Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
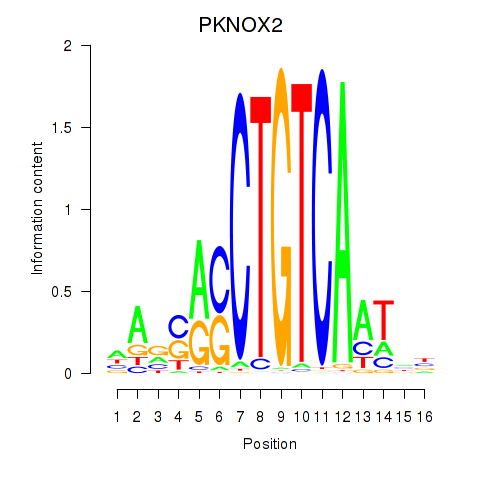
Results for PKNOX2
Z-value: 1.06
Transcription factors associated with PKNOX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PKNOX2
|
ENSG00000165495.11 | PBX/knotted 1 homeobox 2 |
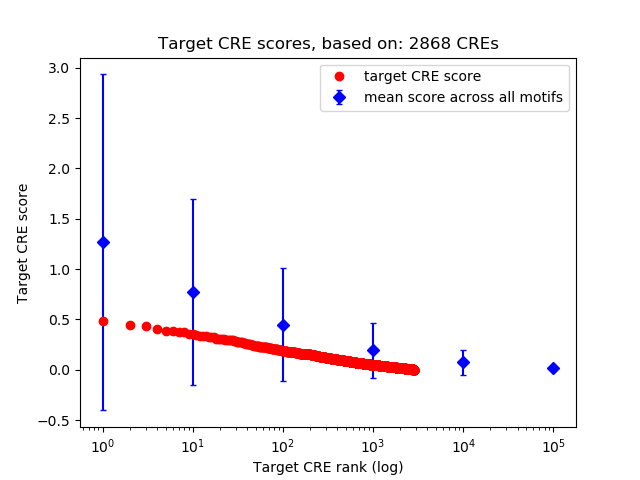
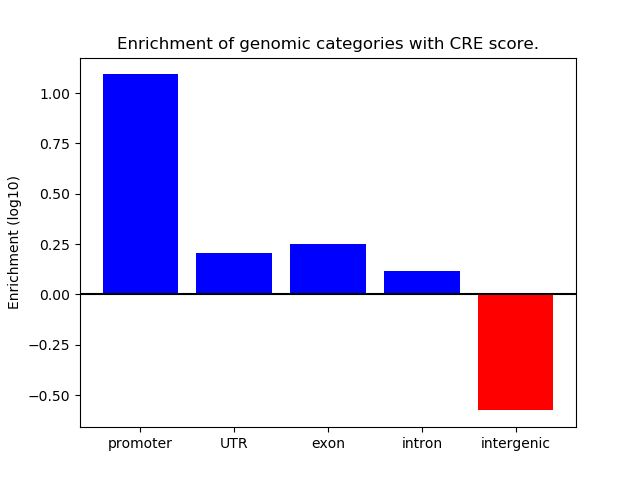
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr11_125035186_125035397 | PKNOX2 | 641 | 0.474796 | 0.59 | 9.3e-02 | Click! |
| chr11_125039995_125040154 | PKNOX2 | 5424 | 0.166772 | 0.57 | 1.1e-01 | Click! |
| chr11_125036574_125036725 | PKNOX2 | 1999 | 0.244857 | 0.54 | 1.3e-01 | Click! |
| chr11_125036127_125036278 | PKNOX2 | 1552 | 0.286129 | 0.51 | 1.6e-01 | Click! |
| chr11_125035763_125035964 | PKNOX2 | 1213 | 0.334760 | 0.48 | 1.9e-01 | Click! |
Activity of the PKNOX2 motif across conditions
Conditions sorted by the z-value of the PKNOX2 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
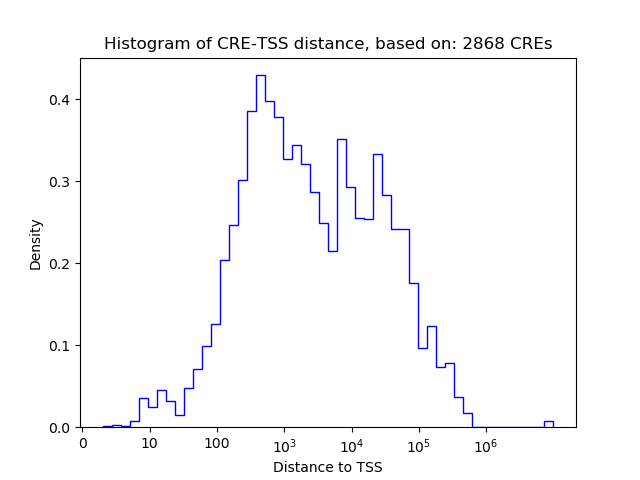
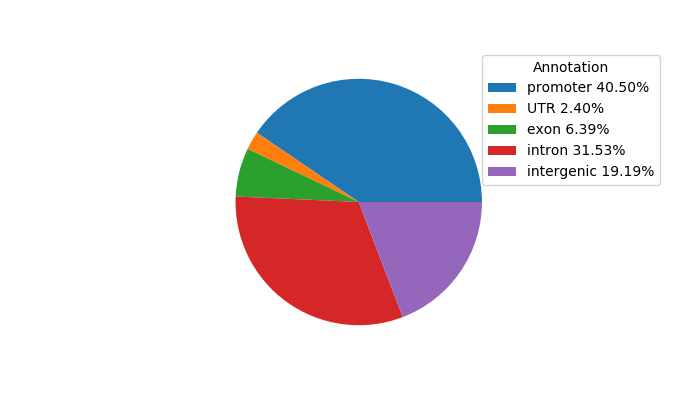
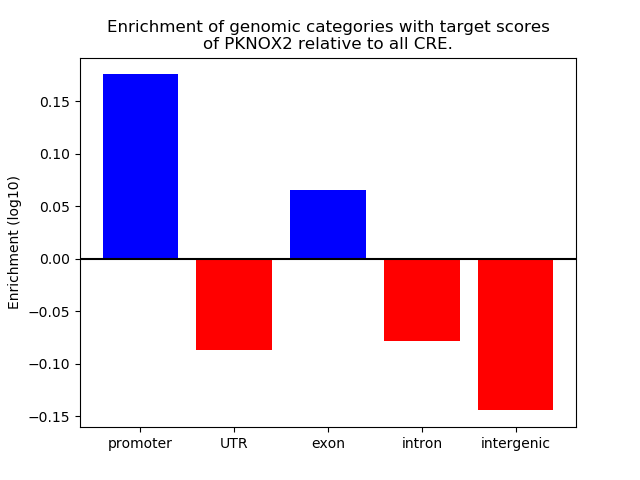
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr1_65530319_65530470 | 0.48 |
ENSG00000199135 |
. |
6203 |
0.2 |
| chr1_66258257_66258503 | 0.44 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
183 |
0.96 |
| chrX_100334236_100334411 | 0.43 |
TMEM35 |
transmembrane protein 35 |
614 |
0.68 |
| chr9_19128575_19128726 | 0.40 |
PLIN2 |
perilipin 2 |
1076 |
0.44 |
| chr6_5498765_5499077 | 0.39 |
RP1-232P20.1 |
|
40613 |
0.2 |
| chr5_67577155_67577306 | 0.38 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
1095 |
0.66 |
| chr5_56112470_56113088 | 0.38 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
1378 |
0.44 |
| chr16_68693087_68693238 | 0.37 |
RP11-615I2.2 |
|
13091 |
0.13 |
| chr1_21859764_21859915 | 0.36 |
ALPL |
alkaline phosphatase, liver/bone/kidney |
17933 |
0.19 |
| chr19_49121963_49122114 | 0.36 |
RPL18 |
ribosomal protein L18 |
346 |
0.53 |
| chr6_82732214_82732365 | 0.35 |
ENSG00000223044 |
. |
187868 |
0.03 |
| chr1_95389567_95389941 | 0.34 |
CNN3 |
calponin 3, acidic |
1583 |
0.36 |
| chr11_3043840_3044115 | 0.34 |
CARS-AS1 |
CARS antisense RNA 1 |
6647 |
0.13 |
| chr14_30054222_30054373 | 0.33 |
PRKD1 |
protein kinase D1 |
49383 |
0.15 |
| chr12_90272914_90273065 | 0.33 |
ENSG00000252823 |
. |
125153 |
0.06 |
| chr16_89381066_89381217 | 0.33 |
AC137932.6 |
|
6400 |
0.14 |
| chr5_157714663_157714814 | 0.32 |
ENSG00000222626 |
. |
310774 |
0.01 |
| chr2_47401669_47401853 | 0.31 |
CALM2 |
calmodulin 2 (phosphorylase kinase, delta) |
1889 |
0.37 |
| chr20_47894397_47894608 | 0.31 |
ZNFX1 |
zinc finger, NFX1-type containing 1 |
76 |
0.67 |
| chr6_24910150_24910301 | 0.30 |
FAM65B |
family with sequence similarity 65, member B |
970 |
0.62 |
| chr1_109740352_109740715 | 0.30 |
ENSG00000238310 |
. |
9937 |
0.15 |
| chr12_92440588_92440837 | 0.30 |
C12orf79 |
chromosome 12 open reading frame 79 |
90085 |
0.08 |
| chr14_92331424_92331867 | 0.30 |
TC2N |
tandem C2 domains, nuclear |
2228 |
0.32 |
| chr22_23282661_23282812 | 0.30 |
IGLJ7 |
immunoglobulin lambda joining 7 |
19174 |
0.07 |
| chr20_8380586_8380742 | 0.30 |
PLCB1-IT1 |
PLCB1 intronic transcript 1 (non-protein coding) |
151292 |
0.04 |
| chr5_163147994_163148145 | 0.29 |
ENSG00000251998 |
. |
75102 |
0.12 |
| chrX_100352542_100352928 | 0.29 |
CENPI |
centromere protein I |
443 |
0.78 |
| chr6_131255225_131255376 | 0.29 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
22325 |
0.27 |
| chr4_3529804_3529955 | 0.29 |
LRPAP1 |
low density lipoprotein receptor-related protein associated protein 1 |
4407 |
0.19 |
| chr7_99516388_99516774 | 0.28 |
TRIM4 |
tripartite motif containing 4 |
147 |
0.91 |
| chr4_141217305_141217524 | 0.28 |
ENSG00000252300 |
. |
21148 |
0.19 |
| chr13_53419960_53420114 | 0.28 |
PCDH8 |
protocadherin 8 |
2603 |
0.34 |
| chr4_139813437_139813623 | 0.28 |
RP11-371F15.3 |
|
45605 |
0.16 |
| chr3_50298349_50298500 | 0.28 |
SEMA3B-AS1 |
SEMA3B antisense RNA 1 (head to head) |
6379 |
0.09 |
| chr1_33804639_33805031 | 0.28 |
ENSG00000222112 |
. |
2370 |
0.19 |
| chr6_82854353_82854866 | 0.27 |
ENSG00000223044 |
. |
65548 |
0.14 |
| chr6_162052389_162052540 | 0.27 |
ENSG00000221021 |
. |
16366 |
0.31 |
| chr19_46171409_46171560 | 0.27 |
GIPR |
gastric inhibitory polypeptide receptor |
18 |
0.94 |
| chr11_111741428_111741733 | 0.26 |
ALG9 |
ALG9, alpha-1,2-mannosyltransferase |
8 |
0.96 |
| chr5_32175143_32175294 | 0.26 |
GOLPH3 |
golgi phosphoprotein 3 (coat-protein) |
762 |
0.67 |
| chr4_178324604_178324755 | 0.25 |
AGA |
aspartylglucosaminidase |
36099 |
0.14 |
| chr8_37728655_37728889 | 0.25 |
RAB11FIP1 |
RAB11 family interacting protein 1 (class I) |
6863 |
0.15 |
| chr1_117910944_117911674 | 0.25 |
MAN1A2 |
mannosidase, alpha, class 1A, member 2 |
1238 |
0.61 |
| chr13_109924709_109925156 | 0.25 |
MYO16-AS1 |
MYO16 antisense RNA 1 |
71101 |
0.13 |
| chr10_82215597_82215808 | 0.25 |
TSPAN14 |
tetraspanin 14 |
1591 |
0.41 |
| chr11_33756310_33756563 | 0.25 |
CD59 |
CD59 molecule, complement regulatory protein |
1518 |
0.33 |
| chr4_779234_779385 | 0.24 |
RP11-440L14.1 |
|
3672 |
0.15 |
| chr18_8755384_8755651 | 0.24 |
SOGA2 |
SOGA family member 2 |
34155 |
0.14 |
| chr12_9216598_9216767 | 0.24 |
A2M-AS1 |
A2M antisense RNA 1 |
1091 |
0.49 |
| chr20_2802410_2802561 | 0.24 |
TMEM239 |
transmembrane protein 239 |
5506 |
0.13 |
| chr5_154229616_154229872 | 0.24 |
FAXDC2 |
fatty acid hydroxylase domain containing 2 |
419 |
0.81 |
| chr2_650542_650693 | 0.24 |
TMEM18 |
transmembrane protein 18 |
25158 |
0.18 |
| chr6_130453758_130453909 | 0.23 |
RP11-73O6.3 |
|
5508 |
0.26 |
| chr11_64013126_64013464 | 0.23 |
PPP1R14B |
protein phosphatase 1, regulatory (inhibitor) subunit 14B |
7 |
0.62 |
| chr5_158407500_158407684 | 0.23 |
CTD-2363C16.1 |
|
2422 |
0.33 |
| chr19_37328825_37329012 | 0.23 |
ZNF790 |
zinc finger protein 790 |
11 |
0.97 |
| chr2_97472579_97472810 | 0.23 |
ENSG00000264157 |
. |
8679 |
0.13 |
| chr1_116293768_116293919 | 0.23 |
CASQ2 |
calsequestrin 2 (cardiac muscle) |
17492 |
0.24 |
| chr11_118489320_118489642 | 0.23 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
11123 |
0.11 |
| chr13_99967074_99967225 | 0.23 |
GPR183 |
G protein-coupled receptor 183 |
7490 |
0.21 |
| chr16_85590478_85590669 | 0.23 |
GSE1 |
Gse1 coiled-coil protein |
54442 |
0.13 |
| chr20_43974638_43974789 | 0.22 |
SDC4 |
syndecan 4 |
2351 |
0.19 |
| chr2_33165553_33165704 | 0.22 |
ENSG00000206951 |
. |
4890 |
0.24 |
| chr11_84028149_84028546 | 0.22 |
DLG2 |
discs, large homolog 2 (Drosophila) |
35 |
0.99 |
| chr12_62654244_62654932 | 0.22 |
USP15 |
ubiquitin specific peptidase 15 |
379 |
0.66 |
| chr16_51180371_51180536 | 0.22 |
AC009166.5 |
|
2697 |
0.32 |
| chr5_1331289_1331440 | 0.22 |
CLPTM1L |
CLPTM1-like |
10489 |
0.17 |
| chr16_2880049_2880363 | 0.22 |
ZG16B |
zymogen granule protein 16B |
36 |
0.94 |
| chr9_130825291_130825442 | 0.22 |
NAIF1 |
nuclear apoptosis inducing factor 1 |
4234 |
0.11 |
| chr1_16122451_16122602 | 0.22 |
FBLIM1 |
filamin binding LIM protein 1 |
31078 |
0.1 |
| chr18_67425087_67425238 | 0.22 |
RP11-543H23.2 |
|
32951 |
0.22 |
| chr6_28949532_28949984 | 0.21 |
HCG15 |
HLA complex group 15 (non-protein coding) |
4222 |
0.13 |
| chr2_163099011_163099583 | 0.21 |
FAP |
fibroblast activation protein, alpha |
261 |
0.94 |
| chr3_73597243_73597428 | 0.21 |
PDZRN3 |
PDZ domain containing ring finger 3 |
13434 |
0.24 |
| chr17_58157127_58157278 | 0.21 |
HEATR6 |
HEAT repeat containing 6 |
910 |
0.49 |
| chr11_46724199_46724350 | 0.21 |
ZNF408 |
zinc finger protein 408 |
1906 |
0.21 |
| chr7_150811251_150811777 | 0.21 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
251 |
0.86 |
| chr1_55370578_55370729 | 0.21 |
RP11-67L3.4 |
|
17248 |
0.14 |
| chr22_42476102_42476312 | 0.21 |
SMDT1 |
single-pass membrane protein with aspartate-rich tail 1 |
508 |
0.6 |
| chr3_42728895_42729046 | 0.21 |
KLHL40 |
kelch-like family member 40 |
1959 |
0.19 |
| chr4_88140632_88141014 | 0.21 |
KLHL8 |
kelch-like family member 8 |
597 |
0.81 |
| chr17_26988932_26989176 | 0.21 |
SUPT6H |
suppressor of Ty 6 homolog (S. cerevisiae) |
55 |
0.59 |
| chr9_12575381_12575532 | 0.21 |
TYRP1 |
tyrosinase-related protein 1 |
109983 |
0.07 |
| chr9_118174215_118174366 | 0.21 |
DEC1 |
deleted in esophageal cancer 1 |
270193 |
0.02 |
| chr6_18397050_18397268 | 0.20 |
ENSG00000238458 |
. |
5124 |
0.24 |
| chr12_81474615_81474766 | 0.20 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
2854 |
0.32 |
| chr18_30349192_30349343 | 0.20 |
AC012123.1 |
Uncharacterized protein |
491 |
0.81 |
| chr5_7869806_7870194 | 0.20 |
MTRR |
5-methyltetrahydrofolate-homocysteine methyltransferase reductase |
374 |
0.74 |
| chr3_194928852_194929003 | 0.20 |
ENSG00000206600 |
. |
6589 |
0.18 |
| chr8_28560557_28560810 | 0.20 |
EXTL3-AS1 |
EXTL3 antisense RNA 1 |
1702 |
0.31 |
| chr11_18344438_18344755 | 0.20 |
GTF2H1 |
general transcription factor IIH, polypeptide 1, 62kDa |
453 |
0.61 |
| chr6_39761310_39761623 | 0.20 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
672 |
0.79 |
| chr8_29498654_29498805 | 0.20 |
ENSG00000221003 |
. |
287392 |
0.01 |
| chr3_71112065_71112645 | 0.19 |
FOXP1 |
forkhead box P1 |
1722 |
0.53 |
| chr4_89741704_89741855 | 0.19 |
FAM13A |
family with sequence similarity 13, member A |
2573 |
0.37 |
| chr16_47178196_47178489 | 0.19 |
RP11-329J18.2 |
|
13 |
0.88 |
| chr7_132260236_132260613 | 0.19 |
PLXNA4 |
plexin A4 |
805 |
0.75 |
| chr11_74659771_74660083 | 0.19 |
XRRA1 |
X-ray radiation resistance associated 1 |
138 |
0.75 |
| chr3_151997671_151997822 | 0.19 |
MBNL1-AS1 |
MBNL1 antisense RNA 1 |
10402 |
0.22 |
| chr16_85768972_85769153 | 0.19 |
ENSG00000222190 |
. |
6244 |
0.13 |
| chr17_12568958_12569229 | 0.19 |
MYOCD |
myocardin |
213 |
0.95 |
| chr4_111084991_111085142 | 0.19 |
ELOVL6 |
ELOVL fatty acid elongase 6 |
34743 |
0.18 |
| chr3_50297269_50297420 | 0.19 |
SEMA3B-AS1 |
SEMA3B antisense RNA 1 (head to head) |
7459 |
0.09 |
| chr6_7312252_7312483 | 0.19 |
SSR1 |
signal sequence receptor, alpha |
1040 |
0.57 |
| chrX_71288852_71289006 | 0.19 |
RGAG4 |
retrotransposon gag domain containing 4 |
62749 |
0.1 |
| chr12_51319016_51319581 | 0.19 |
METTL7A |
methyltransferase like 7A |
764 |
0.59 |
| chr17_76182522_76182694 | 0.19 |
TK1 |
thymidine kinase 1, soluble |
503 |
0.53 |
| chr21_35305075_35305226 | 0.18 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
1632 |
0.32 |
| chr2_160469080_160469419 | 0.18 |
AC009506.1 |
|
2558 |
0.28 |
| chr15_22514117_22514287 | 0.18 |
ENSG00000221641 |
. |
922 |
0.52 |
| chr1_179199050_179199491 | 0.18 |
ABL2 |
c-abl oncogene 2, non-receptor tyrosine kinase |
451 |
0.82 |
| chr15_61266463_61266614 | 0.18 |
RP11-39M21.1 |
|
206910 |
0.02 |
| chr9_136556754_136556905 | 0.18 |
SARDH |
sarcosine dehydrogenase |
11994 |
0.18 |
| chr5_14178346_14178639 | 0.18 |
TRIO |
trio Rho guanine nucleotide exchange factor |
5415 |
0.35 |
| chr1_231177146_231177297 | 0.18 |
FAM89A |
family with sequence similarity 89, member A |
1229 |
0.43 |
| chr1_180199493_180199788 | 0.18 |
LHX4 |
LIM homeobox 4 |
219 |
0.95 |
| chr20_42143535_42143824 | 0.18 |
L3MBTL1 |
l(3)mbt-like 1 (Drosophila) |
513 |
0.75 |
| chr6_143037024_143037175 | 0.18 |
RP1-67K17.3 |
|
32481 |
0.23 |
| chr17_44668558_44668782 | 0.18 |
NSF |
N-ethylmaleimide-sensitive factor |
283 |
0.9 |
| chr11_72145394_72145582 | 0.18 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
62 |
0.97 |
| chr9_128288500_128288651 | 0.18 |
ENSG00000243845 |
. |
13089 |
0.21 |
| chr11_65267780_65268262 | 0.18 |
SCYL1 |
SCY1-like 1 (S. cerevisiae) |
24527 |
0.09 |
| chr17_73852253_73852511 | 0.18 |
WBP2 |
WW domain binding protein 2 |
206 |
0.86 |
| chr17_33868045_33868636 | 0.18 |
SLFN12L |
schlafen family member 12-like |
3460 |
0.14 |
| chr13_109809143_109809440 | 0.18 |
MYO16-AS1 |
MYO16 antisense RNA 1 |
44540 |
0.18 |
| chr6_2901280_2901469 | 0.18 |
SERPINB9 |
serpin peptidase inhibitor, clade B (ovalbumin), member 9 |
2140 |
0.3 |
| chr6_139868311_139868622 | 0.18 |
CITED2 |
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
172709 |
0.03 |
| chr15_34620156_34620427 | 0.18 |
SLC12A6 |
solute carrier family 12 (potassium/chloride transporter), member 6 |
8754 |
0.11 |
| chr1_43826510_43826891 | 0.17 |
CDC20 |
cell division cycle 20 |
2016 |
0.17 |
| chr10_28822829_28823308 | 0.17 |
WAC |
WW domain containing adaptor with coiled-coil |
16 |
0.97 |
| chr1_79306379_79306727 | 0.17 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
77017 |
0.11 |
| chr19_49139505_49139656 | 0.17 |
DBP |
D site of albumin promoter (albumin D-box) binding protein |
1040 |
0.28 |
| chr16_29825122_29825273 | 0.17 |
PRRT2 |
proline-rich transmembrane protein 2 |
1633 |
0.13 |
| chr10_82232604_82232771 | 0.17 |
TSPAN14 |
tetraspanin 14 |
13629 |
0.2 |
| chr22_36753608_36753885 | 0.17 |
MYH9 |
myosin, heavy chain 9, non-muscle |
7410 |
0.2 |
| chr13_34492968_34493208 | 0.17 |
ENSG00000263663 |
. |
6959 |
0.3 |
| chr16_57835623_57835864 | 0.17 |
KIFC3 |
kinesin family member C3 |
618 |
0.65 |
| chr18_5237293_5237735 | 0.17 |
C18orf42 |
chromosome 18 open reading frame 42 |
40012 |
0.18 |
| chr16_70380861_70381493 | 0.17 |
DDX19A |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 19A |
330 |
0.6 |
| chr11_2924254_2924405 | 0.17 |
SLC22A18 |
solute carrier family 22, member 18 |
15 |
0.86 |
| chr9_134297852_134298003 | 0.17 |
PRRC2B |
proline-rich coiled-coil 2B |
7550 |
0.21 |
| chr1_45267568_45268118 | 0.17 |
PLK3 |
polo-like kinase 3 |
1946 |
0.13 |
| chr18_73496416_73496569 | 0.17 |
SMIM21 |
small integral membrane protein 21 |
356834 |
0.01 |
| chr12_85304928_85305252 | 0.17 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
713 |
0.8 |
| chr2_122287274_122287589 | 0.17 |
RP11-204L24.2 |
|
567 |
0.62 |
| chr11_84148388_84148632 | 0.17 |
DLG2 |
discs, large homolog 2 (Drosophila) |
120128 |
0.07 |
| chr2_67095961_67096112 | 0.17 |
MEIS1 |
Meis homeobox 1 |
359977 |
0.01 |
| chr1_151170363_151170514 | 0.17 |
PIP5K1A |
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
13 |
0.95 |
| chr1_193073507_193073986 | 0.16 |
GLRX2 |
glutaredoxin 2 |
815 |
0.53 |
| chr17_44344782_44345197 | 0.16 |
RP11-259G18.1 |
|
211 |
0.93 |
| chr5_139726315_139726693 | 0.16 |
HBEGF |
heparin-binding EGF-like growth factor |
288 |
0.85 |
| chr1_161276676_161276935 | 0.16 |
MPZ |
myelin protein zero |
456 |
0.72 |
| chr7_99868283_99868434 | 0.16 |
GATS |
|
1216 |
0.3 |
| chr6_164530063_164530741 | 0.16 |
ENSG00000266128 |
. |
268809 |
0.02 |
| chr9_33446798_33447442 | 0.16 |
AQP3 |
aquaporin 3 (Gill blood group) |
489 |
0.76 |
| chr6_24775111_24775564 | 0.16 |
GMNN |
geminin, DNA replication inhibitor |
171 |
0.94 |
| chr5_83220316_83220467 | 0.16 |
ENSG00000216125 |
. |
49314 |
0.18 |
| chr1_40847174_40847476 | 0.16 |
SMAP2 |
small ArfGAP2 |
7005 |
0.19 |
| chr3_186743256_186743535 | 0.16 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
124 |
0.97 |
| chr7_75267461_75267701 | 0.16 |
HIP1 |
huntingtin interacting protein 1 |
26420 |
0.17 |
| chr4_30725920_30726277 | 0.16 |
PCDH7 |
protocadherin 7 |
2121 |
0.46 |
| chr15_75085209_75085360 | 0.16 |
CSK |
c-src tyrosine kinase |
101 |
0.94 |
| chr17_74323765_74323916 | 0.16 |
ENSG00000238418 |
. |
3563 |
0.13 |
| chr10_111488296_111488447 | 0.16 |
ENSG00000272160 |
. |
141245 |
0.05 |
| chr15_22482994_22483145 | 0.16 |
IGHV1OR15-4 |
immunoglobulin heavy variable 1/OR15-4 (pseudogene) |
199 |
0.91 |
| chr11_104941226_104941560 | 0.16 |
CARD16 |
caspase recruitment domain family, member 16 |
25290 |
0.15 |
| chr12_121199647_121199798 | 0.16 |
RP11-173P15.7 |
|
327 |
0.84 |
| chr3_111849268_111849419 | 0.16 |
GCSAM |
germinal center-associated, signaling and motility |
2729 |
0.22 |
| chr6_127837776_127838009 | 0.16 |
SOGA3 |
SOGA family member 3 |
133 |
0.97 |
| chr1_27021420_27021612 | 0.16 |
RP5-968P14.2 |
|
894 |
0.39 |
| chr2_121199524_121199751 | 0.16 |
LINC01101 |
long intergenic non-protein coding RNA 1101 |
24060 |
0.24 |
| chr4_111007994_111008259 | 0.16 |
ENSG00000263940 |
. |
30766 |
0.21 |
| chr13_101194913_101195273 | 0.16 |
RP11-151A6.4 |
|
3033 |
0.25 |
| chr3_71354045_71354526 | 0.16 |
FOXP1 |
forkhead box P1 |
374 |
0.9 |
| chr18_53296660_53296811 | 0.16 |
TCF4 |
transcription factor 4 |
1858 |
0.49 |
| chr15_52044209_52044706 | 0.16 |
CTD-2308G16.1 |
|
225 |
0.69 |
| chr1_202428982_202429290 | 0.16 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
2735 |
0.3 |
| chr2_198243514_198243752 | 0.16 |
SF3B1 |
splicing factor 3b, subunit 1, 155kDa |
20119 |
0.14 |
| chr8_59718017_59718168 | 0.16 |
ENSG00000201231 |
. |
9019 |
0.29 |
| chr3_85442393_85442544 | 0.16 |
ENSG00000264084 |
. |
7608 |
0.34 |
| chr7_42276328_42276548 | 0.16 |
GLI3 |
GLI family zinc finger 3 |
174 |
0.98 |
| chr6_45601331_45601597 | 0.16 |
ENSG00000252738 |
. |
12377 |
0.3 |
| chr19_8387111_8387400 | 0.16 |
RPS28 |
ribosomal protein S28 |
871 |
0.33 |
| chr4_54243416_54243714 | 0.16 |
FIP1L1 |
factor interacting with PAPOLA and CPSF1 |
247 |
0.94 |
| chr7_130920122_130920273 | 0.16 |
MKLN1 |
muskelin 1, intracellular mediator containing kelch motifs |
49624 |
0.16 |
| chr7_92860979_92861490 | 0.16 |
CCDC132 |
coiled-coil domain containing 132 |
419 |
0.86 |
| chr18_29522149_29522354 | 0.15 |
RP11-326K13.4 |
|
287 |
0.76 |
| chr4_68410033_68410229 | 0.15 |
CENPC |
centromere protein C |
1193 |
0.53 |
| chr1_244893030_244893263 | 0.15 |
DESI2 |
desumoylating isopeptidase 2 |
76093 |
0.09 |
| chr4_127701440_127701591 | 0.15 |
ENSG00000199862 |
. |
247859 |
0.02 |
| chr5_50040216_50040933 | 0.15 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
77183 |
0.12 |
| chr17_8424688_8424839 | 0.15 |
ENSG00000252363 |
. |
10320 |
0.17 |
| chr1_181002765_181003143 | 0.15 |
MR1 |
major histocompatibility complex, class I-related |
113 |
0.96 |
| chr2_177029952_177030103 | 0.15 |
RP11-387A1.5 |
|
417 |
0.62 |
| chr8_23145924_23146455 | 0.15 |
R3HCC1 |
R3H domain and coiled-coil containing 1 |
254 |
0.9 |
| chr11_1909549_1909755 | 0.15 |
C11orf89 |
chromosome 11 open reading frame 89 |
2432 |
0.16 |
| chr15_38852027_38852178 | 0.15 |
RASGRP1 |
RAS guanyl releasing protein 1 (calcium and DAG-regulated) |
181 |
0.95 |
| chr7_6576295_6576446 | 0.15 |
GRID2IP |
glutamate receptor, ionotropic, delta 2 (Grid2) interacting protein |
158 |
0.94 |
| chr9_45727635_45727849 | 0.15 |
FAM27A |
family with sequence similarity 27, member A |
635 |
0.72 |
| chr17_48797268_48797811 | 0.15 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
531 |
0.69 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.2 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.1 | 0.3 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.1 | 0.2 | GO:0000212 | meiotic spindle organization(GO:0000212) |
| 0.1 | 0.2 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.1 | GO:0035385 | Roundabout signaling pathway(GO:0035385) |
| 0.0 | 0.1 | GO:0060831 | smoothened signaling pathway involved in dorsal/ventral neural tube patterning(GO:0060831) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.2 | GO:0046618 | drug export(GO:0046618) |
| 0.0 | 0.2 | GO:0045217 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0021670 | lateral ventricle development(GO:0021670) |
| 0.0 | 0.0 | GO:0010882 | regulation of cardiac muscle contraction by calcium ion signaling(GO:0010882) |
| 0.0 | 0.0 | GO:0002328 | pro-B cell differentiation(GO:0002328) |
| 0.0 | 0.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.0 | 0.1 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.3 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.1 | GO:0070682 | proteasome regulatory particle assembly(GO:0070682) |
| 0.0 | 0.1 | GO:0003032 | detection of oxygen(GO:0003032) |
| 0.0 | 0.1 | GO:0060292 | long term synaptic depression(GO:0060292) |
| 0.0 | 0.1 | GO:0043923 | positive regulation by host of viral transcription(GO:0043923) |
| 0.0 | 0.1 | GO:0043117 | positive regulation of vascular permeability(GO:0043117) |
| 0.0 | 0.1 | GO:0035588 | adenosine receptor signaling pathway(GO:0001973) G-protein coupled purinergic receptor signaling pathway(GO:0035588) |
| 0.0 | 0.1 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:1901419 | regulation of response to alcohol(GO:1901419) |
| 0.0 | 0.1 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
| 0.0 | 0.2 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.2 | GO:0023058 | desensitization of G-protein coupled receptor protein signaling pathway(GO:0002029) negative adaptation of signaling pathway(GO:0022401) adaptation of signaling pathway(GO:0023058) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.2 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0033539 | fatty acid beta-oxidation using acyl-CoA dehydrogenase(GO:0033539) |
| 0.0 | 0.2 | GO:0033523 | histone H2B ubiquitination(GO:0033523) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.1 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0015840 | urea transport(GO:0015840) |
| 0.0 | 0.1 | GO:0021535 | cell migration in hindbrain(GO:0021535) |
| 0.0 | 0.2 | GO:0032008 | positive regulation of TOR signaling(GO:0032008) |
| 0.0 | 0.1 | GO:0006863 | purine nucleobase transport(GO:0006863) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.0 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.0 | GO:2000192 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) negative regulation of anion transmembrane transport(GO:1903960) negative regulation of fatty acid transport(GO:2000192) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0050847 | progesterone receptor signaling pathway(GO:0050847) |
| 0.0 | 0.1 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.0 | GO:0015959 | diadenosine polyphosphate metabolic process(GO:0015959) |
| 0.0 | 0.1 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.1 | GO:0006670 | sphingosine metabolic process(GO:0006670) |
| 0.0 | 0.0 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.0 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.0 | 0.1 | GO:0010915 | regulation of very-low-density lipoprotein particle clearance(GO:0010915) negative regulation of very-low-density lipoprotein particle clearance(GO:0010916) |
| 0.0 | 0.0 | GO:0046502 | uroporphyrinogen III metabolic process(GO:0046502) |
| 0.0 | 0.0 | GO:0006434 | seryl-tRNA aminoacylation(GO:0006434) |
| 0.0 | 0.3 | GO:0034080 | DNA replication-independent nucleosome assembly(GO:0006336) CENP-A containing nucleosome assembly(GO:0034080) DNA replication-independent nucleosome organization(GO:0034724) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:1901532 | regulation of megakaryocyte differentiation(GO:0045652) regulation of hematopoietic progenitor cell differentiation(GO:1901532) |
| 0.0 | 0.1 | GO:0018101 | protein citrullination(GO:0018101) |
| 0.0 | 0.0 | GO:0043518 | negative regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043518) negative regulation of signal transduction by p53 class mediator(GO:1901797) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.0 | GO:0034085 | establishment of sister chromatid cohesion(GO:0034085) cohesin loading(GO:0071921) regulation of cohesin loading(GO:0071922) |
| 0.0 | 0.2 | GO:0034605 | cellular response to heat(GO:0034605) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0031498 | nucleosome disassembly(GO:0006337) chromatin disassembly(GO:0031498) protein-DNA complex disassembly(GO:0032986) |
| 0.0 | 0.1 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.0 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.0 | GO:1900116 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.0 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0009189 | deoxyribonucleoside diphosphate biosynthetic process(GO:0009189) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0090116 | DNA methylation on cytosine(GO:0032776) C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.0 | GO:0060449 | bud elongation involved in lung branching(GO:0060449) |
| 0.0 | 0.0 | GO:0006530 | asparagine catabolic process(GO:0006530) asparagine catabolic process via L-aspartate(GO:0033345) |
| 0.0 | 0.1 | GO:0045909 | positive regulation of vasodilation(GO:0045909) |
| 0.0 | 0.1 | GO:0008655 | pyrimidine-containing compound salvage(GO:0008655) pyrimidine nucleoside salvage(GO:0043097) |
| 0.0 | 0.1 | GO:0001660 | fever generation(GO:0001660) |
| 0.0 | 0.0 | GO:0046642 | negative regulation of alpha-beta T cell proliferation(GO:0046642) |
| 0.0 | 0.1 | GO:0006734 | NADH metabolic process(GO:0006734) |
| 0.0 | 0.1 | GO:0006743 | ubiquinone metabolic process(GO:0006743) ubiquinone biosynthetic process(GO:0006744) |
| 0.0 | 0.1 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.2 | GO:0070206 | protein trimerization(GO:0070206) |
| 0.0 | 0.0 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.2 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0031305 | integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.0 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.1 | GO:0008540 | proteasome regulatory particle, base subcomplex(GO:0008540) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0000791 | euchromatin(GO:0000791) |
| 0.0 | 0.1 | GO:0045240 | dihydrolipoyl dehydrogenase complex(GO:0045240) |
| 0.0 | 0.0 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.5 | GO:0005626 | obsolete insoluble fraction(GO:0005626) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.1 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.1 | GO:0005678 | obsolete chromatin assembly complex(GO:0005678) |
| 0.0 | 0.1 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0035145 | exon-exon junction complex(GO:0035145) |
| 0.0 | 0.1 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.0 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.0 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.0 | GO:0005851 | eukaryotic translation initiation factor 2B complex(GO:0005851) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.0 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001099 | basal transcription machinery binding(GO:0001098) basal RNA polymerase II transcription machinery binding(GO:0001099) |
| 0.1 | 0.3 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0008545 | JUN kinase kinase activity(GO:0008545) |
| 0.1 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) |
| 0.0 | 0.2 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0015204 | urea transmembrane transporter activity(GO:0015204) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0015520 | tetracycline:proton antiporter activity(GO:0015520) |
| 0.0 | 0.1 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.4 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0016723 | oxidoreductase activity, oxidizing metal ions, NAD or NADP as acceptor(GO:0016723) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.1 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) adenyl deoxyribonucleotide binding(GO:0032558) |
| 0.0 | 0.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.2 | GO:0004176 | ATP-dependent peptidase activity(GO:0004176) |
| 0.0 | 0.2 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.1 | GO:0005131 | growth hormone receptor binding(GO:0005131) |
| 0.0 | 0.2 | GO:0004690 | cyclic nucleotide-dependent protein kinase activity(GO:0004690) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.0 | 0.1 | GO:0048039 | ubiquinone binding(GO:0048039) |
| 0.0 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.0 | 0.1 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.1 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.1 | GO:0050656 | 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.0 | 0.1 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.0 | 0.2 | GO:0046030 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.1 | GO:0004551 | nucleotide diphosphatase activity(GO:0004551) |
| 0.0 | 0.1 | GO:0005346 | purine ribonucleotide transmembrane transporter activity(GO:0005346) |
| 0.0 | 0.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0004583 | dolichyl-phosphate-glucose-glycolipid alpha-glucosyltransferase activity(GO:0004583) |
| 0.0 | 0.0 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 0.0 | GO:0004828 | serine-tRNA ligase activity(GO:0004828) |
| 0.0 | 0.2 | GO:0060590 | ATPase regulator activity(GO:0060590) |
| 0.0 | 0.1 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.0 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0061630 | ubiquitin-ubiquitin ligase activity(GO:0034450) ubiquitin protein ligase activity(GO:0061630) ubiquitin-like protein ligase activity(GO:0061659) |
| 0.0 | 0.2 | GO:0070491 | repressing transcription factor binding(GO:0070491) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.1 | GO:0033558 | protein deacetylase activity(GO:0033558) |
| 0.0 | 0.0 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.1 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.0 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0032393 | MHC class I receptor activity(GO:0032393) |
| 0.0 | 0.0 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.1 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.2 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.2 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.1 | GO:0004415 | hyalurononglucosaminidase activity(GO:0004415) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0003711 | obsolete transcription elongation regulator activity(GO:0003711) |
| 0.0 | 0.0 | GO:0070492 | oligosaccharide binding(GO:0070492) |
| 0.0 | 0.0 | GO:0000739 | obsolete DNA strand annealing activity(GO:0000739) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.4 | ST T CELL SIGNAL TRANSDUCTION | T Cell Signal Transduction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.6 | REACTOME DARPP 32 EVENTS | Genes involved in DARPP-32 events |
| 0.0 | 0.3 | REACTOME RAP1 SIGNALLING | Genes involved in Rap1 signalling |
| 0.0 | 0.1 | REACTOME ADP SIGNALLING THROUGH P2RY1 | Genes involved in ADP signalling through P2Y purinoceptor 1 |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.3 | REACTOME INTEGRIN ALPHAIIB BETA3 SIGNALING | Genes involved in Integrin alphaIIb beta3 signaling |
| 0.0 | 0.2 | REACTOME ORGANIC CATION ANION ZWITTERION TRANSPORT | Genes involved in Organic cation/anion/zwitterion transport |
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.0 | 0.1 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.0 | REACTOME FORMATION OF TUBULIN FOLDING INTERMEDIATES BY CCT TRIC | Genes involved in Formation of tubulin folding intermediates by CCT/TriC |
| 0.0 | 0.2 | REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | Genes involved in Transport to the Golgi and subsequent modification |
| 0.0 | 0.1 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.0 | REACTOME DAG AND IP3 SIGNALING | Genes involved in DAG and IP3 signaling |
| 0.0 | 0.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.3 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.0 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |