Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
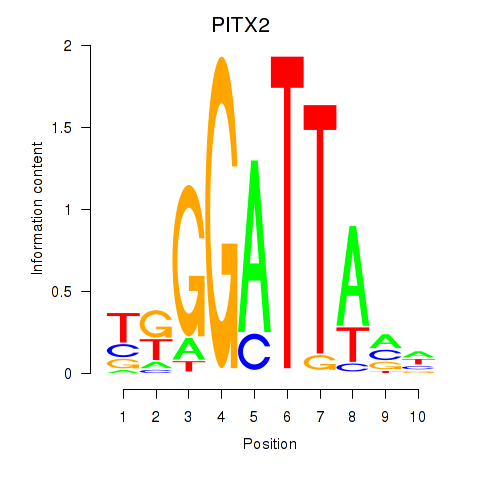
Results for PITX2
Z-value: 1.47
Transcription factors associated with PITX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PITX2
|
ENSG00000164093.11 | paired like homeodomain 2 |
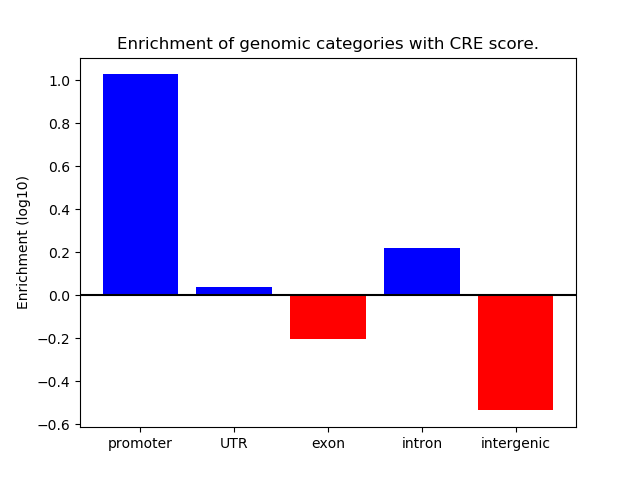
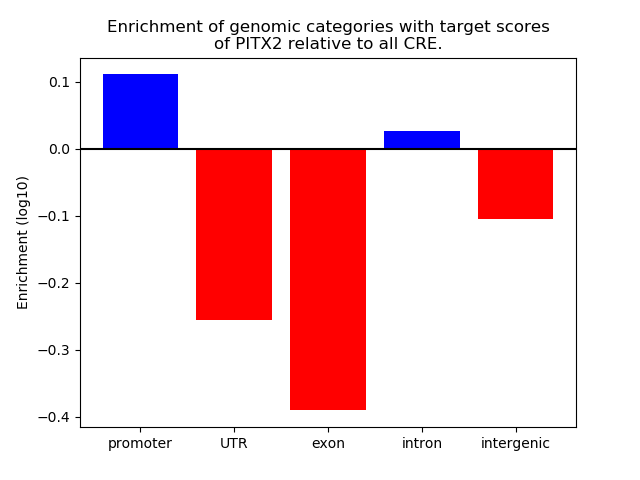
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr4_111557687_111558154 | PITX2 | 256 | 0.954379 | -0.79 | 1.2e-02 | Click! |
| chr4_111557385_111557536 | PITX2 | 716 | 0.790624 | -0.77 | 1.6e-02 | Click! |
| chr4_111532964_111533115 | PITX2 | 11168 | 0.276361 | -0.76 | 1.9e-02 | Click! |
| chr4_111558170_111558322 | PITX2 | 64 | 0.985579 | -0.75 | 2.1e-02 | Click! |
| chr4_111507244_111507551 | PITX2 | 36810 | 0.189272 | -0.73 | 2.7e-02 | Click! |
Activity of the PITX2 motif across conditions
Conditions sorted by the z-value of the PITX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
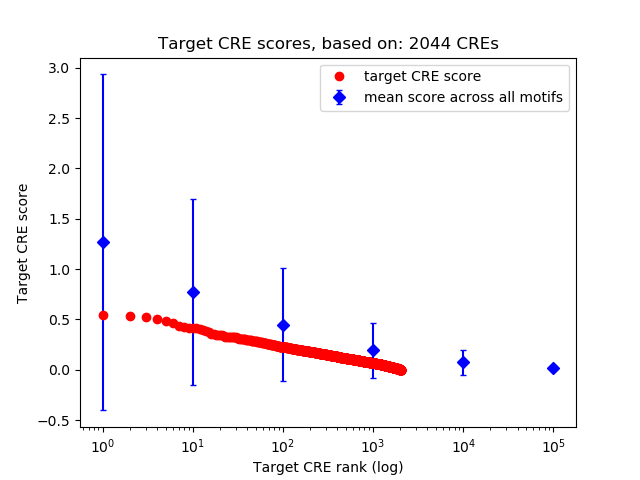
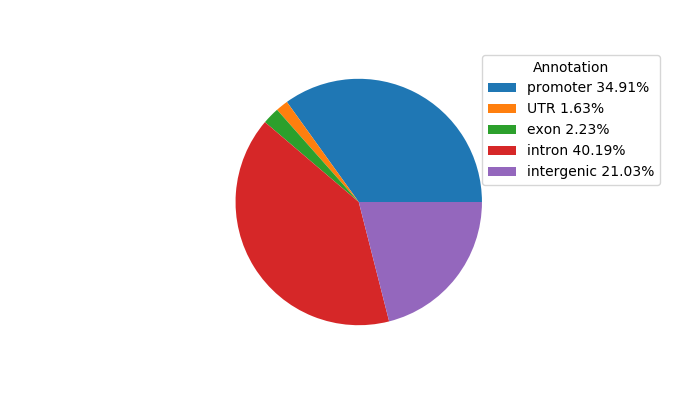
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr16_30484007_30484505 | 0.55 |
ITGAL |
integrin, alpha L (antigen CD11A (p180), lymphocyte function-associated antigen 1; alpha polypeptide) |
184 |
0.81 |
| chr19_2084975_2085126 | 0.54 |
MOB3A |
MOB kinase activator 3A |
341 |
0.8 |
| chr12_122287932_122288083 | 0.53 |
RP11-7M8.2 |
|
6158 |
0.15 |
| chr19_2084034_2084229 | 0.51 |
MOB3A |
MOB kinase activator 3A |
1260 |
0.31 |
| chr17_29640289_29640482 | 0.48 |
EVI2B |
ecotropic viral integration site 2B |
717 |
0.57 |
| chr5_130588828_130589214 | 0.46 |
CDC42SE2 |
CDC42 small effector 2 |
10681 |
0.28 |
| chr12_9911780_9911931 | 0.44 |
CD69 |
CD69 molecule |
1642 |
0.33 |
| chr13_46751053_46751538 | 0.42 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
5164 |
0.18 |
| chr10_101414814_101414965 | 0.41 |
RP11-483F11.7 |
|
1227 |
0.38 |
| chr19_8642522_8642673 | 0.41 |
MYO1F |
myosin IF |
136 |
0.93 |
| chr4_40201423_40201705 | 0.41 |
RHOH |
ras homolog family member H |
400 |
0.87 |
| chr1_226073126_226073382 | 0.40 |
TMEM63A |
transmembrane protein 63A |
3185 |
0.16 |
| chr9_117147390_117147560 | 0.40 |
AKNA |
AT-hook transcription factor |
2768 |
0.28 |
| chr7_73624740_73625015 | 0.39 |
LAT2 |
linker for activation of T cells family, member 2 |
524 |
0.74 |
| chr6_154568354_154568864 | 0.37 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
53 |
0.99 |
| chr1_209942353_209942874 | 0.36 |
TRAF3IP3 |
TRAF3 interacting protein 3 |
653 |
0.63 |
| chr16_11678677_11678828 | 0.35 |
LITAF |
lipopolysaccharide-induced TNF factor |
1477 |
0.42 |
| chr2_101916869_101917041 | 0.35 |
RNF149 |
ring finger protein 149 |
8203 |
0.17 |
| chr15_40599321_40599893 | 0.35 |
PLCB2 |
phospholipase C, beta 2 |
419 |
0.68 |
| chr1_244488212_244488377 | 0.34 |
C1orf100 |
chromosome 1 open reading frame 100 |
27643 |
0.22 |
| chr6_159465648_159465970 | 0.34 |
TAGAP |
T-cell activation RhoGTPase activating protein |
241 |
0.93 |
| chrX_118825241_118825467 | 0.34 |
SEPT6 |
septin 6 |
1438 |
0.4 |
| chr2_161993917_161994236 | 0.33 |
TANK |
TRAF family member-associated NFKB activator |
610 |
0.79 |
| chr1_175158346_175158497 | 0.33 |
KIAA0040 |
KIAA0040 |
3469 |
0.33 |
| chr5_95155660_95155831 | 0.33 |
GLRX |
glutaredoxin (thioltransferase) |
2670 |
0.22 |
| chr8_23083253_23083404 | 0.33 |
ENSG00000246582 |
. |
594 |
0.43 |
| chr9_71590240_71590555 | 0.33 |
PRKACG |
protein kinase, cAMP-dependent, catalytic, gamma |
38642 |
0.15 |
| chr10_28622520_28622887 | 0.33 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
712 |
0.72 |
| chr17_33863104_33863360 | 0.32 |
SLFN12L |
schlafen family member 12-like |
1648 |
0.24 |
| chr1_248069833_248069987 | 0.32 |
OR2W3 |
olfactory receptor, family 2, subfamily W, member 3 |
11021 |
0.1 |
| chr17_47818679_47819212 | 0.32 |
FAM117A |
family with sequence similarity 117, member A |
17056 |
0.14 |
| chr15_40366035_40366186 | 0.31 |
RP11-521C20.3 |
|
14923 |
0.14 |
| chr1_150849308_150849506 | 0.31 |
ARNT |
aryl hydrocarbon receptor nuclear translocator |
163 |
0.93 |
| chr17_33863414_33863594 | 0.31 |
SLFN12L |
schlafen family member 12-like |
1376 |
0.29 |
| chr4_170000682_170000833 | 0.31 |
RP11-483A20.3 |
|
69176 |
0.1 |
| chr2_24114630_24115019 | 0.31 |
ATAD2B |
ATPase family, AAA domain containing 2B |
24067 |
0.18 |
| chr18_9084010_9084262 | 0.30 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
18492 |
0.17 |
| chr12_32113291_32114105 | 0.30 |
KIAA1551 |
KIAA1551 |
1345 |
0.51 |
| chr17_76776568_76776719 | 0.30 |
CYTH1 |
cytohesin 1 |
1711 |
0.35 |
| chr1_160766445_160766727 | 0.30 |
LY9 |
lymphocyte antigen 9 |
609 |
0.72 |
| chr12_50937322_50937473 | 0.30 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
38507 |
0.16 |
| chr3_32502059_32502315 | 0.30 |
CMTM6 |
CKLF-like MARVEL transmembrane domain containing 6 |
42713 |
0.15 |
| chr2_75067589_75067763 | 0.30 |
HK2 |
hexokinase 2 |
5379 |
0.28 |
| chr1_39499733_39499961 | 0.29 |
NDUFS5 |
NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (NADH-coenzyme Q reductase) |
7857 |
0.18 |
| chr2_106417256_106417407 | 0.29 |
NCK2 |
NCK adaptor protein 2 |
15684 |
0.26 |
| chr3_95425254_95425405 | 0.29 |
ENSG00000221477 |
. |
460417 |
0.01 |
| chr12_51714054_51714205 | 0.29 |
BIN2 |
bridging integrator 2 |
3770 |
0.19 |
| chr6_167461487_167461638 | 0.29 |
FGFR1OP |
FGFR1 oncogene partner |
48666 |
0.1 |
| chr2_198071025_198071176 | 0.29 |
ANKRD44 |
ankyrin repeat domain 44 |
8338 |
0.21 |
| chr1_206749362_206749620 | 0.29 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
18998 |
0.14 |
| chr2_238605925_238606076 | 0.28 |
LRRFIP1 |
leucine rich repeat (in FLII) interacting protein 1 |
5018 |
0.26 |
| chr5_39274254_39274635 | 0.28 |
FYB |
FYN binding protein |
186 |
0.97 |
| chr19_59049459_59049613 | 0.28 |
ZBTB45 |
zinc finger and BTB domain containing 45 |
742 |
0.4 |
| chr18_66429429_66429580 | 0.28 |
RP11-106E15.1 |
|
7684 |
0.22 |
| chr20_3996304_3996483 | 0.28 |
RNF24 |
ring finger protein 24 |
164 |
0.96 |
| chr20_55975682_55975833 | 0.27 |
RP4-800J21.3 |
|
7639 |
0.16 |
| chr13_30949154_30949340 | 0.27 |
KATNAL1 |
katanin p60 subunit A-like 1 |
67626 |
0.12 |
| chr19_16441170_16441321 | 0.27 |
KLF2 |
Kruppel-like factor 2 |
5594 |
0.16 |
| chr14_93123309_93123460 | 0.27 |
RIN3 |
Ras and Rab interactor 3 |
4538 |
0.3 |
| chr17_29136154_29136502 | 0.27 |
CRLF3 |
cytokine receptor-like factor 3 |
15361 |
0.12 |
| chr17_28048860_28049057 | 0.27 |
RP11-82O19.1 |
|
39163 |
0.1 |
| chr8_131330583_131330734 | 0.27 |
ASAP1 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
22525 |
0.27 |
| chr14_65006652_65007080 | 0.26 |
RP11-973N13.4 |
|
220 |
0.6 |
| chr3_50656380_50656531 | 0.26 |
MAPKAPK3 |
mitogen-activated protein kinase-activated protein kinase 3 |
1634 |
0.31 |
| chr1_150976139_150976311 | 0.26 |
FAM63A |
family with sequence similarity 63, member A |
2736 |
0.12 |
| chr7_72407870_72408021 | 0.26 |
RP11-313P13.5 |
|
12282 |
0.1 |
| chr11_64532141_64532616 | 0.26 |
SF1 |
splicing factor 1 |
2711 |
0.16 |
| chr1_117299964_117300188 | 0.26 |
CD2 |
CD2 molecule |
2987 |
0.29 |
| chr1_246740599_246740764 | 0.26 |
CNST |
consortin, connexin sorting protein |
10865 |
0.17 |
| chr18_10290164_10290429 | 0.26 |
ENSG00000239031 |
. |
99701 |
0.08 |
| chr5_126094875_126095026 | 0.25 |
ENSG00000252185 |
. |
3841 |
0.27 |
| chr3_32997666_32997928 | 0.25 |
CCR4 |
chemokine (C-C motif) receptor 4 |
4731 |
0.29 |
| chr4_40211316_40211467 | 0.25 |
RHOH |
ras homolog family member H |
9427 |
0.22 |
| chrX_118813168_118813330 | 0.25 |
SEPT6 |
septin 6 |
13543 |
0.17 |
| chr16_21656376_21656527 | 0.25 |
ENSG00000207042 |
. |
2836 |
0.16 |
| chr7_72705892_72706043 | 0.25 |
GTF2IRD2P1 |
GTF2I repeat domain containing 2 pseudogene 1 |
11824 |
0.13 |
| chr19_17377966_17378117 | 0.25 |
BABAM1 |
BRISC and BRCA1 A complex member 1 |
118 |
0.9 |
| chr2_71299479_71299745 | 0.25 |
NAGK |
N-acetylglucosamine kinase |
691 |
0.54 |
| chr6_90122617_90122768 | 0.25 |
RRAGD |
Ras-related GTP binding D |
703 |
0.69 |
| chr1_150976711_150976969 | 0.24 |
FAM63A |
family with sequence similarity 63, member A |
2121 |
0.14 |
| chrX_70331856_70332007 | 0.24 |
IL2RG |
interleukin 2 receptor, gamma |
27 |
0.95 |
| chr11_128171806_128172037 | 0.24 |
ETS1 |
v-ets avian erythroblastosis virus E26 oncogene homolog 1 |
203368 |
0.03 |
| chr16_3627461_3627612 | 0.24 |
NLRC3 |
NLR family, CARD domain containing 3 |
135 |
0.95 |
| chr19_8567892_8568043 | 0.24 |
PRAM1 |
PML-RARA regulated adaptor molecule 1 |
29 |
0.96 |
| chr13_41554921_41555161 | 0.24 |
ELF1 |
E74-like factor 1 (ets domain transcription factor) |
1377 |
0.45 |
| chr2_65173224_65173438 | 0.24 |
ENSG00000238696 |
. |
37511 |
0.12 |
| chr2_197127548_197127913 | 0.24 |
AC020571.3 |
|
2573 |
0.29 |
| chrX_70415693_70415844 | 0.24 |
RP5-1091N2.9 |
|
2257 |
0.22 |
| chr10_14601825_14601976 | 0.24 |
FAM107B |
family with sequence similarity 107, member B |
3721 |
0.3 |
| chr15_64984890_64985206 | 0.24 |
AC100830.4 |
|
2192 |
0.18 |
| chr1_150540056_150540997 | 0.23 |
ENSG00000264508 |
. |
1190 |
0.22 |
| chr1_198646318_198646469 | 0.23 |
RP11-553K8.5 |
|
10203 |
0.25 |
| chr19_54881402_54882266 | 0.23 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
224 |
0.88 |
| chr11_65129750_65129901 | 0.23 |
RP11-867O8.5 |
|
5084 |
0.11 |
| chr6_2941043_2941270 | 0.23 |
SERPINB6 |
serpin peptidase inhibitor, clade B (ovalbumin), member 6 |
20405 |
0.14 |
| chr10_69146333_69146514 | 0.23 |
CTNNA3 |
catenin (cadherin-associated protein), alpha 3 |
206222 |
0.03 |
| chr1_239882649_239882800 | 0.23 |
CHRM3 |
cholinergic receptor, muscarinic 3 |
119 |
0.79 |
| chr19_14886284_14886435 | 0.23 |
EMR2 |
egf-like module containing, mucin-like, hormone receptor-like 2 |
1209 |
0.4 |
| chr11_118086318_118086469 | 0.23 |
AMICA1 |
adhesion molecule, interacts with CXADR antigen 1 |
787 |
0.57 |
| chr20_34331618_34331769 | 0.23 |
RBM39 |
RNA binding motif protein 39 |
1459 |
0.29 |
| chr14_35016119_35016807 | 0.23 |
ENSG00000206596 |
. |
543 |
0.71 |
| chr22_51059908_51060313 | 0.22 |
ARSA |
arylsulfatase A |
6473 |
0.1 |
| chr12_76662658_76662809 | 0.22 |
ENSG00000223273 |
. |
46798 |
0.16 |
| chr12_122065838_122066280 | 0.22 |
ORAI1 |
ORAI calcium release-activated calcium modulator 1 |
1338 |
0.42 |
| chr8_67341017_67341168 | 0.22 |
RRS1 |
RRS1 ribosome biogenesis regulator homolog (S. cerevisiae) |
171 |
0.93 |
| chr7_114566088_114566321 | 0.22 |
MDFIC |
MyoD family inhibitor domain containing |
3073 |
0.4 |
| chr10_6095725_6095937 | 0.22 |
IL2RA |
interleukin 2 receptor, alpha |
8422 |
0.15 |
| chr3_176916851_176917002 | 0.22 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
1665 |
0.48 |
| chr6_11727513_11727664 | 0.22 |
ADTRP |
androgen-dependent TFPI-regulating protein |
8301 |
0.25 |
| chr21_15921648_15921799 | 0.22 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
3059 |
0.32 |
| chr5_14582511_14582672 | 0.22 |
FAM105A |
family with sequence similarity 105, member A |
707 |
0.77 |
| chr19_55009541_55009776 | 0.22 |
LAIR2 |
leukocyte-associated immunoglobulin-like receptor 2 |
558 |
0.61 |
| chr9_132565219_132565403 | 0.22 |
TOR1B |
torsin family 1, member B (torsin B) |
121 |
0.95 |
| chr1_145382873_145383278 | 0.22 |
ENSG00000201558 |
. |
318 |
0.87 |
| chr16_85793355_85793506 | 0.22 |
C16orf74 |
chromosome 16 open reading frame 74 |
8695 |
0.11 |
| chr10_30818638_30818892 | 0.22 |
ENSG00000239744 |
. |
26068 |
0.21 |
| chr13_28001325_28001476 | 0.22 |
GTF3A |
general transcription factor IIIA |
2695 |
0.24 |
| chr10_26731274_26731437 | 0.21 |
APBB1IP |
amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein |
4001 |
0.33 |
| chr3_5231543_5231696 | 0.21 |
EDEM1 |
ER degradation enhancer, mannosidase alpha-like 1 |
1585 |
0.26 |
| chr2_106776864_106777181 | 0.21 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
18 |
0.98 |
| chr12_50908112_50908263 | 0.21 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
9297 |
0.23 |
| chr16_89449371_89449542 | 0.21 |
RP1-168P16.2 |
|
29516 |
0.11 |
| chr14_91865794_91865945 | 0.21 |
CCDC88C |
coiled-coil domain containing 88C |
17821 |
0.22 |
| chr17_36452726_36452901 | 0.21 |
MRPL45 |
mitochondrial ribosomal protein L45 |
176 |
0.94 |
| chr17_41446028_41446236 | 0.21 |
ENSG00000188825 |
. |
18653 |
0.11 |
| chr17_33376713_33376864 | 0.21 |
ENSG00000238858 |
. |
4725 |
0.13 |
| chr22_40299734_40299885 | 0.21 |
GRAP2 |
GRB2-related adaptor protein 2 |
2696 |
0.25 |
| chr20_821090_821241 | 0.21 |
FAM110A |
family with sequence similarity 110, member A |
4120 |
0.27 |
| chr15_38854702_38854853 | 0.21 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
2059 |
0.32 |
| chr1_45769094_45769262 | 0.21 |
HPDL |
4-hydroxyphenylpyruvate dioxygenase-like |
23367 |
0.14 |
| chrX_129225529_129225680 | 0.21 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
18732 |
0.19 |
| chr22_41033042_41033281 | 0.21 |
MKL1 |
megakaryoblastic leukemia (translocation) 1 |
466 |
0.84 |
| chr4_89534455_89534711 | 0.21 |
HERC3 |
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
7638 |
0.19 |
| chr21_34776986_34777191 | 0.21 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
1316 |
0.43 |
| chr22_31139652_31139831 | 0.21 |
ENSG00000264661 |
. |
12197 |
0.17 |
| chr1_28879171_28879359 | 0.20 |
TRNAU1AP |
tRNA selenocysteine 1 associated protein 1 |
332 |
0.8 |
| chr19_6425167_6425318 | 0.20 |
KHSRP |
KH-type splicing regulatory protein |
437 |
0.66 |
| chr10_73529493_73529671 | 0.20 |
C10orf54 |
chromosome 10 open reading frame 54 |
3673 |
0.23 |
| chr15_34609507_34609658 | 0.20 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
1398 |
0.29 |
| chr12_47611625_47611801 | 0.20 |
PCED1B |
PC-esterase domain containing 1B |
1332 |
0.49 |
| chr15_75230580_75230787 | 0.20 |
COX5A |
cytochrome c oxidase subunit Va |
174 |
0.92 |
| chrX_123096191_123096863 | 0.20 |
STAG2 |
stromal antigen 2 |
493 |
0.85 |
| chr17_76771796_76772004 | 0.20 |
CYTH1 |
cytohesin 1 |
6454 |
0.2 |
| chr21_46300234_46300404 | 0.20 |
PTTG1IP |
pituitary tumor-transforming 1 interacting protein |
6567 |
0.13 |
| chr1_212461608_212461798 | 0.20 |
PPP2R5A |
protein phosphatase 2, regulatory subunit B', alpha |
2824 |
0.24 |
| chr19_10706452_10706725 | 0.20 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
6545 |
0.1 |
| chr15_93458767_93458918 | 0.20 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
11139 |
0.19 |
| chr11_60213804_60213975 | 0.20 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
9336 |
0.16 |
| chr4_39429086_39429237 | 0.20 |
ENSG00000238797 |
. |
14124 |
0.11 |
| chr19_54875923_54876565 | 0.20 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
170 |
0.91 |
| chr1_231666043_231666200 | 0.20 |
TSNAX |
translin-associated factor X |
1720 |
0.31 |
| chr6_34283894_34284092 | 0.20 |
C6orf1 |
chromosome 6 open reading frame 1 |
66746 |
0.09 |
| chr14_77330807_77330958 | 0.20 |
ENSG00000223174 |
. |
25724 |
0.16 |
| chr16_84603491_84603642 | 0.20 |
TLDC1 |
TBC/LysM-associated domain containing 1 |
15927 |
0.16 |
| chr4_1195622_1195773 | 0.20 |
SPON2 |
spondin 2, extracellular matrix protein |
3179 |
0.18 |
| chrX_8150685_8150836 | 0.20 |
VCX2 |
variable charge, X-linked 2 |
11452 |
0.28 |
| chr7_99070141_99070303 | 0.20 |
ZNF789 |
zinc finger protein 789 |
242 |
0.83 |
| chr2_190442967_190443214 | 0.20 |
SLC40A1 |
solute carrier family 40 (iron-regulated transporter), member 1 |
2523 |
0.35 |
| chr15_80262648_80263745 | 0.20 |
BCL2A1 |
BCL2-related protein A1 |
315 |
0.89 |
| chr2_37762882_37763076 | 0.20 |
AC006369.2 |
|
64300 |
0.12 |
| chr20_2311261_2311412 | 0.19 |
TGM3 |
transglutaminase 3 |
34689 |
0.16 |
| chr12_58230016_58230207 | 0.19 |
RP11-620J15.1 |
|
764 |
0.44 |
| chr2_223290336_223290619 | 0.19 |
SGPP2 |
sphingosine-1-phosphate phosphatase 2 |
1241 |
0.5 |
| chr11_118213153_118213331 | 0.19 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
104 |
0.93 |
| chr12_124155262_124155441 | 0.19 |
TCTN2 |
tectonic family member 2 |
309 |
0.86 |
| chr21_36420108_36421159 | 0.19 |
RUNX1 |
runt-related transcription factor 1 |
829 |
0.77 |
| chr19_46179944_46180134 | 0.19 |
ENSG00000207773 |
. |
1853 |
0.16 |
| chr17_45570068_45570247 | 0.19 |
MRPL45P2 |
mitochondrial ribosomal protein L45 pseudogene 2 |
332 |
0.88 |
| chr1_243430515_243430666 | 0.19 |
SDCCAG8 |
serologically defined colon cancer antigen 8 |
11218 |
0.19 |
| chr2_136504991_136505142 | 0.19 |
UBXN4 |
UBX domain protein 4 |
5762 |
0.21 |
| chr12_94517592_94517768 | 0.19 |
PLXNC1 |
plexin C1 |
24819 |
0.2 |
| chr1_22351357_22351739 | 0.19 |
ENSG00000201273 |
. |
14070 |
0.09 |
| chr21_47035540_47035741 | 0.19 |
PCBP3 |
poly(rC) binding protein 3 |
27968 |
0.2 |
| chr12_132469658_132469809 | 0.19 |
ENSG00000271905 |
. |
16587 |
0.15 |
| chr13_40397451_40397602 | 0.19 |
ENSG00000212553 |
. |
33838 |
0.19 |
| chr7_76828015_76828166 | 0.19 |
FGL2 |
fibrinogen-like 2 |
1053 |
0.59 |
| chrX_109708973_109709124 | 0.19 |
RGAG1 |
retrotransposon gag domain containing 1 |
15255 |
0.23 |
| chr6_53209536_53209824 | 0.19 |
ELOVL5 |
ELOVL fatty acid elongase 5 |
3907 |
0.26 |
| chr17_56419518_56419935 | 0.19 |
BZRAP1-AS1 |
BZRAP1 antisense RNA 1 |
5007 |
0.12 |
| chr8_65607910_65608061 | 0.19 |
RP11-1D12.1 |
|
39031 |
0.2 |
| chr12_65058254_65058405 | 0.19 |
RP11-338E21.3 |
|
9530 |
0.14 |
| chr19_59055180_59055577 | 0.19 |
TRIM28 |
tripartite motif containing 28 |
80 |
0.82 |
| chr17_78991849_78992000 | 0.19 |
AC127496.1 |
Uncharacterized protein |
14789 |
0.11 |
| chr14_81629657_81629808 | 0.18 |
RP11-114N19.3 |
|
7026 |
0.24 |
| chr12_40012002_40012194 | 0.18 |
ABCD2 |
ATP-binding cassette, sub-family D (ALD), member 2 |
1455 |
0.48 |
| chr4_185741986_185742137 | 0.18 |
ACSL1 |
acyl-CoA synthetase long-chain family member 1 |
5127 |
0.2 |
| chr10_120891057_120891670 | 0.18 |
FAM45A |
family with sequence similarity 45, member A |
27734 |
0.11 |
| chr1_212927626_212927881 | 0.18 |
NSL1 |
NSL1, MIS12 kinetochore complex component |
37170 |
0.12 |
| chr1_154581072_154581232 | 0.18 |
ADAR |
adenosine deaminase, RNA-specific |
470 |
0.73 |
| chr1_155946704_155947008 | 0.18 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
1095 |
0.29 |
| chr1_147994918_147995202 | 0.18 |
ENSG00000212456 |
. |
914 |
0.59 |
| chr17_18966559_18966712 | 0.18 |
ENSG00000262074 |
. |
814 |
0.4 |
| chr2_175456985_175457136 | 0.18 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
5433 |
0.19 |
| chrY_21906454_21906814 | 0.18 |
KDM5D |
lysine (K)-specific demethylase 5D |
13 |
0.99 |
| chr2_220117396_220117589 | 0.18 |
TUBA4A |
tubulin, alpha 4a |
223 |
0.82 |
| chr14_32366460_32366611 | 0.18 |
RP11-187E13.1 |
Uncharacterized protein |
47524 |
0.17 |
| chr21_35885755_35885906 | 0.18 |
KCNE1 |
potassium voltage-gated channel, Isk-related family, member 1 |
1257 |
0.46 |
| chr19_4975873_4976024 | 0.18 |
KDM4B |
lysine (K)-specific demethylase 4B |
6805 |
0.19 |
| chr10_64915626_64915777 | 0.18 |
NRBF2 |
nuclear receptor binding factor 2 |
22616 |
0.18 |
| chr1_148766073_148766315 | 0.18 |
ENSG00000206968 |
. |
908 |
0.66 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.1 | 0.1 | GO:0043618 | regulation of transcription from RNA polymerase II promoter in response to stress(GO:0043618) |
| 0.1 | 0.2 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.1 | 0.2 | GO:0051138 | NK T cell differentiation(GO:0001865) regulation of NK T cell differentiation(GO:0051136) positive regulation of NK T cell differentiation(GO:0051138) |
| 0.0 | 0.4 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.0 | 0.1 | GO:0042494 | detection of bacterial lipoprotein(GO:0042494) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.3 | GO:0032515 | negative regulation of phosphoprotein phosphatase activity(GO:0032515) |
| 0.0 | 0.2 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.3 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0072239 | metanephric glomerulus development(GO:0072224) metanephric glomerulus vasculature development(GO:0072239) |
| 0.0 | 0.3 | GO:0001580 | detection of chemical stimulus involved in sensory perception of bitter taste(GO:0001580) |
| 0.0 | 0.2 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.1 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.2 | GO:0032876 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0016139 | glycoside metabolic process(GO:0016137) glycoside catabolic process(GO:0016139) |
| 0.0 | 0.2 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.2 | GO:0042159 | lipoprotein catabolic process(GO:0042159) |
| 0.0 | 0.2 | GO:0019987 | obsolete negative regulation of anti-apoptosis(GO:0019987) |
| 0.0 | 0.1 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0032418 | lysosome localization(GO:0032418) |
| 0.0 | 0.1 | GO:0046931 | pore complex assembly(GO:0046931) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0010834 | obsolete telomere maintenance via telomere shortening(GO:0010834) |
| 0.0 | 0.1 | GO:0032025 | response to cobalt ion(GO:0032025) |
| 0.0 | 0.5 | GO:0002448 | mast cell mediated immunity(GO:0002448) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.3 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0042483 | negative regulation of odontogenesis(GO:0042483) |
| 0.0 | 0.1 | GO:0001911 | negative regulation of leukocyte mediated cytotoxicity(GO:0001911) |
| 0.0 | 0.5 | GO:0050798 | activated T cell proliferation(GO:0050798) |
| 0.0 | 0.1 | GO:0050685 | positive regulation of mRNA 3'-end processing(GO:0031442) positive regulation of mRNA processing(GO:0050685) positive regulation of mRNA metabolic process(GO:1903313) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.1 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0050687 | negative regulation of defense response to virus(GO:0050687) |
| 0.0 | 0.2 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.2 | GO:0007080 | mitotic metaphase plate congression(GO:0007080) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0016246 | RNA interference(GO:0016246) |
| 0.0 | 0.1 | GO:0000389 | mRNA 3'-splice site recognition(GO:0000389) |
| 0.0 | 0.1 | GO:0072698 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.1 | GO:0000098 | sulfur amino acid catabolic process(GO:0000098) |
| 0.0 | 0.1 | GO:0034755 | iron ion transmembrane transport(GO:0034755) |
| 0.0 | 0.1 | GO:0045343 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.3 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.2 | GO:0009650 | UV protection(GO:0009650) |
| 0.0 | 0.3 | GO:0070536 | protein K63-linked deubiquitination(GO:0070536) |
| 0.0 | 0.5 | GO:0043124 | negative regulation of I-kappaB kinase/NF-kappaB signaling(GO:0043124) |
| 0.0 | 0.1 | GO:0009299 | mRNA transcription(GO:0009299) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.0 | GO:0001866 | NK T cell proliferation(GO:0001866) |
| 0.0 | 0.0 | GO:0071922 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.1 | GO:0090005 | negative regulation of establishment of protein localization to plasma membrane(GO:0090005) negative regulation of protein localization to plasma membrane(GO:1903077) negative regulation of protein localization to cell periphery(GO:1904376) |
| 0.0 | 0.3 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) regulation of interferon-gamma-mediated signaling pathway(GO:0060334) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.1 | GO:0045601 | regulation of endothelial cell differentiation(GO:0045601) |
| 0.0 | 0.1 | GO:0009128 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.1 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.1 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0042832 | defense response to protozoan(GO:0042832) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.2 | GO:0071230 | cellular response to amino acid stimulus(GO:0071230) |
| 0.0 | 0.1 | GO:0000730 | DNA recombinase assembly(GO:0000730) double-strand break repair via synthesis-dependent strand annealing(GO:0045003) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.0 | GO:0001672 | regulation of chromatin assembly or disassembly(GO:0001672) |
| 0.0 | 0.1 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0046835 | carbohydrate phosphorylation(GO:0046835) |
| 0.0 | 0.1 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.1 | GO:0009298 | GDP-mannose biosynthetic process(GO:0009298) |
| 0.0 | 0.1 | GO:0006978 | DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:0006978) |
| 0.0 | 0.0 | GO:0032968 | positive regulation of transcription elongation from RNA polymerase II promoter(GO:0032968) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.0 | GO:0045586 | regulation of gamma-delta T cell differentiation(GO:0045586) positive regulation of gamma-delta T cell differentiation(GO:0045588) |
| 0.0 | 0.0 | GO:0046606 | negative regulation of centrosome duplication(GO:0010826) negative regulation of centrosome cycle(GO:0046606) |
| 0.0 | 0.0 | GO:0050765 | negative regulation of phagocytosis(GO:0050765) |
| 0.0 | 0.0 | GO:0007144 | female meiosis I(GO:0007144) |
| 0.0 | 0.0 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.1 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.0 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0032988 | ribonucleoprotein complex disassembly(GO:0032988) |
| 0.0 | 0.0 | GO:0070266 | necroptotic process(GO:0070266) programmed necrotic cell death(GO:0097300) |
| 0.0 | 0.1 | GO:0000301 | retrograde transport, vesicle recycling within Golgi(GO:0000301) |
| 0.0 | 0.1 | GO:0046827 | positive regulation of protein export from nucleus(GO:0046827) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.1 | GO:0098927 | early endosome to late endosome transport(GO:0045022) vesicle-mediated transport between endosomal compartments(GO:0098927) |
| 0.0 | 0.1 | GO:0015939 | pantothenate metabolic process(GO:0015939) |
| 0.0 | 0.0 | GO:0070933 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.1 | GO:0072531 | pyrimidine-containing compound transmembrane transport(GO:0072531) |
| 0.0 | 0.1 | GO:0018106 | peptidyl-histidine phosphorylation(GO:0018106) |
| 0.0 | 0.0 | GO:0009173 | UMP biosynthetic process(GO:0006222) pyrimidine ribonucleoside monophosphate metabolic process(GO:0009173) pyrimidine ribonucleoside monophosphate biosynthetic process(GO:0009174) UMP metabolic process(GO:0046049) |
| 0.0 | 0.0 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.0 | GO:0034067 | protein localization to Golgi apparatus(GO:0034067) |
| 0.0 | 0.1 | GO:0032770 | positive regulation of monooxygenase activity(GO:0032770) |
| 0.0 | 0.0 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.0 | 0.0 | GO:0008608 | attachment of spindle microtubules to kinetochore(GO:0008608) |
| 0.0 | 0.0 | GO:0033522 | histone H2A ubiquitination(GO:0033522) |
| 0.0 | 0.0 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 0.0 | GO:0090201 | negative regulation of mitochondrion organization(GO:0010823) negative regulation of release of cytochrome c from mitochondria(GO:0090201) negative regulation of apoptotic signaling pathway(GO:2001234) |
| 0.0 | 0.0 | GO:0046549 | retinal cone cell differentiation(GO:0042670) retinal cone cell development(GO:0046549) |
| 0.0 | 0.0 | GO:0032966 | negative regulation of collagen biosynthetic process(GO:0032966) |
| 0.0 | 0.1 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0032372 | negative regulation of sterol transport(GO:0032372) negative regulation of cholesterol transport(GO:0032375) |
| 0.0 | 0.2 | GO:0051225 | spindle assembly(GO:0051225) |
| 0.0 | 0.0 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0032715 | negative regulation of interleukin-6 production(GO:0032715) |
| 0.0 | 0.1 | GO:0048261 | negative regulation of receptor-mediated endocytosis(GO:0048261) |
| 0.0 | 0.0 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.0 | GO:0048670 | regulation of collateral sprouting(GO:0048670) |
| 0.0 | 0.0 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:0008216 | spermidine metabolic process(GO:0008216) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.1 | 0.4 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.5 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0005719 | nuclear euchromatin(GO:0005719) |
| 0.0 | 0.1 | GO:0031371 | ubiquitin conjugating enzyme complex(GO:0031371) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.2 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.2 | GO:0032279 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.2 | GO:0031105 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.2 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.2 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.1 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0000242 | pericentriolar material(GO:0000242) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.1 | GO:0001739 | sex chromatin(GO:0001739) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0070688 | MLL5-L complex(GO:0070688) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0000125 | PCAF complex(GO:0000125) |
| 0.0 | 0.2 | GO:0001673 | male germ cell nucleus(GO:0001673) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.0 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.0 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.2 | GO:0030530 | obsolete heterogeneous nuclear ribonucleoprotein complex(GO:0030530) |
| 0.0 | 0.0 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0070652 | HAUS complex(GO:0070652) |
| 0.0 | 0.2 | GO:0005669 | transcription factor TFIID complex(GO:0005669) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.3 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.2 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.2 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.2 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.4 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.0 | GO:0043142 | single-stranded DNA-dependent ATPase activity(GO:0043142) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0005381 | iron ion transmembrane transporter activity(GO:0005381) |
| 0.0 | 0.2 | GO:0004904 | interferon receptor activity(GO:0004904) |
| 0.0 | 0.4 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0043139 | 5'-3' DNA helicase activity(GO:0043139) |
| 0.0 | 0.1 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.1 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.5 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0043047 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:0045309 | protein phosphorylated amino acid binding(GO:0045309) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.1 | GO:0004340 | glucokinase activity(GO:0004340) |
| 0.0 | 0.1 | GO:0004677 | DNA-dependent protein kinase activity(GO:0004677) |
| 0.0 | 0.1 | GO:0071723 | lipopeptide binding(GO:0071723) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.1 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.1 | GO:0051733 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) |
| 0.0 | 0.1 | GO:0047045 | testosterone 17-beta-dehydrogenase (NADP+) activity(GO:0047045) |
| 0.0 | 0.2 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0010340 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0052659 | inositol-1,4,5-trisphosphate 5-phosphatase activity(GO:0052658) inositol-1,3,4,5-tetrakisphosphate 5-phosphatase activity(GO:0052659) |
| 0.0 | 0.1 | GO:0016670 | oxidoreductase activity, acting on a sulfur group of donors, oxygen as acceptor(GO:0016670) |
| 0.0 | 0.1 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.1 | GO:0008898 | S-adenosylmethionine-homocysteine S-methyltransferase activity(GO:0008898) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.5 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0080025 | phosphatidylinositol-3,5-bisphosphate binding(GO:0080025) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0051880 | G-quadruplex DNA binding(GO:0051880) |
| 0.0 | 0.0 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0016675 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors(GO:0016675) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.0 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.1 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0016530 | metallochaperone activity(GO:0016530) |
| 0.0 | 0.2 | GO:0019200 | carbohydrate kinase activity(GO:0019200) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0016889 | endodeoxyribonuclease activity, producing 3'-phosphomonoesters(GO:0016889) |
| 0.0 | 0.2 | GO:0016251 | obsolete general RNA polymerase II transcription factor activity(GO:0016251) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
| 0.0 | 0.0 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0003906 | DNA-(apurinic or apyrimidinic site) lyase activity(GO:0003906) |
| 0.0 | 0.1 | GO:0015924 | mannosyl-oligosaccharide mannosidase activity(GO:0015924) |
| 0.0 | 0.1 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0017069 | snRNA binding(GO:0017069) |
| 0.0 | 0.0 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0051018 | protein kinase A binding(GO:0051018) |
| 0.0 | 0.0 | GO:0004607 | phosphatidylcholine-sterol O-acyltransferase activity(GO:0004607) |
| 0.0 | 0.1 | GO:0036041 | long-chain fatty acid binding(GO:0036041) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 1.0 | PID TCR PATHWAY | TCR signaling in naïve CD4+ T cells |
| 0.0 | 0.4 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
| 0.0 | 0.3 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 0.5 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.2 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.2 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.4 | PID ATM PATHWAY | ATM pathway |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.3 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.3 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.2 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 0.2 | PID MYC PATHWAY | C-MYC pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME SIGNALING BY FGFR IN DISEASE | Genes involved in Signaling by FGFR in disease |
| 0.0 | 0.4 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.3 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNA SIGNALING | Genes involved in Regulation of IFNA signaling |
| 0.0 | 0.1 | REACTOME MICRORNA MIRNA BIOGENESIS | Genes involved in MicroRNA (miRNA) Biogenesis |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR1 FUSION MUTANTS | Genes involved in Signaling by FGFR1 fusion mutants |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.2 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.3 | REACTOME REGULATION OF HYPOXIA INDUCIBLE FACTOR HIF BY OXYGEN | Genes involved in Regulation of Hypoxia-inducible Factor (HIF) by Oxygen |
| 0.0 | 0.1 | REACTOME REGULATION OF THE FANCONI ANEMIA PATHWAY | Genes involved in Regulation of the Fanconi anemia pathway |
| 0.0 | 0.9 | REACTOME MEIOTIC SYNAPSIS | Genes involved in Meiotic Synapsis |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME ARMS MEDIATED ACTIVATION | Genes involved in ARMS-mediated activation |
| 0.0 | 0.2 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.6 | REACTOME RNA POL II PRE TRANSCRIPTION EVENTS | Genes involved in RNA Polymerase II Pre-transcription Events |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.2 | REACTOME APC C CDC20 MEDIATED DEGRADATION OF CYCLIN B | Genes involved in APC/C:Cdc20 mediated degradation of Cyclin B |
| 0.0 | 0.1 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.1 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME CYCLIN A B1 ASSOCIATED EVENTS DURING G2 M TRANSITION | Genes involved in Cyclin A/B1 associated events during G2/M transition |
| 0.0 | 0.4 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |
| 0.0 | 0.3 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |