| 0.0 | 0.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.1 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.2 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0006498 | N-terminal protein lipidation(GO:0006498) |
| 0.0 | 0.1 | GO:0061030 | epithelial cell differentiation involved in mammary gland alveolus development(GO:0061030) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.1 | GO:0002830 | positive regulation of type 2 immune response(GO:0002830) |
| 0.0 | 0.2 | GO:0051491 | positive regulation of filopodium assembly(GO:0051491) |
| 0.0 | 0.5 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.0 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.1 | GO:0045603 | positive regulation of endothelial cell differentiation(GO:0045603) |
| 0.0 | 0.1 | GO:0070672 | response to interleukin-15(GO:0070672) |
| 0.0 | 0.2 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.2 | GO:0046033 | AMP metabolic process(GO:0046033) |
| 0.0 | 0.2 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.2 | GO:0060263 | regulation of respiratory burst(GO:0060263) |
| 0.0 | 0.4 | GO:0050853 | B cell receptor signaling pathway(GO:0050853) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.6 | GO:0030101 | natural killer cell activation(GO:0030101) |
| 0.0 | 1.7 | GO:0031295 | lymphocyte costimulation(GO:0031294) T cell costimulation(GO:0031295) |
| 0.0 | 0.1 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.2 | GO:0036230 | granulocyte activation(GO:0036230) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0071636 | positive regulation of transforming growth factor beta production(GO:0071636) |
| 0.0 | 0.1 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.2 | GO:0032703 | negative regulation of interleukin-2 production(GO:0032703) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0046719 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) regulation by virus of viral protein levels in host cell(GO:0046719) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0002478 | antigen processing and presentation of exogenous peptide antigen(GO:0002478) |
| 0.0 | 0.6 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.1 | GO:0072180 | mesonephric duct morphogenesis(GO:0072180) |
| 0.0 | 0.1 | GO:0042661 | regulation of mesodermal cell fate specification(GO:0042661) regulation of mesoderm development(GO:2000380) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0045091 | regulation of single stranded viral RNA replication via double stranded DNA intermediate(GO:0045091) |
| 0.0 | 0.3 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 0.4 | GO:0045576 | mast cell activation(GO:0045576) |
| 0.0 | 0.1 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.0 | 0.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.3 | GO:0009309 | amine biosynthetic process(GO:0009309) |
| 0.0 | 0.3 | GO:0016578 | histone deubiquitination(GO:0016578) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0019673 | GDP-mannose metabolic process(GO:0019673) |
| 0.0 | 0.0 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.0 | GO:0032621 | interleukin-18 production(GO:0032621) regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.1 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) positive regulation of membrane depolarization(GO:1904181) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.2 | GO:0006349 | regulation of gene expression by genetic imprinting(GO:0006349) |
| 0.0 | 0.2 | GO:0048208 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0060017 | parathyroid gland development(GO:0060017) |
| 0.0 | 0.0 | GO:0046137 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) negative regulation of vitamin D biosynthetic process(GO:0010957) negative regulation of vitamin metabolic process(GO:0046137) |
| 0.0 | 0.1 | GO:0032494 | response to peptidoglycan(GO:0032494) |
| 0.0 | 0.1 | GO:0006030 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.0 | 0.1 | GO:0071168 | protein localization to chromatin(GO:0071168) |
| 0.0 | 0.1 | GO:0051290 | protein heterotetramerization(GO:0051290) |
| 0.0 | 0.0 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.0 | GO:0042362 | fat-soluble vitamin biosynthetic process(GO:0042362) |
| 0.0 | 0.1 | GO:0010738 | regulation of protein kinase A signaling(GO:0010738) |
| 0.0 | 0.0 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.1 | GO:0030502 | negative regulation of bone mineralization(GO:0030502) |
| 0.0 | 0.1 | GO:0032747 | interleukin-23 production(GO:0032627) regulation of interleukin-23 production(GO:0032667) positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.0 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0050665 | hydrogen peroxide biosynthetic process(GO:0050665) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.4 | GO:0046847 | filopodium assembly(GO:0046847) |
| 0.0 | 0.2 | GO:0097191 | extrinsic apoptotic signaling pathway via death domain receptors(GO:0008625) extrinsic apoptotic signaling pathway(GO:0097191) |
| 0.0 | 0.0 | GO:0048669 | collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.0 | GO:1901341 | activation of store-operated calcium channel activity(GO:0032237) regulation of store-operated calcium channel activity(GO:1901339) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.1 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.0 | GO:0045618 | positive regulation of keratinocyte differentiation(GO:0045618) |
| 0.0 | 0.0 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.0 | GO:0032467 | positive regulation of cytokinesis(GO:0032467) |
| 0.0 | 0.2 | GO:0045540 | regulation of cholesterol biosynthetic process(GO:0045540) |
| 0.0 | 0.2 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.1 | GO:0002286 | T cell activation involved in immune response(GO:0002286) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.0 | GO:0072131 | kidney mesenchyme morphogenesis(GO:0072131) metanephric mesenchyme morphogenesis(GO:0072133) |
| 0.0 | 0.0 | GO:1901522 | positive regulation of transcription from RNA polymerase II promoter involved in cellular response to chemical stimulus(GO:1901522) |
| 0.0 | 0.0 | GO:0014805 | smooth muscle adaptation(GO:0014805) |
| 0.0 | 0.0 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.0 | GO:0006222 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0006863 | purine nucleobase transport(GO:0006863) |
| 0.0 | 0.1 | GO:0033151 | V(D)J recombination(GO:0033151) |
| 0.0 | 0.0 | GO:0090382 | phagolysosome assembly(GO:0001845) phagosome maturation(GO:0090382) |
| 0.0 | 0.0 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.2 | GO:0015804 | neutral amino acid transport(GO:0015804) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0014002 | astrocyte development(GO:0014002) |
| 0.0 | 0.1 | GO:0018206 | peptidyl-methionine modification(GO:0018206) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.2 | GO:0030903 | notochord development(GO:0030903) |
| 0.0 | 0.0 | GO:0014742 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.0 | 0.1 | GO:0050655 | dermatan sulfate proteoglycan metabolic process(GO:0050655) |
| 0.0 | 0.1 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) positive regulation of keratinocyte migration(GO:0051549) |
| 0.0 | 0.1 | GO:1903077 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.0 | GO:0042940 | D-amino acid transport(GO:0042940) |
| 0.0 | 0.0 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0010288 | response to lead ion(GO:0010288) |
| 0.0 | 0.0 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.0 | GO:0060253 | regulation of Schwann cell proliferation(GO:0010624) negative regulation of Schwann cell proliferation(GO:0010626) regulation of glial cell proliferation(GO:0060251) negative regulation of glial cell proliferation(GO:0060253) |
| 0.0 | 0.0 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.1 | GO:0003057 | regulation of the force of heart contraction by chemical signal(GO:0003057) |
| 0.0 | 0.0 | GO:0032801 | receptor catabolic process(GO:0032801) |
| 0.0 | 0.1 | GO:0072498 | embryonic skeletal joint morphogenesis(GO:0060272) embryonic skeletal joint development(GO:0072498) |
| 0.0 | 0.0 | GO:0006681 | galactosylceramide metabolic process(GO:0006681) |
| 0.0 | 0.1 | GO:0048873 | homeostasis of number of cells within a tissue(GO:0048873) |
| 0.0 | 0.1 | GO:0018065 | protein-cofactor linkage(GO:0018065) |
| 0.0 | 0.0 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.2 | GO:0006903 | vesicle targeting(GO:0006903) |
| 0.0 | 0.0 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0015669 | gas transport(GO:0015669) |
| 0.0 | 0.0 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.0 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.0 | GO:0060426 | lung vasculature development(GO:0060426) |
| 0.0 | 0.1 | GO:0007096 | regulation of exit from mitosis(GO:0007096) |
| 0.0 | 0.1 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.0 | GO:0045672 | positive regulation of osteoclast differentiation(GO:0045672) |
| 0.0 | 0.0 | GO:2000644 | regulation of low-density lipoprotein particle receptor catabolic process(GO:0032803) regulation of receptor catabolic process(GO:2000644) |
| 0.0 | 0.0 | GO:0019441 | tryptophan catabolic process to kynurenine(GO:0019441) |
| 0.0 | 0.0 | GO:0071360 | cellular response to exogenous dsRNA(GO:0071360) |
| 0.0 | 0.0 | GO:0070874 | negative regulation of glycogen metabolic process(GO:0070874) |
| 0.0 | 0.1 | GO:0046838 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.3 | GO:0032480 | negative regulation of type I interferon production(GO:0032480) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0042726 | flavin-containing compound metabolic process(GO:0042726) |
| 0.0 | 0.2 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.0 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0043983 | histone H4-K12 acetylation(GO:0043983) |
| 0.0 | 0.0 | GO:0034219 | carbohydrate transmembrane transport(GO:0034219) |
| 0.0 | 0.1 | GO:0045022 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.0 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) positive regulation of hematopoietic progenitor cell differentiation(GO:1901534) |
| 0.0 | 0.0 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.1 | GO:0008535 | respiratory chain complex IV assembly(GO:0008535) |
| 0.0 | 0.0 | GO:0001543 | ovarian follicle rupture(GO:0001543) |
| 0.0 | 0.1 | GO:0042088 | T-helper 1 type immune response(GO:0042088) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.0 | GO:0000019 | regulation of mitotic recombination(GO:0000019) |
| 0.0 | 0.0 | GO:0046501 | protoporphyrinogen IX metabolic process(GO:0046501) |
| 0.0 | 0.0 | GO:0010988 | regulation of low-density lipoprotein particle clearance(GO:0010988) |
| 0.0 | 0.1 | GO:0000085 | mitotic G2 phase(GO:0000085) G2 phase(GO:0051319) |
| 0.0 | 0.2 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
| 0.0 | 0.1 | GO:0032007 | negative regulation of TOR signaling(GO:0032007) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.5 | GO:0000346 | transcription export complex(GO:0000346) |
| 0.1 | 0.2 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.1 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.2 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.1 | 0.4 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.1 | 0.2 | GO:0000124 | SAGA complex(GO:0000124) |
| 0.1 | 0.2 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0042629 | mast cell granule(GO:0042629) |
| 0.0 | 0.2 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.3 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.3 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.2 | GO:0031254 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.2 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.2 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.1 | GO:0033011 | perinuclear theca(GO:0033011) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.1 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.7 | GO:0030864 | cortical actin cytoskeleton(GO:0030864) |
| 0.0 | 0.1 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:0045275 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0071256 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0071204 | histone pre-mRNA 3'end processing complex(GO:0071204) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.6 | GO:0030175 | filopodium(GO:0030175) |
| 0.0 | 0.1 | GO:0042598 | obsolete vesicular fraction(GO:0042598) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.8 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0042582 | primary lysosome(GO:0005766) azurophil granule(GO:0042582) |
| 0.0 | 0.7 | GO:0000118 | histone deacetylase complex(GO:0000118) |
| 0.0 | 0.1 | GO:0061200 | clathrin-sculpted gamma-aminobutyric acid transport vesicle(GO:0061200) clathrin-sculpted gamma-aminobutyric acid transport vesicle membrane(GO:0061202) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.1 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0031080 | nuclear pore outer ring(GO:0031080) |
| 0.0 | 0.9 | GO:0030027 | lamellipodium(GO:0030027) |
| 0.0 | 0.1 | GO:0051233 | spindle midzone(GO:0051233) |
| 0.0 | 0.1 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.1 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.0 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.0 | 0.1 | GO:0000300 | obsolete peripheral to membrane of membrane fraction(GO:0000300) |
| 0.0 | 0.0 | GO:0044462 | external encapsulating structure part(GO:0044462) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0005779 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.0 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.1 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.0 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.1 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.0 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.0 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.0 | GO:0001740 | Barr body(GO:0001740) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.1 | 0.3 | GO:1901567 | icosanoid binding(GO:0050542) fatty acid derivative binding(GO:1901567) |
| 0.1 | 0.6 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.5 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.1 | 0.3 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.1 | 0.4 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.1 | 0.4 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.1 | 0.2 | GO:0004525 | ribonuclease III activity(GO:0004525) double-stranded RNA-specific ribonuclease activity(GO:0032296) |
| 0.1 | 0.3 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.1 | 0.2 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.1 | 0.2 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.1 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.1 | 0.2 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.1 | 0.2 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.1 | 0.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 1.2 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.2 | GO:0044020 | histone methyltransferase activity (H4-R3 specific)(GO:0044020) |
| 0.0 | 0.3 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.3 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.4 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 1.6 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
| 0.0 | 0.1 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.3 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.1 | GO:0004476 | mannose-6-phosphate isomerase activity(GO:0004476) |
| 0.0 | 0.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0034713 | type I transforming growth factor beta receptor binding(GO:0034713) |
| 0.0 | 0.8 | GO:0003823 | antigen binding(GO:0003823) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.1 | GO:0001537 | N-acetylgalactosamine 4-O-sulfotransferase activity(GO:0001537) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0016885 | ligase activity, forming carbon-carbon bonds(GO:0016885) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.1 | GO:0003964 | telomerase activity(GO:0003720) RNA-directed DNA polymerase activity(GO:0003964) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.2 | GO:0005283 | sodium:amino acid symporter activity(GO:0005283) |
| 0.0 | 0.0 | GO:0032558 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.0 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.0 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.1 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0030343 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.1 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.1 | GO:0003701 | obsolete RNA polymerase I transcription factor activity(GO:0003701) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.3 | GO:0008656 | cysteine-type endopeptidase activator activity involved in apoptotic process(GO:0008656) |
| 0.0 | 0.1 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.2 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.2 | GO:0042162 | telomeric DNA binding(GO:0042162) |
| 0.0 | 0.4 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.5 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.0 | 0.3 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.0 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0008934 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.0 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.0 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.0 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.3 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.1 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.1 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0004566 | beta-glucuronidase activity(GO:0004566) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.0 | GO:0042808 | obsolete neuronal Cdc2-like kinase binding(GO:0042808) |
| 0.0 | 0.0 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.0 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.2 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.0 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.2 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0003993 | acid phosphatase activity(GO:0003993) |
| 0.0 | 0.1 | GO:0005345 | purine nucleobase transmembrane transporter activity(GO:0005345) |
| 0.0 | 0.1 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.0 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.0 | GO:0016004 | phospholipase activator activity(GO:0016004) |
| 0.0 | 0.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.1 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.2 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:0032552 | deoxyribonucleotide binding(GO:0032552) |
| 0.0 | 0.4 | GO:0003697 | single-stranded DNA binding(GO:0003697) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.0 | GO:0004449 | isocitrate dehydrogenase (NAD+) activity(GO:0004449) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.1 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.0 | 0.0 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.0 | GO:0016635 | oxidoreductase activity, acting on the CH-CH group of donors, quinone or related compound as acceptor(GO:0016635) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 1.0 | GO:0004519 | endonuclease activity(GO:0004519) |
| 0.0 | 0.0 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.4 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.0 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.3 | GO:0097472 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) cyclin-dependent protein kinase activity(GO:0097472) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.0 | GO:0003873 | 6-phosphofructo-2-kinase activity(GO:0003873) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.3 | GO:0004869 | cysteine-type endopeptidase inhibitor activity(GO:0004869) |
| 0.0 | 0.0 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.1 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0051766 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) inositol trisphosphate kinase activity(GO:0051766) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 2.5 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.2 | PID CD8 TCR PATHWAY | TCR signaling in naïve CD8+ T cells |
| 0.0 | 0.3 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 1.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 0.2 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.1 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.8 | PID INTEGRIN A4B1 PATHWAY | Alpha4 beta1 integrin signaling events |
| 0.0 | 0.6 | PID BCR 5PATHWAY | BCR signaling pathway |
| 0.0 | 0.6 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.4 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.3 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.2 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.2 | SA TRKA RECEPTOR | The TrkA receptor binds nerve growth factor to activate MAP kinase pathways and promote cell growth. |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.0 | PID P38 ALPHA BETA PATHWAY | Regulation of p38-alpha and p38-beta |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | PID HDAC CLASSII PATHWAY | Signaling events mediated by HDAC Class II |
| 0.0 | 0.2 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | PID GMCSF PATHWAY | GMCSF-mediated signaling events |
| 0.0 | 0.1 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.4 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.1 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 0.2 | PID CD40 PATHWAY | CD40/CD40L signaling |
| 0.0 | 0.1 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.5 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.5 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 0.1 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.2 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.2 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.1 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.2 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.3 | PID IL4 2PATHWAY | IL4-mediated signaling events |
| 0.0 | 0.2 | PID IL12 2PATHWAY | IL12-mediated signaling events |
| 0.0 | 0.2 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.1 | PID ER NONGENOMIC PATHWAY | Plasma membrane estrogen receptor signaling |
| 0.0 | 0.2 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 2.0 | REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | Genes involved in Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell |
| 0.0 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.0 | REACTOME SCFSKP2 MEDIATED DEGRADATION OF P27 P21 | Genes involved in SCF(Skp2)-mediated degradation of p27/p21 |
| 0.0 | 0.3 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.0 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME GPVI MEDIATED ACTIVATION CASCADE | Genes involved in GPVI-mediated activation cascade |
| 0.0 | 0.8 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.9 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.4 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.2 | REACTOME ACTIVATION OF IRF3 IRF7 MEDIATED BY TBK1 IKK EPSILON | Genes involved in Activation of IRF3/IRF7 mediated by TBK1/IKK epsilon |
| 0.0 | 0.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 0.2 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.0 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.2 | REACTOME IL 2 SIGNALING | Genes involved in Interleukin-2 signaling |
| 0.0 | 0.1 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.0 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.5 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.2 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF THE APC C | Genes involved in Phosphorylation of the APC/C |
| 0.0 | 0.3 | REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | Genes involved in Antigen Activates B Cell Receptor Leading to Generation of Second Messengers |
| 0.0 | 0.1 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 0.2 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.1 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.1 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.0 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.1 | REACTOME APC C CDH1 MEDIATED DEGRADATION OF CDC20 AND OTHER APC C CDH1 TARGETED PROTEINS IN LATE MITOSIS EARLY G1 | Genes involved in APC/C:Cdh1 mediated degradation of Cdc20 and other APC/C:Cdh1 targeted proteins in late mitosis/early G1 |
| 0.0 | 0.1 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.2 | REACTOME HOMOLOGOUS RECOMBINATION REPAIR OF REPLICATION INDEPENDENT DOUBLE STRAND BREAKS | Genes involved in Homologous recombination repair of replication-independent double-strand breaks |
| 0.0 | 0.0 | REACTOME REGULATION OF AMPK ACTIVITY VIA LKB1 | Genes involved in Regulation of AMPK activity via LKB1 |
| 0.0 | 0.2 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.1 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.1 | REACTOME TCR SIGNALING | Genes involved in TCR signaling |
| 0.0 | 0.2 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.1 | REACTOME METABOLISM OF POLYAMINES | Genes involved in Metabolism of polyamines |
| 0.0 | 0.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.0 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.2 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.2 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.1 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.0 | REACTOME SHC RELATED EVENTS | Genes involved in SHC-related events |
| 0.0 | 0.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.1 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.2 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |