Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
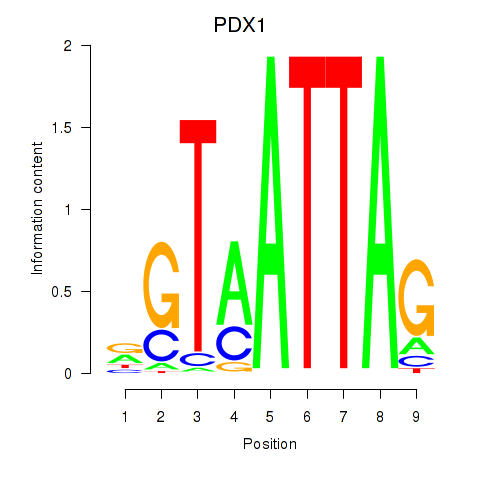
Results for PDX1
Z-value: 0.76
Transcription factors associated with PDX1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PDX1
|
ENSG00000139515.5 | pancreatic and duodenal homeobox 1 |
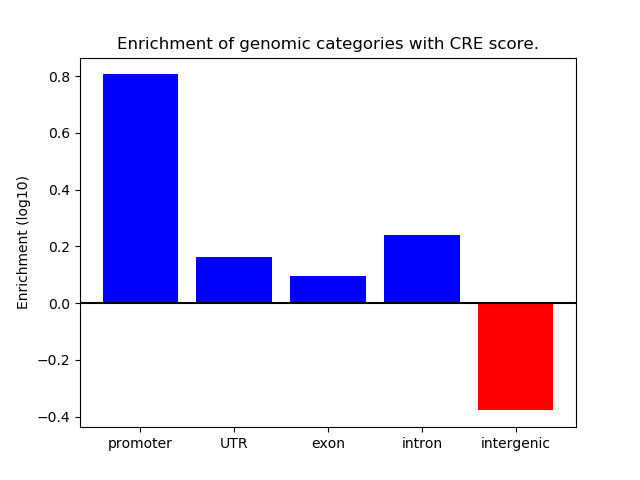
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_28494142_28494415 | PDX1 | 121 | 0.934648 | -0.53 | 1.4e-01 | Click! |
| chr13_28491022_28491173 | PDX1 | 3060 | 0.201109 | 0.50 | 1.7e-01 | Click! |
| chr13_28493957_28494140 | PDX1 | 109 | 0.944493 | -0.41 | 2.8e-01 | Click! |
| chr13_28493585_28493736 | PDX1 | 497 | 0.716519 | -0.17 | 6.6e-01 | Click! |
Activity of the PDX1 motif across conditions
Conditions sorted by the z-value of the PDX1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
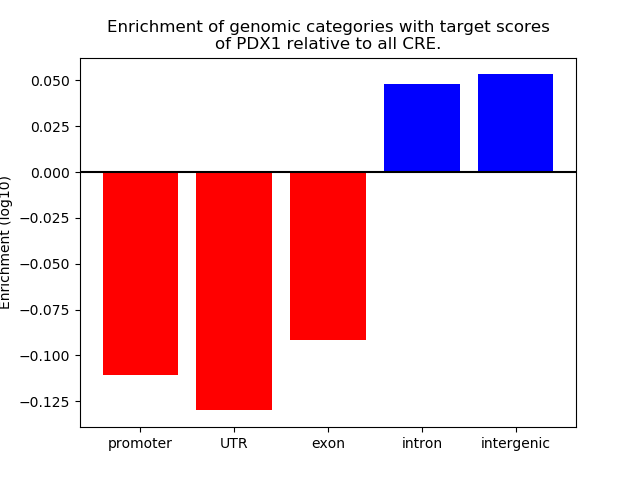
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr11_106239153_106239583 | 0.44 |
RP11-680E19.1 |
|
104326 |
0.08 |
| chr7_93550787_93551002 | 0.38 |
GNG11 |
guanine nucleotide binding protein (G protein), gamma 11 |
117 |
0.96 |
| chr12_78336292_78336638 | 0.33 |
NAV3 |
neuron navigator 3 |
23591 |
0.28 |
| chr16_54968513_54968925 | 0.32 |
IRX5 |
iroquois homeobox 5 |
2735 |
0.43 |
| chr12_49661445_49661747 | 0.32 |
TUBA1C |
tubulin, alpha 1c |
2732 |
0.15 |
| chr7_90012947_90013805 | 0.31 |
CLDN12 |
claudin 12 |
341 |
0.9 |
| chr6_56715748_56715936 | 0.31 |
DST |
dystonin |
907 |
0.63 |
| chr12_56917781_56918152 | 0.30 |
RBMS2 |
RNA binding motif, single stranded interacting protein 2 |
2183 |
0.23 |
| chr16_89990965_89991193 | 0.30 |
TUBB3 |
Tubulin beta-3 chain |
1305 |
0.26 |
| chr10_33620428_33620906 | 0.29 |
NRP1 |
neuropilin 1 |
2643 |
0.37 |
| chr15_96878465_96878729 | 0.29 |
ENSG00000222651 |
. |
2107 |
0.24 |
| chr8_42398666_42399077 | 0.28 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
1802 |
0.28 |
| chr1_196577911_196578299 | 0.28 |
KCNT2 |
potassium channel, subfamily T, member 2 |
250 |
0.94 |
| chr21_17442999_17443232 | 0.27 |
ENSG00000252273 |
. |
35286 |
0.24 |
| chr13_24146066_24146217 | 0.27 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
1338 |
0.59 |
| chr10_6763782_6764322 | 0.27 |
PRKCQ |
protein kinase C, theta |
141789 |
0.05 |
| chr3_150089259_150089633 | 0.27 |
TSC22D2 |
TSC22 domain family, member 2 |
36676 |
0.21 |
| chr15_96878987_96879612 | 0.26 |
ENSG00000222651 |
. |
2809 |
0.21 |
| chr17_2117226_2117470 | 0.26 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
295 |
0.52 |
| chr7_95107815_95108198 | 0.25 |
ASB4 |
ankyrin repeat and SOCS box containing 4 |
7207 |
0.2 |
| chr1_151964634_151965021 | 0.25 |
S100A10 |
S100 calcium binding protein A10 |
227 |
0.9 |
| chr13_95619763_95620412 | 0.25 |
ENSG00000252335 |
. |
51402 |
0.18 |
| chr10_29010683_29010909 | 0.25 |
ENSG00000201001 |
. |
38936 |
0.14 |
| chr3_112356169_112356393 | 0.25 |
CCDC80 |
coiled-coil domain containing 80 |
663 |
0.77 |
| chr22_36232810_36232961 | 0.24 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
3380 |
0.35 |
| chr11_65324098_65324348 | 0.24 |
LTBP3 |
latent transforming growth factor beta binding protein 3 |
1013 |
0.31 |
| chr10_119303944_119304573 | 0.24 |
EMX2OS |
EMX2 opposite strand/antisense RNA |
321 |
0.88 |
| chr3_87036535_87037613 | 0.23 |
VGLL3 |
vestigial like 3 (Drosophila) |
2778 |
0.42 |
| chr4_41215270_41215769 | 0.23 |
APBB2 |
amyloid beta (A4) precursor protein-binding, family B, member 2 |
956 |
0.59 |
| chr4_95917210_95917447 | 0.23 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
55 |
0.99 |
| chr5_148786412_148786563 | 0.23 |
ENSG00000208035 |
. |
21994 |
0.11 |
| chr8_41165312_41165463 | 0.23 |
SFRP1 |
secreted frizzled-related protein 1 |
878 |
0.57 |
| chr11_126456116_126456330 | 0.23 |
KIRREL3-AS1 |
KIRREL3 antisense RNA 1 |
42381 |
0.16 |
| chr3_115342492_115342792 | 0.23 |
GAP43 |
growth associated protein 43 |
285 |
0.94 |
| chr11_31830603_31830754 | 0.23 |
PAX6 |
paired box 6 |
1175 |
0.49 |
| chr2_200446222_200446758 | 0.23 |
SATB2 |
SATB homeobox 2 |
110501 |
0.07 |
| chr5_3597777_3597928 | 0.23 |
IRX1 |
iroquois homeobox 1 |
1684 |
0.41 |
| chr18_25754335_25754486 | 0.22 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
3000 |
0.42 |
| chr17_70113469_70114161 | 0.22 |
SOX9 |
SRY (sex determining region Y)-box 9 |
3346 |
0.37 |
| chr10_33621519_33621932 | 0.22 |
NRP1 |
neuropilin 1 |
1585 |
0.49 |
| chr11_121966638_121966789 | 0.22 |
ENSG00000207971 |
. |
3839 |
0.19 |
| chr9_94710782_94711296 | 0.22 |
ROR2 |
receptor tyrosine kinase-like orphan receptor 2 |
123 |
0.98 |
| chr10_33622034_33622426 | 0.22 |
NRP1 |
neuropilin 1 |
1080 |
0.63 |
| chr2_203244162_203244384 | 0.22 |
BMPR2 |
bone morphogenetic protein receptor, type II (serine/threonine kinase) |
2116 |
0.28 |
| chr20_36040358_36040543 | 0.22 |
SRC |
v-src avian sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog |
27897 |
0.21 |
| chr14_74300817_74300968 | 0.22 |
PTGR2 |
prostaglandin reductase 2 |
17655 |
0.1 |
| chr6_130686119_130686489 | 0.21 |
SAMD3 |
sterile alpha motif domain containing 3 |
266 |
0.75 |
| chr15_96881380_96881711 | 0.21 |
ENSG00000222651 |
. |
5055 |
0.17 |
| chr15_60388257_60388408 | 0.21 |
FOXB1 |
forkhead box B1 |
91911 |
0.09 |
| chr13_84456002_84456225 | 0.21 |
SLITRK1 |
SLIT and NTRK-like family, member 1 |
415 |
0.92 |
| chr5_16713576_16713796 | 0.21 |
MYO10 |
myosin X |
24810 |
0.21 |
| chr4_169553357_169553551 | 0.21 |
PALLD |
palladin, cytoskeletal associated protein |
686 |
0.73 |
| chr2_216283171_216283492 | 0.21 |
FN1 |
fibronectin 1 |
17459 |
0.2 |
| chr7_94025506_94025990 | 0.20 |
COL1A2 |
collagen, type I, alpha 2 |
1875 |
0.46 |
| chr14_52247052_52247252 | 0.20 |
RP11-280K24.4 |
|
15158 |
0.18 |
| chr11_24518160_24518510 | 0.20 |
LUZP2 |
leucine zipper protein 2 |
389 |
0.91 |
| chr13_95003800_95004010 | 0.20 |
ENSG00000212057 |
. |
113739 |
0.06 |
| chr16_69854762_69854918 | 0.20 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
17800 |
0.14 |
| chr3_120215962_120216316 | 0.20 |
FSTL1 |
follistatin-like 1 |
46039 |
0.16 |
| chr7_98029134_98029377 | 0.20 |
BAIAP2L1 |
BAI1-associated protein 2-like 1 |
1125 |
0.59 |
| chr5_71406834_71406985 | 0.20 |
MAP1B |
microtubule-associated protein 1B |
3596 |
0.33 |
| chr14_36409550_36409712 | 0.20 |
RP11-116N8.1 |
|
42443 |
0.16 |
| chr8_42063953_42065062 | 0.20 |
PLAT |
plasminogen activator, tissue |
576 |
0.71 |
| chr8_42358808_42359006 | 0.20 |
SLC20A2 |
solute carrier family 20 (phosphate transporter), member 2 |
71 |
0.97 |
| chr7_80546522_80546706 | 0.19 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
1885 |
0.51 |
| chr11_31831142_31831293 | 0.19 |
PAX6 |
paired box 6 |
636 |
0.72 |
| chr7_129933276_129933885 | 0.19 |
CPA4 |
carboxypeptidase A4 |
461 |
0.76 |
| chrX_102585134_102585599 | 0.19 |
TCEAL7 |
transcription elongation factor A (SII)-like 7 |
209 |
0.92 |
| chr7_100777700_100777901 | 0.19 |
SERPINE1 |
serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 1 |
7421 |
0.1 |
| chr11_122311916_122312418 | 0.19 |
ENSG00000252776 |
. |
7751 |
0.23 |
| chr12_59311566_59311750 | 0.19 |
LRIG3 |
leucine-rich repeats and immunoglobulin-like domains 3 |
1669 |
0.41 |
| chr4_81191052_81191243 | 0.19 |
FGF5 |
fibroblast growth factor 5 |
3354 |
0.31 |
| chr11_12697523_12698245 | 0.19 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
834 |
0.75 |
| chr3_44063258_44064101 | 0.18 |
ENSG00000252980 |
. |
48900 |
0.17 |
| chr10_29228474_29228881 | 0.18 |
ENSG00000199402 |
. |
65241 |
0.13 |
| chr4_71997150_71997347 | 0.18 |
SLC4A4 |
solute carrier family 4 (sodium bicarbonate cotransporter), member 4 |
55755 |
0.16 |
| chr1_164529940_164530511 | 0.18 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
1165 |
0.61 |
| chr9_100069620_100070257 | 0.18 |
CCDC180 |
coiled-coil domain containing 180 |
3 |
0.98 |
| chr7_116660690_116660934 | 0.18 |
ST7 |
suppression of tumorigenicity 7 |
481 |
0.81 |
| chr9_133722351_133722531 | 0.18 |
ABL1 |
c-abl oncogene 1, non-receptor tyrosine kinase |
11988 |
0.21 |
| chr8_22023419_22024083 | 0.18 |
BMP1 |
bone morphogenetic protein 1 |
951 |
0.4 |
| chr18_53069042_53069242 | 0.18 |
TCF4 |
transcription factor 4 |
60 |
0.98 |
| chr7_110732102_110732377 | 0.18 |
LRRN3 |
leucine rich repeat neuronal 3 |
1140 |
0.59 |
| chr20_10844541_10844810 | 0.18 |
RP11-103J8.1 |
|
38257 |
0.22 |
| chr9_101014697_101014848 | 0.18 |
TBC1D2 |
TBC1 domain family, member 2 |
3090 |
0.33 |
| chr18_30310087_30311054 | 0.18 |
AC012123.1 |
Uncharacterized protein |
39188 |
0.17 |
| chr13_61987939_61988245 | 0.18 |
PCDH20 |
protocadherin 20 |
1563 |
0.57 |
| chr3_110791913_110792064 | 0.18 |
PVRL3 |
poliovirus receptor-related 3 |
973 |
0.53 |
| chr5_92921648_92921875 | 0.18 |
ENSG00000237187 |
. |
407 |
0.85 |
| chr16_17931379_17931530 | 0.17 |
XYLT1 |
xylosyltransferase I |
366716 |
0.01 |
| chr10_102761690_102761841 | 0.17 |
LZTS2 |
leucine zipper, putative tumor suppressor 2 |
2155 |
0.16 |
| chr2_98962646_98962882 | 0.17 |
CNGA3 |
cyclic nucleotide gated channel alpha 3 |
113 |
0.9 |
| chrX_86773476_86773748 | 0.17 |
KLHL4 |
kelch-like family member 4 |
795 |
0.74 |
| chr15_37402857_37403086 | 0.17 |
MEIS2 |
Meis homeobox 2 |
9467 |
0.23 |
| chr12_91575190_91575391 | 0.17 |
DCN |
decorin |
537 |
0.85 |
| chr1_30179791_30179986 | 0.17 |
ENSG00000221126 |
. |
62363 |
0.15 |
| chr13_27927443_27927612 | 0.17 |
ENSG00000252247 |
. |
9644 |
0.16 |
| chr14_50996670_50996889 | 0.17 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
2411 |
0.26 |
| chr6_116833589_116833880 | 0.17 |
TRAPPC3L |
trafficking protein particle complex 3-like |
228 |
0.83 |
| chr4_15471989_15472140 | 0.17 |
CC2D2A |
coiled-coil and C2 domain containing 2A |
427 |
0.85 |
| chr5_140864017_140864278 | 0.17 |
PCDHGC4 |
protocadherin gamma subfamily C, 4 |
594 |
0.53 |
| chr12_72724242_72724393 | 0.17 |
ENSG00000236333 |
. |
55630 |
0.15 |
| chr15_63673071_63673237 | 0.17 |
CA12 |
carbonic anhydrase XII |
880 |
0.7 |
| chr14_54687357_54687695 | 0.17 |
CDKN3 |
cyclin-dependent kinase inhibitor 3 |
176147 |
0.03 |
| chr10_49734913_49735076 | 0.17 |
ARHGAP22 |
Rho GTPase activating protein 22 |
2713 |
0.31 |
| chr6_116834038_116834189 | 0.17 |
TRAPPC3L |
trafficking protein particle complex 3-like |
607 |
0.55 |
| chr16_2315946_2316097 | 0.16 |
RNPS1 |
RNA binding protein S1, serine-rich domain |
503 |
0.49 |
| chr9_69882113_69882264 | 0.16 |
ENSG00000206804 |
. |
10705 |
0.26 |
| chr5_15620950_15621349 | 0.16 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
5058 |
0.28 |
| chr1_30559030_30559253 | 0.16 |
ENSG00000222787 |
. |
201392 |
0.03 |
| chr8_33457544_33457712 | 0.16 |
DUSP26 |
dual specificity phosphatase 26 (putative) |
4 |
0.98 |
| chr14_52822199_52822380 | 0.16 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
41176 |
0.18 |
| chr6_82600909_82601087 | 0.16 |
ENSG00000206886 |
. |
127257 |
0.05 |
| chr12_105651079_105651410 | 0.16 |
APPL2 |
adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 2 |
21228 |
0.19 |
| chr22_18640512_18640663 | 0.16 |
USP18 |
ubiquitin specific peptidase 18 |
7921 |
0.17 |
| chr16_69959859_69960167 | 0.16 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
1120 |
0.44 |
| chr4_186853985_186854481 | 0.16 |
ENSG00000239034 |
. |
13004 |
0.21 |
| chr9_18474095_18474958 | 0.16 |
ADAMTSL1 |
ADAMTS-like 1 |
295 |
0.95 |
| chr5_87780535_87780686 | 0.16 |
TMEM161B-AS1 |
TMEM161B antisense RNA 1 |
58098 |
0.14 |
| chr3_159483781_159484285 | 0.16 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
1141 |
0.44 |
| chr5_3197978_3198129 | 0.16 |
IRX1 |
iroquois homeobox 1 |
398115 |
0.01 |
| chr7_55142263_55142544 | 0.16 |
EGFR |
epidermal growth factor receptor |
35013 |
0.22 |
| chr21_17792302_17792515 | 0.16 |
ENSG00000207638 |
. |
119001 |
0.06 |
| chr12_30906652_30906803 | 0.16 |
CAPRIN2 |
caprin family member 2 |
518 |
0.86 |
| chr4_95635906_95636061 | 0.16 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
43136 |
0.21 |
| chr2_317550_317840 | 0.16 |
FAM150B |
family with sequence similarity 150, member B |
28844 |
0.16 |
| chr10_33630285_33630504 | 0.16 |
NRP1 |
neuropilin 1 |
5204 |
0.3 |
| chr12_120246427_120246712 | 0.16 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
5382 |
0.27 |
| chr2_192416956_192417327 | 0.16 |
NABP1 |
nucleic acid binding protein 1 |
125721 |
0.05 |
| chr10_26846446_26846738 | 0.16 |
ENSG00000199733 |
. |
48074 |
0.17 |
| chr11_19585849_19586032 | 0.15 |
ENSG00000265210 |
. |
10917 |
0.19 |
| chr15_77710885_77711123 | 0.15 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
1438 |
0.39 |
| chr17_48124761_48124984 | 0.15 |
ITGA3 |
integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) |
8467 |
0.14 |
| chr7_151297260_151297484 | 0.15 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
32067 |
0.18 |
| chr6_19691587_19692480 | 0.15 |
ENSG00000200957 |
. |
49273 |
0.18 |
| chr14_53416976_53417370 | 0.15 |
FERMT2 |
fermitin family member 2 |
113 |
0.98 |
| chr11_8290078_8290567 | 0.15 |
LMO1 |
LIM domain only 1 (rhombotin 1) |
59 |
0.98 |
| chr17_2151727_2151878 | 0.15 |
SMG6 |
SMG6 nonsense mediated mRNA decay factor |
3596 |
0.15 |
| chr9_66699115_66699266 | 0.15 |
ENSG00000206946 |
. |
13544 |
0.3 |
| chr10_25008347_25008498 | 0.15 |
ARHGAP21 |
Rho GTPase activating protein 21 |
2373 |
0.41 |
| chr6_37105124_37105275 | 0.15 |
PIM1 |
pim-1 oncogene |
32780 |
0.15 |
| chr9_21555895_21556299 | 0.15 |
MIR31HG |
MIR31 host gene (non-protein coding) |
3571 |
0.22 |
| chr10_112296305_112296649 | 0.15 |
SMC3 |
structural maintenance of chromosomes 3 |
30972 |
0.13 |
| chr4_126311194_126311396 | 0.15 |
FAT4 |
FAT atypical cadherin 4 |
3796 |
0.32 |
| chr1_78474827_78475270 | 0.15 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
4510 |
0.17 |
| chr6_140859694_140859893 | 0.15 |
ENSG00000221336 |
. |
122057 |
0.06 |
| chr4_81192022_81192173 | 0.15 |
FGF5 |
fibroblast growth factor 5 |
4304 |
0.29 |
| chr1_52456055_52456657 | 0.15 |
RAB3B |
RAB3B, member RAS oncogene family |
80 |
0.95 |
| chr8_56901426_56901577 | 0.15 |
ENSG00000240905 |
. |
8405 |
0.16 |
| chr7_134465735_134465936 | 0.15 |
CALD1 |
caldesmon 1 |
1406 |
0.57 |
| chr10_18453155_18453355 | 0.14 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
23146 |
0.25 |
| chr3_111717643_111718087 | 0.14 |
TAGLN3 |
transgelin 3 |
171 |
0.95 |
| chr1_39877379_39877579 | 0.14 |
KIAA0754 |
KIAA0754 |
1328 |
0.41 |
| chr8_38855409_38855789 | 0.14 |
ADAM9 |
ADAM metallopeptidase domain 9 |
1094 |
0.34 |
| chr5_3560421_3560572 | 0.14 |
IRX1 |
iroquois homeobox 1 |
35672 |
0.2 |
| chr2_238318512_238318719 | 0.14 |
COL6A3 |
collagen, type VI, alpha 3 |
4176 |
0.24 |
| chr4_4576417_4576570 | 0.14 |
STX18-AS1 |
STX18 antisense RNA 1 (head to head) |
32348 |
0.18 |
| chr15_39985088_39985239 | 0.14 |
RP11-37C7.3 |
|
89609 |
0.07 |
| chr15_52675291_52675442 | 0.14 |
MYO5A |
myosin VA (heavy chain 12, myoxin) |
31688 |
0.19 |
| chr1_170639404_170639633 | 0.14 |
PRRX1 |
paired related homeobox 1 |
6440 |
0.31 |
| chr5_58883529_58883730 | 0.14 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
1304 |
0.62 |
| chr15_96886136_96886425 | 0.14 |
ENSG00000222651 |
. |
9790 |
0.16 |
| chr6_99796840_99797257 | 0.14 |
FAXC |
failed axon connections homolog (Drosophila) |
483 |
0.83 |
| chr1_215178346_215178497 | 0.14 |
KCNK2 |
potassium channel, subfamily K, member 2 |
777 |
0.8 |
| chr1_85873190_85873383 | 0.14 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
3106 |
0.28 |
| chr8_6456469_6456620 | 0.14 |
ANGPT2 |
angiopoietin 2 |
35614 |
0.18 |
| chr2_239337309_239337646 | 0.14 |
AC016999.2 |
|
1107 |
0.42 |
| chr2_202929676_202929827 | 0.14 |
AC079354.1 |
uncharacterized protein KIAA2012 |
8227 |
0.16 |
| chr7_129934048_129934199 | 0.14 |
CPA4 |
carboxypeptidase A4 |
1004 |
0.45 |
| chr3_61546098_61546562 | 0.14 |
PTPRG |
protein tyrosine phosphatase, receptor type, G |
1255 |
0.64 |
| chr3_105657205_105657471 | 0.14 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
68942 |
0.15 |
| chr7_18549207_18549425 | 0.14 |
HDAC9 |
histone deacetylase 9 |
380 |
0.92 |
| chr10_128250315_128250594 | 0.14 |
ENSG00000221717 |
. |
4821 |
0.29 |
| chr19_15361458_15361910 | 0.14 |
EPHX3 |
epoxide hydrolase 3 |
17438 |
0.15 |
| chr5_155409653_155409804 | 0.14 |
SGCD |
sarcoglycan, delta (35kDa dystrophin-associated glycoprotein) |
112374 |
0.07 |
| chr6_9116214_9116470 | 0.14 |
SLC35B3 |
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B3 |
680548 |
0.0 |
| chr10_116474996_116475239 | 0.14 |
ABLIM1 |
actin binding LIM protein 1 |
30703 |
0.21 |
| chr12_79916269_79916420 | 0.14 |
ENSG00000243714 |
. |
2606 |
0.3 |
| chr11_33399282_33399433 | 0.14 |
ENSG00000223134 |
. |
23346 |
0.23 |
| chr5_171986318_171986532 | 0.14 |
AC027309.1 |
|
50011 |
0.13 |
| chr3_55521292_55522040 | 0.14 |
WNT5A-AS1 |
WNT5A antisense RNA 1 |
61 |
0.83 |
| chr4_87373379_87373575 | 0.13 |
MAPK10 |
mitogen-activated protein kinase 10 |
806 |
0.74 |
| chr19_1361544_1361801 | 0.13 |
MUM1 |
melanoma associated antigen (mutated) 1 |
5309 |
0.1 |
| chr10_119302142_119302346 | 0.13 |
EMX2 |
empty spiracles homeobox 2 |
265 |
0.64 |
| chr5_110252790_110252941 | 0.13 |
TSLP |
thymic stromal lymphopoietin |
152895 |
0.04 |
| chr7_130589596_130590032 | 0.13 |
ENSG00000226380 |
. |
27516 |
0.22 |
| chr8_87188438_87188625 | 0.13 |
CTD-3118D11.3 |
|
4116 |
0.24 |
| chr1_182932139_182932350 | 0.13 |
ENSG00000264768 |
. |
3534 |
0.19 |
| chr14_33776607_33776786 | 0.13 |
NPAS3 |
neuronal PAS domain protein 3 |
92178 |
0.1 |
| chr15_86162614_86163207 | 0.13 |
RP11-815J21.3 |
|
8010 |
0.15 |
| chr10_92679511_92679800 | 0.13 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
1378 |
0.39 |
| chr15_71054590_71054766 | 0.13 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
1091 |
0.55 |
| chr1_86079084_86079255 | 0.13 |
ENSG00000199934 |
. |
21206 |
0.18 |
| chr11_126506918_126507382 | 0.13 |
RP11-115C10.1 |
|
15597 |
0.26 |
| chr9_18472965_18473116 | 0.13 |
ADAMTSL1 |
ADAMTS-like 1 |
852 |
0.75 |
| chr9_91791007_91791172 | 0.13 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
2593 |
0.36 |
| chr5_2750019_2750705 | 0.13 |
IRX2 |
iroquois homeobox 2 |
1407 |
0.43 |
| chr10_102761020_102761171 | 0.13 |
LZTS2 |
leucine zipper, putative tumor suppressor 2 |
1485 |
0.23 |
| chr3_99989145_99989333 | 0.13 |
TBC1D23 |
TBC1 domain family, member 23 |
3203 |
0.26 |
| chr7_126848587_126848738 | 0.13 |
AC000099.1 |
|
6519 |
0.26 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.1 | 0.3 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.1 | 0.3 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.1 | 0.2 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.2 | GO:0030538 | embryonic genitalia morphogenesis(GO:0030538) |
| 0.1 | 0.2 | GO:0071504 | cellular response to heparin(GO:0071504) |
| 0.1 | 0.2 | GO:0072047 | proximal/distal pattern formation involved in nephron development(GO:0072047) specification of nephron tubule identity(GO:0072081) |
| 0.1 | 0.3 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) positive regulation of sprouting angiogenesis(GO:1903672) |
| 0.1 | 0.3 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.1 | 0.3 | GO:0045662 | negative regulation of myoblast differentiation(GO:0045662) |
| 0.1 | 0.2 | GO:0060013 | righting reflex(GO:0060013) |
| 0.1 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.2 | GO:0009158 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) positive regulation of signal transduction by p53 class mediator(GO:1901798) |
| 0.0 | 0.2 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.3 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.1 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0021542 | dentate gyrus development(GO:0021542) |
| 0.0 | 0.1 | GO:0010815 | bradykinin catabolic process(GO:0010815) |
| 0.0 | 0.3 | GO:0045880 | positive regulation of smoothened signaling pathway(GO:0045880) |
| 0.0 | 0.1 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0051967 | negative regulation of synaptic transmission, glutamatergic(GO:0051967) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:2000054 | negative regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000054) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.1 | GO:0032236 | obsolete positive regulation of calcium ion transport via store-operated calcium channel activity(GO:0032236) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0097709 | connective tissue replacement involved in inflammatory response wound healing(GO:0002248) connective tissue replacement(GO:0097709) |
| 0.0 | 0.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0033688 | regulation of osteoblast proliferation(GO:0033688) |
| 0.0 | 0.1 | GO:0060676 | ureteric bud formation(GO:0060676) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.0 | GO:0042091 | interleukin-10 biosynthetic process(GO:0042091) regulation of interleukin-10 biosynthetic process(GO:0045074) |
| 0.0 | 0.0 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.0 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) |
| 0.0 | 0.1 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.1 | GO:0048875 | surfactant homeostasis(GO:0043129) chemical homeostasis within a tissue(GO:0048875) |
| 0.0 | 0.0 | GO:0032898 | neurotrophin production(GO:0032898) |
| 0.0 | 0.1 | GO:0048343 | paraxial mesodermal cell differentiation(GO:0048342) paraxial mesodermal cell fate commitment(GO:0048343) |
| 0.0 | 0.1 | GO:0071694 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0002634 | regulation of germinal center formation(GO:0002634) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.0 | GO:0060916 | mesenchymal cell proliferation involved in lung development(GO:0060916) |
| 0.0 | 0.0 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.0 | 0.0 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.3 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.0 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.0 | 0.1 | GO:0060174 | limb bud formation(GO:0060174) |
| 0.0 | 0.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.0 | 0.0 | GO:0061430 | bone trabecula formation(GO:0060346) bone trabecula morphogenesis(GO:0061430) |
| 0.0 | 0.1 | GO:0045749 | obsolete negative regulation of S phase of mitotic cell cycle(GO:0045749) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0048311 | mitochondrion distribution(GO:0048311) |
| 0.0 | 0.0 | GO:1903053 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.0 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.1 | GO:0051016 | barbed-end actin filament capping(GO:0051016) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.2 | GO:0098743 | cartilage condensation(GO:0001502) cell aggregation(GO:0098743) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0045627 | positive regulation of T-helper 1 cell differentiation(GO:0045627) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.2 | GO:0035329 | hippo signaling(GO:0035329) |
| 0.0 | 0.0 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.0 | GO:0022617 | extracellular matrix disassembly(GO:0022617) |
| 0.0 | 0.1 | GO:0006546 | glycine catabolic process(GO:0006546) |
| 0.0 | 0.0 | GO:0048808 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.0 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.0 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.0 | GO:0033004 | negative regulation of mast cell activation(GO:0033004) |
| 0.0 | 0.0 | GO:0051665 | membrane raft localization(GO:0051665) |
| 0.0 | 0.0 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.0 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.0 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.3 | GO:0051101 | regulation of DNA binding(GO:0051101) |
| 0.0 | 0.1 | GO:0002052 | positive regulation of neuroblast proliferation(GO:0002052) |
| 0.0 | 0.1 | GO:0060638 | mesenchymal-epithelial cell signaling(GO:0060638) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.3 | GO:0071804 | cellular potassium ion transport(GO:0071804) potassium ion transmembrane transport(GO:0071805) |
| 0.0 | 0.0 | GO:0009215 | purine deoxyribonucleoside triphosphate metabolic process(GO:0009215) |
| 0.0 | 0.4 | GO:0071377 | cellular response to glucagon stimulus(GO:0071377) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.0 | 0.0 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0012510 | trans-Golgi network transport vesicle membrane(GO:0012510) |
| 0.0 | 0.2 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.2 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 1.0 | GO:0030426 | growth cone(GO:0030426) |
| 0.0 | 0.0 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0071437 | invadopodium(GO:0071437) |
| 0.0 | 0.1 | GO:0005697 | telomerase holoenzyme complex(GO:0005697) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.3 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0001098 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.1 | 0.5 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.1 | 0.3 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.1 | 0.3 | GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding(GO:0000977) RNA polymerase II regulatory region DNA binding(GO:0001012) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.3 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0016933 | extracellular-glycine-gated ion channel activity(GO:0016933) extracellular-glycine-gated chloride channel activity(GO:0016934) |
| 0.0 | 0.2 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.4 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.0 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.3 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.0 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.3 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0001085 | RNA polymerase II transcription factor binding(GO:0001085) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.0 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0042910 | xenobiotic-transporting ATPase activity(GO:0008559) xenobiotic transporter activity(GO:0042910) |
| 0.0 | 0.1 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0070064 | proline-rich region binding(GO:0070064) |
| 0.0 | 0.0 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.0 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0051378 | serotonin binding(GO:0051378) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.1 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.1 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.0 | 0.1 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.0 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) |
| 0.0 | 0.0 | GO:0031994 | insulin-like growth factor I binding(GO:0031994) |
| 0.0 | 0.1 | GO:0005523 | tropomyosin binding(GO:0005523) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.2 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.2 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.4 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.5 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.3 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.4 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.8 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.3 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.1 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.2 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME NETRIN1 SIGNALING | Genes involved in Netrin-1 signaling |
| 0.0 | 0.4 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME PROSTACYCLIN SIGNALLING THROUGH PROSTACYCLIN RECEPTOR | Genes involved in Prostacyclin signalling through prostacyclin receptor |
| 0.0 | 0.1 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.0 | REACTOME SIGNALING BY ERBB4 | Genes involved in Signaling by ERBB4 |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |
| 0.0 | 0.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.0 | REACTOME NEURONAL SYSTEM | Genes involved in Neuronal System |
| 0.0 | 0.2 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.1 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |