Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
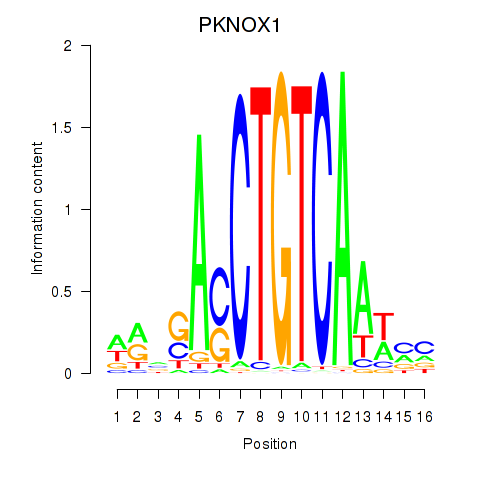
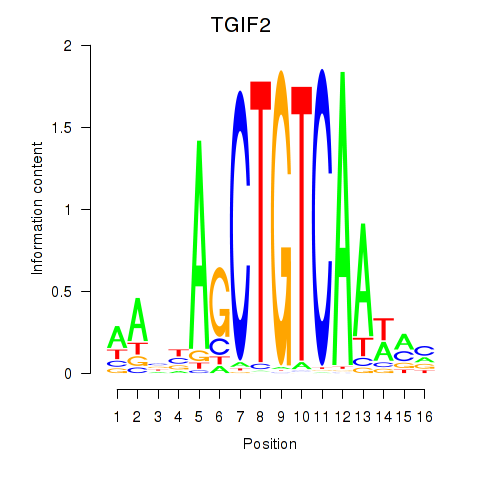
Results for PKNOX1_TGIF2
Z-value: 0.92
Transcription factors associated with PKNOX1_TGIF2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX1
|
ENSG00000160199.10 | PBX/knotted 1 homeobox 1 |
|
TGIF2
|
ENSG00000118707.5 | TGFB induced factor homeobox 2 |
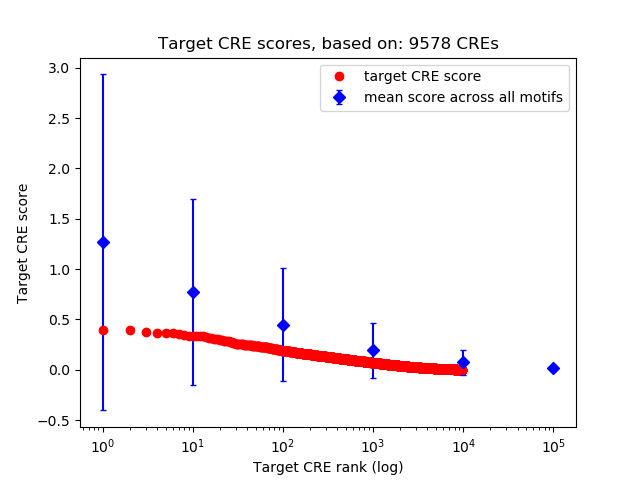
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr21_44395763_44396208 | PKNOX1 | 1234 | 0.425688 | 0.23 | 5.4e-01 | Click! |
| chr21_44394961_44395127 | PKNOX1 | 293 | 0.888993 | 0.18 | 6.4e-01 | Click! |
| chr21_44395511_44395752 | PKNOX1 | 880 | 0.556819 | 0.01 | 9.8e-01 | Click! |
| chr21_44393727_44394254 | PKNOX1 | 630 | 0.687024 | -0.00 | 9.9e-01 | Click! |
| chr20_35202977_35203807 | TGIF2 | 317 | 0.552898 | 0.40 | 2.9e-01 | Click! |
| chr20_35201957_35202480 | TGIF2 | 208 | 0.648461 | 0.30 | 4.3e-01 | Click! |
| chr20_35203812_35203967 | TGIF2 | 814 | 0.358401 | 0.25 | 5.2e-01 | Click! |
| chr20_35204810_35204961 | TGIF2 | 1810 | 0.204835 | -0.09 | 8.3e-01 | Click! |
Activity of the PKNOX1_TGIF2 motif across conditions
Conditions sorted by the z-value of the PKNOX1_TGIF2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr13_50144599_50144750 | 0.39 |
RCBTB1 |
regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 1 |
3215 |
0.28 |
| chr19_1856469_1856846 | 0.39 |
CTB-31O20.8 |
|
3592 |
0.1 |
| chrX_78621891_78622920 | 0.38 |
ITM2A |
integral membrane protein 2A |
451 |
0.91 |
| chr17_17726041_17726867 | 0.37 |
SREBF1 |
sterol regulatory element binding transcription factor 1 |
478 |
0.73 |
| chr7_104908860_104909472 | 0.37 |
SRPK2 |
SRSF protein kinase 2 |
296 |
0.92 |
| chr8_60028293_60029059 | 0.36 |
RP11-25K19.1 |
|
2923 |
0.25 |
| chr1_193154702_193155811 | 0.36 |
B3GALT2 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 |
528 |
0.8 |
| chr9_33446798_33447442 | 0.35 |
AQP3 |
aquaporin 3 (Gill blood group) |
489 |
0.76 |
| chr20_39677452_39677945 | 0.34 |
TOP1 |
topoisomerase (DNA) I |
20240 |
0.19 |
| chr3_132349108_132349259 | 0.34 |
UBA5 |
ubiquitin-like modifier activating enzyme 5 |
24107 |
0.16 |
| chr20_58512577_58512765 | 0.33 |
PPP1R3D |
protein phosphatase 1, regulatory subunit 3D |
2681 |
0.21 |
| chr4_157692249_157692489 | 0.33 |
RP11-154F14.2 |
|
70142 |
0.11 |
| chr4_30725920_30726277 | 0.33 |
PCDH7 |
protocadherin 7 |
2121 |
0.46 |
| chr21_35305075_35305226 | 0.33 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
1632 |
0.32 |
| chr6_64972397_64972548 | 0.31 |
RP11-349P19.1 |
|
115216 |
0.07 |
| chr1_19278015_19278216 | 0.31 |
IFFO2 |
intermediate filament family orphan 2 |
4077 |
0.23 |
| chr5_146257425_146257826 | 0.31 |
PPP2R2B |
protein phosphatase 2, regulatory subunit B, beta |
580 |
0.82 |
| chr11_46581548_46581763 | 0.31 |
AMBRA1 |
autophagy/beclin-1 regulator 1 |
31259 |
0.15 |
| chr10_99442034_99442185 | 0.30 |
AVPI1 |
arginine vasopressin-induced 1 |
4971 |
0.16 |
| chr17_64464860_64465690 | 0.29 |
RP11-4F22.2 |
|
52303 |
0.15 |
| chr9_33447649_33448227 | 0.29 |
AQP3 |
aquaporin 3 (Gill blood group) |
329 |
0.86 |
| chr1_78468921_78469158 | 0.29 |
RP11-386I14.4 |
|
1199 |
0.35 |
| chr13_22956879_22957030 | 0.29 |
ENSG00000253094 |
. |
420330 |
0.01 |
| chr6_82732214_82732365 | 0.29 |
ENSG00000223044 |
. |
187868 |
0.03 |
| chr18_55930027_55930178 | 0.28 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
41299 |
0.16 |
| chr2_161994699_161994850 | 0.28 |
TANK |
TRAF family member-associated NFKB activator |
1308 |
0.52 |
| chr3_31575511_31576107 | 0.27 |
STT3B |
STT3B, subunit of the oligosaccharyltransferase complex (catalytic) |
1527 |
0.55 |
| chr13_28494142_28494415 | 0.27 |
PDX1 |
pancreatic and duodenal homeobox 1 |
121 |
0.93 |
| chr4_54244645_54244895 | 0.27 |
FIP1L1 |
factor interacting with PAPOLA and CPSF1 |
764 |
0.7 |
| chr1_108584009_108584286 | 0.26 |
ENSG00000264753 |
. |
22754 |
0.22 |
| chr22_25508803_25509083 | 0.26 |
CTA-221G9.11 |
|
284 |
0.93 |
| chr16_27413508_27414237 | 0.26 |
IL21R |
interleukin 21 receptor |
377 |
0.88 |
| chr12_54891563_54891796 | 0.26 |
NCKAP1L |
NCK-associated protein 1-like |
184 |
0.93 |
| chr3_15482302_15482608 | 0.26 |
EAF1-AS1 |
EAF1 antisense RNA 1 |
354 |
0.48 |
| chr17_18949450_18949786 | 0.25 |
GRAP |
GRB2-related adaptor protein |
692 |
0.54 |
| chr14_93674231_93674490 | 0.25 |
UBR7 |
ubiquitin protein ligase E3 component n-recognin 7 (putative) |
721 |
0.44 |
| chr17_63060035_63060186 | 0.25 |
GNA13 |
guanine nucleotide binding protein (G protein), alpha 13 |
7153 |
0.19 |
| chrX_72434024_72434767 | 0.25 |
NAP1L2 |
nucleosome assembly protein 1-like 2 |
273 |
0.94 |
| chr19_42636826_42637026 | 0.25 |
CTC-378H22.1 |
|
219 |
0.52 |
| chr18_54318050_54319471 | 0.25 |
TXNL1 |
thioredoxin-like 1 |
64 |
0.61 |
| chr17_19031223_19031746 | 0.25 |
GRAPL |
GRB2-related adaptor protein-like |
636 |
0.51 |
| chr10_54759512_54759663 | 0.24 |
ENSG00000201196 |
. |
147220 |
0.05 |
| chr16_30710458_30711178 | 0.24 |
SRCAP |
Snf2-related CREBBP activator protein |
356 |
0.48 |
| chr17_74749100_74749251 | 0.24 |
ENSG00000200257 |
. |
181 |
0.89 |
| chr2_99955142_99955293 | 0.24 |
EIF5B |
eukaryotic translation initiation factor 5B |
1401 |
0.34 |
| chr7_142425930_142426124 | 0.24 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
31292 |
0.16 |
| chr21_45660106_45660362 | 0.24 |
ICOSLG |
inducible T-cell co-stimulator ligand |
489 |
0.69 |
| chr1_26453661_26453812 | 0.24 |
PDIK1L |
PDLIM1 interacting kinase 1 like |
15396 |
0.1 |
| chr1_185529229_185529380 | 0.24 |
ENSG00000207108 |
. |
70356 |
0.11 |
| chr9_92111480_92111784 | 0.24 |
SEMA4D |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4D |
715 |
0.59 |
| chr11_64013126_64013464 | 0.24 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
7 |
0.62 |
| chr1_40851828_40852273 | 0.24 |
SMAP2 |
small ArfGAP2 |
10457 |
0.17 |
| chr1_161195725_161196723 | 0.23 |
TOMM40L |
translocase of outer mitochondrial membrane 40 homolog (yeast)-like |
130 |
0.84 |
| chr18_28779454_28779605 | 0.23 |
DSC1 |
desmocollin 1 |
36710 |
0.15 |
| chr1_208417858_208418217 | 0.23 |
PLXNA2 |
plexin A2 |
372 |
0.93 |
| chr5_162930555_162931177 | 0.23 |
MAT2B |
methionine adenosyltransferase II, beta |
746 |
0.62 |
| chr6_90896871_90897022 | 0.23 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
109515 |
0.06 |
| chr18_18823015_18823269 | 0.23 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
823 |
0.71 |
| chr16_85673169_85673320 | 0.23 |
GSE1 |
Gse1 coiled-coil protein |
14794 |
0.16 |
| chr6_149386973_149387124 | 0.23 |
RP11-162J8.3 |
|
33339 |
0.2 |
| chr6_27858007_27858288 | 0.23 |
HIST1H3J |
histone cluster 1, H3j |
423 |
0.61 |
| chr10_35089896_35090047 | 0.23 |
PARD3 |
par-3 family cell polarity regulator |
14278 |
0.22 |
| chr9_18569385_18569536 | 0.22 |
ENSG00000264638 |
. |
3844 |
0.33 |
| chr20_48989399_48989550 | 0.22 |
ENSG00000244376 |
. |
56546 |
0.13 |
| chr1_17379619_17379770 | 0.22 |
SDHB |
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
971 |
0.54 |
| chr2_4455409_4455560 | 0.22 |
ENSG00000215960 |
. |
179376 |
0.03 |
| chr1_228362327_228362478 | 0.22 |
IBA57 |
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
151 |
0.92 |
| chr7_75933552_75933703 | 0.22 |
HSPB1 |
heat shock 27kDa protein 1 |
733 |
0.62 |
| chr1_16122451_16122602 | 0.22 |
FBLIM1 |
filamin binding LIM protein 1 |
31078 |
0.1 |
| chr1_121159703_121159854 | 0.22 |
FCGR1B |
Fc fragment of IgG, high affinity Ib, receptor (CD64) |
223841 |
0.02 |
| chr6_139868311_139868622 | 0.22 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
172709 |
0.03 |
| chr20_33867276_33867427 | 0.22 |
RP4-614O4.11 |
|
495 |
0.56 |
| chr18_53296660_53296811 | 0.21 |
TCF4 |
transcription factor 4 |
1858 |
0.49 |
| chr8_23145924_23146455 | 0.21 |
R3HCC1 |
R3H domain and coiled-coil containing 1 |
254 |
0.9 |
| chr13_30528667_30528971 | 0.21 |
LINC00572 |
long intergenic non-protein coding RNA 572 |
28031 |
0.25 |
| chr6_167536422_167536640 | 0.21 |
CCR6 |
chemokine (C-C motif) receptor 6 |
272 |
0.91 |
| chr1_205462241_205462485 | 0.21 |
CDK18 |
cyclin-dependent kinase 18 |
11360 |
0.17 |
| chr9_126234353_126234504 | 0.21 |
RP11-230L22.4 |
|
19735 |
0.22 |
| chr18_22911937_22912088 | 0.21 |
ZNF521 |
zinc finger protein 521 |
19145 |
0.24 |
| chr22_18640172_18640391 | 0.21 |
USP18 |
ubiquitin specific peptidase 18 |
7615 |
0.17 |
| chr6_128580481_128581088 | 0.21 |
PTPRK |
protein tyrosine phosphatase, receptor type, K |
62542 |
0.14 |
| chr15_80354539_80354690 | 0.21 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
1848 |
0.41 |
| chr1_22463869_22464020 | 0.20 |
WNT4 |
wingless-type MMTV integration site family, member 4 |
5515 |
0.23 |
| chr12_89855825_89855993 | 0.20 |
POC1B |
POC1 centriolar protein B |
35125 |
0.15 |
| chr10_73532392_73533248 | 0.20 |
C10orf54 |
chromosome 10 open reading frame 54 |
435 |
0.83 |
| chr15_33300884_33301035 | 0.20 |
ENSG00000212415 |
. |
38031 |
0.18 |
| chr8_68514800_68514982 | 0.20 |
ENSG00000221660 |
. |
108976 |
0.07 |
| chr15_36872415_36872566 | 0.20 |
C15orf41 |
chromosome 15 open reading frame 41 |
441 |
0.9 |
| chr2_11887384_11888228 | 0.20 |
LPIN1 |
lipin 1 |
1042 |
0.41 |
| chr15_40532008_40532159 | 0.20 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
12 |
0.96 |
| chr1_72470618_72470769 | 0.20 |
NEGR1 |
neuronal growth regulator 1 |
95920 |
0.09 |
| chr1_201290547_201290698 | 0.20 |
PKP1 |
plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) |
37791 |
0.13 |
| chr4_52708225_52709084 | 0.20 |
DCUN1D4 |
DCN1, defective in cullin neddylation 1, domain containing 4 |
575 |
0.86 |
| chr3_16522365_16522516 | 0.19 |
RFTN1 |
raftlin, lipid raft linker 1 |
1932 |
0.44 |
| chr18_32660928_32661079 | 0.19 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
39389 |
0.2 |
| chr17_38803171_38803349 | 0.19 |
SMARCE1 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 |
633 |
0.63 |
| chr14_22988565_22988896 | 0.19 |
TRAJ15 |
T cell receptor alpha joining 15 |
9850 |
0.11 |
| chr7_29233941_29234476 | 0.19 |
CHN2 |
chimerin 2 |
87 |
0.9 |
| chr20_56275354_56275505 | 0.19 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
8543 |
0.26 |
| chr10_25869445_25869596 | 0.19 |
ENSG00000222543 |
. |
186747 |
0.03 |
| chr10_134261699_134261957 | 0.19 |
C10orf91 |
chromosome 10 open reading frame 91 |
3135 |
0.24 |
| chr17_40274740_40274891 | 0.19 |
HSPB9 |
heat shock protein, alpha-crystallin-related, B9 |
59 |
0.94 |
| chr19_34717882_34718033 | 0.19 |
KIAA0355 |
KIAA0355 |
27485 |
0.19 |
| chr1_208418253_208418520 | 0.19 |
PLXNA2 |
plexin A2 |
721 |
0.82 |
| chr2_204801338_204802214 | 0.19 |
ICOS |
inducible T-cell co-stimulator |
273 |
0.95 |
| chr14_100842881_100843420 | 0.19 |
WARS |
tryptophanyl-tRNA synthetase |
8 |
0.78 |
| chr5_40685855_40686084 | 0.19 |
PTGER4 |
prostaglandin E receptor 4 (subtype EP4) |
6369 |
0.22 |
| chr4_78743157_78743308 | 0.19 |
CNOT6L |
CCR4-NOT transcription complex, subunit 6-like |
2463 |
0.37 |
| chr5_176237821_176238145 | 0.19 |
UNC5A |
unc-5 homolog A (C. elegans) |
505 |
0.84 |
| chr17_72756037_72756203 | 0.19 |
SLC9A3R1 |
solute carrier family 9, subfamily A (NHE3, cation proton antiporter 3), member 3 regulator 1 |
2022 |
0.18 |
| chr6_106961305_106961653 | 0.19 |
AIM1 |
absent in melanoma 1 |
1749 |
0.38 |
| chr8_26249046_26249197 | 0.19 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
1243 |
0.5 |
| chr2_129365909_129366162 | 0.19 |
ENSG00000238379 |
. |
163263 |
0.04 |
| chr17_4762663_4762814 | 0.18 |
ENSG00000263599 |
. |
16183 |
0.08 |
| chr6_13761461_13761614 | 0.18 |
MCUR1 |
mitochondrial calcium uniporter regulator 1 |
40040 |
0.15 |
| chr1_117910944_117911674 | 0.18 |
MAN1A2 |
mannosidase, alpha, class 1A, member 2 |
1238 |
0.61 |
| chr8_119452205_119452425 | 0.18 |
SAMD12 |
sterile alpha motif domain containing 12 |
140806 |
0.05 |
| chr12_92440588_92440837 | 0.18 |
C12orf79 |
chromosome 12 open reading frame 79 |
90085 |
0.08 |
| chr8_59718017_59718168 | 0.18 |
ENSG00000201231 |
. |
9019 |
0.29 |
| chr22_27441616_27441767 | 0.18 |
ENSG00000200443 |
. |
9841 |
0.32 |
| chr12_14525729_14525880 | 0.18 |
ATF7IP |
activating transcription factor 7 interacting protein |
5497 |
0.26 |
| chr15_71300550_71300701 | 0.18 |
LRRC49 |
leucine rich repeat containing 49 |
71751 |
0.1 |
| chr1_27028870_27029082 | 0.18 |
ARID1A |
AT rich interactive domain 1A (SWI-like) |
6081 |
0.16 |
| chr1_215020458_215020609 | 0.18 |
KCNK2 |
potassium channel, subfamily K, member 2 |
158665 |
0.04 |
| chr4_34195647_34195839 | 0.18 |
ENSG00000263814 |
. |
192716 |
0.03 |
| chr22_31335429_31335580 | 0.18 |
MORC2 |
MORC family CW-type zinc finger 2 |
6623 |
0.16 |
| chr7_45614612_45614823 | 0.18 |
ADCY1 |
adenylate cyclase 1 (brain) |
596 |
0.84 |
| chr5_139283271_139283734 | 0.18 |
NRG2 |
neuregulin 2 |
480 |
0.84 |
| chr4_83956375_83957237 | 0.18 |
COPS4 |
COP9 signalosome subunit 4 |
428 |
0.82 |
| chr1_4606573_4606724 | 0.18 |
AJAP1 |
adherens junctions associated protein 1 |
108144 |
0.08 |
| chr16_89381066_89381217 | 0.18 |
AC137932.6 |
|
6400 |
0.14 |
| chr2_229949087_229949238 | 0.18 |
PID1 |
phosphotyrosine interaction domain containing 1 |
147639 |
0.05 |
| chr1_17541747_17541898 | 0.18 |
PADI1 |
peptidyl arginine deiminase, type I |
10201 |
0.17 |
| chr8_129005826_129005977 | 0.18 |
ENSG00000207110 |
. |
5476 |
0.19 |
| chr1_6453925_6454377 | 0.17 |
ACOT7 |
acyl-CoA thioesterase 7 |
300 |
0.65 |
| chr11_77062251_77062402 | 0.17 |
PAK1 |
p21 protein (Cdc42/Rac)-activated kinase 1 |
1994 |
0.4 |
| chr2_23736447_23736598 | 0.17 |
AC011239.1 |
Uncharacterized protein |
10692 |
0.3 |
| chr1_9653728_9653879 | 0.17 |
TMEM201 |
transmembrane protein 201 |
3152 |
0.26 |
| chr19_31830955_31831409 | 0.17 |
AC007796.1 |
|
8605 |
0.25 |
| chr7_48675933_48676084 | 0.17 |
ABCA13 |
ATP-binding cassette, sub-family A (ABC1), member 13 |
128540 |
0.06 |
| chr19_42626071_42626364 | 0.17 |
POU2F2 |
POU class 2 homeobox 2 |
515 |
0.63 |
| chr17_16944854_16945024 | 0.17 |
MPRIP |
myosin phosphatase Rho interacting protein |
920 |
0.58 |
| chr11_59473933_59474084 | 0.17 |
ENSG00000223223 |
. |
152 |
0.93 |
| chr7_127023381_127023532 | 0.17 |
ZNF800 |
zinc finger protein 800 |
8716 |
0.24 |
| chr19_54676940_54677234 | 0.17 |
TMC4 |
transmembrane channel-like 4 |
143 |
0.89 |
| chr14_56662445_56662672 | 0.17 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
76731 |
0.11 |
| chr12_118455272_118455463 | 0.17 |
RFC5 |
replication factor C (activator 1) 5, 36.5kDa |
833 |
0.64 |
| chr18_13567514_13567726 | 0.17 |
RP11-53B2.4 |
|
2586 |
0.2 |
| chr16_56945671_56945822 | 0.17 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
20214 |
0.11 |
| chr18_30049396_30050442 | 0.17 |
GAREM |
GRB2 associated, regulator of MAPK1 |
476 |
0.87 |
| chr11_9272154_9272305 | 0.17 |
RP11-5L12.1 |
|
5173 |
0.21 |
| chr3_58012091_58012242 | 0.17 |
FLNB |
filamin B, beta |
18039 |
0.25 |
| chr11_62321822_62322177 | 0.17 |
AHNAK |
AHNAK nucleoprotein |
1708 |
0.18 |
| chr11_44588275_44588433 | 0.16 |
CD82 |
CD82 molecule |
1081 |
0.58 |
| chr7_115850994_115851250 | 0.16 |
TES |
testis derived transcript (3 LIM domains) |
519 |
0.82 |
| chr9_100748941_100749092 | 0.16 |
RP11-535C21.3 |
|
922 |
0.5 |
| chr2_96812420_96812577 | 0.16 |
DUSP2 |
dual specificity phosphatase 2 |
1319 |
0.38 |
| chr3_107317640_107318380 | 0.16 |
BBX |
bobby sox homolog (Drosophila) |
150 |
0.98 |
| chr11_69079389_69079561 | 0.16 |
MYEOV |
myeloma overexpressed |
17850 |
0.27 |
| chrX_118071502_118071653 | 0.16 |
LONRF3 |
LON peptidase N-terminal domain and ring finger 3 |
37004 |
0.2 |
| chr16_88490951_88491102 | 0.16 |
ZNF469 |
zinc finger protein 469 |
2853 |
0.26 |
| chr3_182983612_182983806 | 0.16 |
B3GNT5 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
577 |
0.77 |
| chr13_30267514_30267665 | 0.16 |
SLC7A1 |
solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 |
97764 |
0.08 |
| chr19_30792538_30792689 | 0.16 |
ZNF536 |
zinc finger protein 536 |
70706 |
0.12 |
| chr20_43150070_43150299 | 0.16 |
SERINC3 |
serine incorporator 3 |
467 |
0.76 |
| chrX_22153236_22153476 | 0.16 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
37534 |
0.16 |
| chr16_57831320_57831471 | 0.16 |
KIFC3 |
kinesin family member C3 |
281 |
0.88 |
| chr20_47757335_47757486 | 0.16 |
STAU1 |
staufen double-stranded RNA binding protein 1 |
33390 |
0.16 |
| chr2_86114637_86114901 | 0.16 |
ST3GAL5 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 5 |
97 |
0.97 |
| chr12_51015115_51015266 | 0.16 |
ENSG00000207136 |
. |
11866 |
0.21 |
| chr17_8907022_8907441 | 0.16 |
NTN1 |
netrin 1 |
17628 |
0.22 |
| chr2_175197056_175197207 | 0.16 |
SP9 |
Sp9 transcription factor |
2543 |
0.24 |
| chr16_31076129_31076313 | 0.16 |
ZNF668 |
zinc finger protein 668 |
69 |
0.86 |
| chr2_64441847_64441998 | 0.16 |
AC074289.1 |
|
7813 |
0.27 |
| chr20_39786785_39786936 | 0.16 |
RP1-1J6.2 |
|
20217 |
0.18 |
| chr5_153416653_153416804 | 0.16 |
FAM114A2 |
family with sequence similarity 114, member A2 |
142 |
0.95 |
| chr4_40517752_40517903 | 0.16 |
RBM47 |
RNA binding motif protein 47 |
163 |
0.96 |
| chrX_130020204_130020367 | 0.16 |
ENOX2 |
ecto-NOX disulfide-thiol exchanger 2 |
16893 |
0.28 |
| chr8_62621050_62621416 | 0.16 |
ASPH |
aspartate beta-hydroxylase |
5851 |
0.24 |
| chr12_53781982_53782414 | 0.16 |
SP1 |
Sp1 transcription factor |
7112 |
0.13 |
| chr10_126314631_126314782 | 0.16 |
FAM53B-AS1 |
FAM53B antisense RNA 1 |
77488 |
0.09 |
| chr1_156785141_156785328 | 0.16 |
NTRK1 |
neurotrophic tyrosine kinase, receptor, type 1 |
214 |
0.87 |
| chr13_103251895_103252050 | 0.16 |
TPP2 |
tripeptidyl peptidase II |
2599 |
0.29 |
| chr4_31171719_31171870 | 0.16 |
RP11-619J20.1 |
|
376202 |
0.01 |
| chr11_1909549_1909755 | 0.16 |
C11orf89 |
chromosome 11 open reading frame 89 |
2432 |
0.16 |
| chr9_43133207_43133798 | 0.16 |
ANKRD20A3 |
ankyrin repeat domain 20 family, member A3 |
42 |
0.97 |
| chr1_36173086_36173270 | 0.16 |
ENSG00000239859 |
. |
1303 |
0.39 |
| chr8_143443109_143443260 | 0.16 |
TSNARE1 |
t-SNARE domain containing 1 |
7060 |
0.23 |
| chr21_36562046_36562197 | 0.16 |
RUNX1 |
runt-related transcription factor 1 |
140480 |
0.05 |
| chr18_11487781_11487932 | 0.15 |
NPIPB1P |
nuclear pore complex interacting protein family, member B1, pseudogene |
151604 |
0.04 |
| chr17_3289151_3289302 | 0.15 |
RP11-64J4.2 |
|
361 |
0.82 |
| chr4_1228146_1228426 | 0.15 |
CTBP1 |
C-terminal binding protein 1 |
6214 |
0.15 |
| chr18_68267310_68267461 | 0.15 |
GTSCR1 |
Gilles de la Tourette syndrome chromosome region, candidate 1 |
50562 |
0.17 |
| chr5_112824477_112824726 | 0.15 |
MCC |
mutated in colorectal cancers |
74 |
0.97 |
| chr2_131798149_131798300 | 0.15 |
ARHGEF4 |
Rho guanine nucleotide exchange factor (GEF) 4 |
626 |
0.76 |
| chr1_67893569_67893720 | 0.15 |
SERBP1 |
SERPINE1 mRNA binding protein 1 |
2411 |
0.31 |
| chr8_1791296_1791447 | 0.15 |
ARHGEF10 |
Rho guanine nucleotide exchange factor (GEF) 10 |
17094 |
0.2 |
| chr10_104420612_104420763 | 0.15 |
TRIM8 |
tripartite motif containing 8 |
16043 |
0.16 |
| chr9_18689231_18689382 | 0.15 |
ENSG00000252960 |
. |
38161 |
0.2 |
| chr3_150059920_150060071 | 0.15 |
TSC22D2 |
TSC22 domain family, member 2 |
66127 |
0.13 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.4 | GO:0036037 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.1 | 0.3 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.1 | 0.3 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.1 | 0.4 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.1 | 0.2 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.1 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.1 | 0.2 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.1 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0045341 | MHC class I biosynthetic process(GO:0045341) regulation of MHC class I biosynthetic process(GO:0045343) |
| 0.0 | 0.1 | GO:0006922 | obsolete cleavage of lamin involved in execution phase of apoptosis(GO:0006922) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.2 | GO:0043922 | negative regulation by host of viral transcription(GO:0043922) |
| 0.0 | 0.3 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.0 | 0.1 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.0 | 0.1 | GO:0001921 | positive regulation of receptor recycling(GO:0001921) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.2 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.1 | GO:0043578 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:2000114 | regulation of establishment of cell polarity(GO:2000114) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0060197 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.0 | 0.1 | GO:0043985 | histone H4-R3 methylation(GO:0043985) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.1 | GO:0032959 | inositol trisphosphate biosynthetic process(GO:0032959) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0046880 | regulation of follicle-stimulating hormone secretion(GO:0046880) |
| 0.0 | 0.2 | GO:0006337 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0033261 | obsolete regulation of S phase(GO:0033261) |
| 0.0 | 0.0 | GO:0002890 | negative regulation of B cell mediated immunity(GO:0002713) negative regulation of immunoglobulin mediated immune response(GO:0002890) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.0 | GO:0033091 | positive regulation of immature T cell proliferation(GO:0033091) |
| 0.0 | 0.0 | GO:0060633 | negative regulation of transcription initiation from RNA polymerase II promoter(GO:0060633) negative regulation of DNA-templated transcription, initiation(GO:2000143) |
| 0.0 | 0.1 | GO:0044380 | protein localization to cytoskeleton(GO:0044380) protein localization to microtubule cytoskeleton(GO:0072698) |
| 0.0 | 0.0 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0032753 | positive regulation of interleukin-4 production(GO:0032753) |
| 0.0 | 0.1 | GO:0006166 | purine ribonucleoside salvage(GO:0006166) |
| 0.0 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.0 | 0.2 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.1 | GO:0035092 | sperm chromatin condensation(GO:0035092) |
| 0.0 | 0.1 | GO:0072583 | synaptic vesicle recycling(GO:0036465) synaptic vesicle endocytosis(GO:0048488) clathrin-mediated endocytosis(GO:0072583) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.0 | GO:0042482 | positive regulation of odontogenesis(GO:0042482) |
| 0.0 | 0.1 | GO:0045906 | negative regulation of vasoconstriction(GO:0045906) |
| 0.0 | 0.2 | GO:0050858 | negative regulation of antigen receptor-mediated signaling pathway(GO:0050858) negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0040016 | embryonic cleavage(GO:0040016) |
| 0.0 | 0.0 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0045217 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0018216 | peptidyl-arginine methylation(GO:0018216) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0034723 | DNA replication-dependent nucleosome organization(GO:0034723) |
| 0.0 | 0.1 | GO:0006855 | drug transmembrane transport(GO:0006855) |
| 0.0 | 0.1 | GO:0060028 | convergent extension involved in axis elongation(GO:0060028) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.1 | GO:0033160 | positive regulation of protein import into nucleus, translocation(GO:0033160) |
| 0.0 | 0.1 | GO:0000090 | mitotic anaphase(GO:0000090) |
| 0.0 | 0.1 | GO:0006538 | glutamate catabolic process(GO:0006538) |
| 0.0 | 0.2 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:0048711 | positive regulation of astrocyte differentiation(GO:0048711) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0060391 | positive regulation of SMAD protein import into nucleus(GO:0060391) |
| 0.0 | 0.0 | GO:0070293 | renal absorption(GO:0070293) |
| 0.0 | 0.0 | GO:0071569 | protein ufmylation(GO:0071569) |
| 0.0 | 0.0 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.0 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.0 | 0.0 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.0 | 0.0 | GO:0032351 | negative regulation of hormone metabolic process(GO:0032351) negative regulation of hormone biosynthetic process(GO:0032353) |
| 0.0 | 0.0 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0021799 | cerebral cortex radially oriented cell migration(GO:0021799) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) |
| 0.0 | 0.3 | GO:0009267 | cellular response to starvation(GO:0009267) |
| 0.0 | 0.0 | GO:0006562 | proline catabolic process(GO:0006562) |
| 0.0 | 0.5 | GO:0000245 | spliceosomal complex assembly(GO:0000245) |
| 0.0 | 0.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.0 | GO:0045607 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.0 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:0051085 | chaperone mediated protein folding requiring cofactor(GO:0051085) |
| 0.0 | 0.1 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0071883 | adrenergic receptor signaling pathway(GO:0071875) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.0 | GO:0019372 | lipoxygenase pathway(GO:0019372) |
| 0.0 | 0.1 | GO:0001706 | endoderm formation(GO:0001706) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:0008054 | negative regulation of cyclin-dependent protein serine/threonine kinase by cyclin degradation(GO:0008054) |
| 0.0 | 0.0 | GO:0001778 | plasma membrane repair(GO:0001778) |
| 0.0 | 0.1 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0090598 | male genitalia morphogenesis(GO:0048808) male anatomical structure morphogenesis(GO:0090598) |
| 0.0 | 0.1 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.1 | GO:0017182 | peptidyl-diphthamide metabolic process(GO:0017182) peptidyl-diphthamide biosynthetic process from peptidyl-histidine(GO:0017183) |
| 0.0 | 0.0 | GO:0050667 | homocysteine metabolic process(GO:0050667) |
| 0.0 | 0.1 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:0009067 | aspartate family amino acid biosynthetic process(GO:0009067) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.0 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.0 | GO:1901722 | regulation of cell proliferation involved in kidney development(GO:1901722) |
| 0.0 | 0.2 | GO:0046834 | lipid phosphorylation(GO:0046834) |
| 0.0 | 0.0 | GO:0070634 | transepithelial ammonium transport(GO:0070634) |
| 0.0 | 0.1 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.0 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.0 | GO:0050703 | interleukin-1 alpha secretion(GO:0050703) |
| 0.0 | 0.0 | GO:0006549 | isoleucine metabolic process(GO:0006549) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0040023 | establishment of nucleus localization(GO:0040023) |
| 0.0 | 0.1 | GO:0006972 | hyperosmotic response(GO:0006972) |
| 0.0 | 0.0 | GO:0021571 | rhombomere 5 development(GO:0021571) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.0 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0006105 | succinate metabolic process(GO:0006105) |
| 0.0 | 0.0 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0030812 | negative regulation of nucleotide catabolic process(GO:0030812) |
| 0.0 | 0.0 | GO:0001821 | histamine secretion(GO:0001821) |
| 0.0 | 0.0 | GO:0045046 | protein import into peroxisome membrane(GO:0045046) |
| 0.0 | 0.0 | GO:0042501 | serine phosphorylation of STAT protein(GO:0042501) |
| 0.0 | 0.0 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.0 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.0 | 0.1 | GO:0031115 | negative regulation of microtubule polymerization(GO:0031115) |
| 0.0 | 0.0 | GO:2000095 | regulation of non-canonical Wnt signaling pathway(GO:2000050) regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000095) |
| 0.0 | 0.0 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.3 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0045257 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.2 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0002141 | stereocilia coupling link(GO:0002139) stereocilia ankle link(GO:0002141) stereocilia ankle link complex(GO:0002142) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0033270 | paranode region of axon(GO:0033270) |
| 0.0 | 0.2 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0002178 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.1 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.0 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.0 | GO:0070062 | extracellular exosome(GO:0070062) extracellular vesicle(GO:1903561) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.0 | GO:0070188 | obsolete Stn1-Ten1 complex(GO:0070188) |
| 0.0 | 0.1 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.3 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.1 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.1 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.0 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.1 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.1 | GO:0005849 | mRNA cleavage factor complex(GO:0005849) |
| 0.0 | 0.0 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0046930 | pore complex(GO:0046930) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.0 | GO:0071818 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.2 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.2 | GO:0015288 | porin activity(GO:0015288) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0016151 | nickel cation binding(GO:0016151) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0008294 | calcium- and calmodulin-responsive adenylate cyclase activity(GO:0008294) |
| 0.0 | 0.1 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.2 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.2 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0042608 | T cell receptor binding(GO:0042608) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.2 | GO:0016273 | arginine N-methyltransferase activity(GO:0016273) protein-arginine N-methyltransferase activity(GO:0016274) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.1 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0016362 | activin receptor activity, type II(GO:0016362) |
| 0.0 | 0.1 | GO:0042975 | peroxisome proliferator activated receptor binding(GO:0042975) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.2 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.4 | GO:0008373 | sialyltransferase activity(GO:0008373) |
| 0.0 | 0.1 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.3 | GO:0008601 | protein phosphatase type 2A regulator activity(GO:0008601) |
| 0.0 | 0.1 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.0 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.0 | 0.1 | GO:0005536 | glucose binding(GO:0005536) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0051380 | norepinephrine binding(GO:0051380) |
| 0.0 | 0.0 | GO:0004367 | glycerol-3-phosphate dehydrogenase [NAD+] activity(GO:0004367) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0003689 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.1 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.0 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0031849 | olfactory receptor binding(GO:0031849) |
| 0.0 | 0.0 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.0 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.1 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) ubiquitin-like protein conjugating enzyme binding(GO:0044390) |
| 0.0 | 0.0 | GO:0098847 | single-stranded telomeric DNA binding(GO:0043047) sequence-specific single stranded DNA binding(GO:0098847) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0016307 | phosphatidylinositol phosphate kinase activity(GO:0016307) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.1 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.0 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.0 | GO:0051379 | alpha2-adrenergic receptor activity(GO:0004938) epinephrine binding(GO:0051379) |
| 0.0 | 0.0 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.0 | GO:0061731 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.0 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.0 | GO:0010485 | H4 histone acetyltransferase activity(GO:0010485) |
| 0.0 | 0.2 | GO:0005484 | SNAP receptor activity(GO:0005484) |
| 0.0 | 0.0 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.1 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.0 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0070569 | uridylyltransferase activity(GO:0070569) |
| 0.0 | 0.1 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0035197 | siRNA binding(GO:0035197) |
| 0.0 | 0.0 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.0 | GO:0004477 | methenyltetrahydrofolate cyclohydrolase activity(GO:0004477) |
| 0.0 | 0.1 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.4 | GO:0001067 | regulatory region DNA binding(GO:0000975) regulatory region nucleic acid binding(GO:0001067) transcription regulatory region DNA binding(GO:0044212) |
| 0.0 | 0.0 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.0 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.0 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.4 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.3 | PID SMAD2 3PATHWAY | Regulation of cytoplasmic and nuclear SMAD2/3 signaling |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.6 | PID ECADHERIN STABILIZATION PATHWAY | Stabilization and expansion of the E-cadherin adherens junction |
| 0.0 | 0.1 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.5 | PID ATR PATHWAY | ATR signaling pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.1 | REACTOME P75NTR RECRUITS SIGNALLING COMPLEXES | Genes involved in p75NTR recruits signalling complexes |
| 0.0 | 0.1 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.3 | REACTOME RORA ACTIVATES CIRCADIAN EXPRESSION | Genes involved in RORA Activates Circadian Expression |
| 0.0 | 0.1 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.2 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.2 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 0.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.2 | REACTOME PROLONGED ERK ACTIVATION EVENTS | Genes involved in Prolonged ERK activation events |
| 0.0 | 0.2 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.1 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.1 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.0 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.1 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |