Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
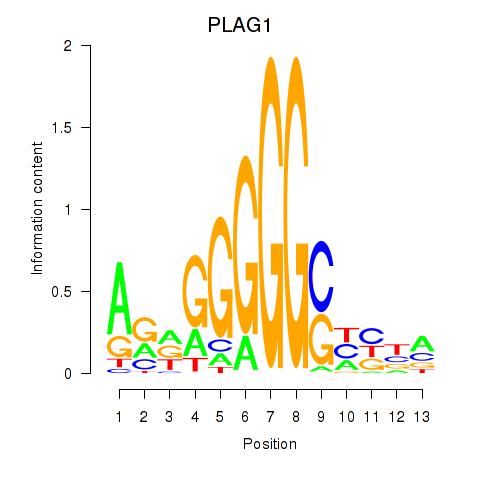
Results for PLAG1
Z-value: 0.51
Transcription factors associated with PLAG1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PLAG1
|
ENSG00000181690.3 | PLAG1 zinc finger |
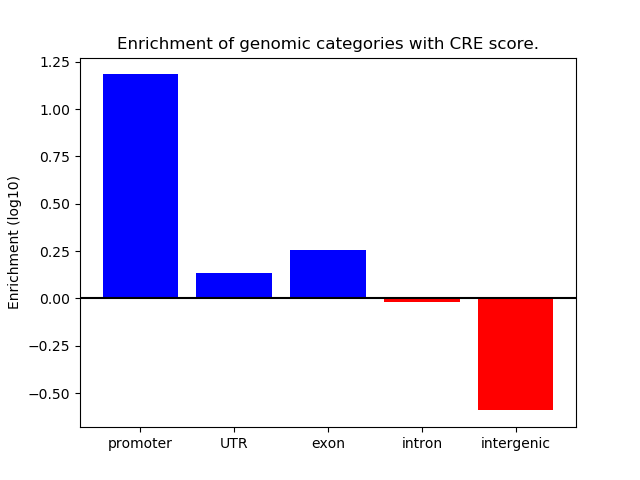
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr8_57086542_57086822 | PLAG1 | 37156 | 0.121655 | 0.59 | 9.3e-02 | Click! |
| chr8_57122306_57122457 | PLAG1 | 1457 | 0.353600 | 0.57 | 1.1e-01 | Click! |
| chr8_57123539_57123769 | PLAG1 | 184 | 0.830733 | -0.52 | 1.5e-01 | Click! |
| chr8_57086200_57086351 | PLAG1 | 37563 | 0.120665 | 0.51 | 1.6e-01 | Click! |
| chr8_57122958_57123494 | PLAG1 | 612 | 0.588802 | -0.41 | 2.7e-01 | Click! |
Activity of the PLAG1 motif across conditions
Conditions sorted by the z-value of the PLAG1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
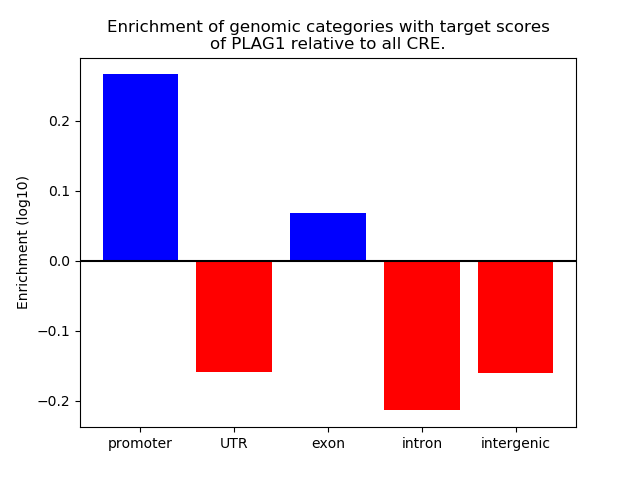
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chrX_54947220_54948504 | 0.29 |
TRO |
trophinin |
41 |
0.97 |
| chr14_105436556_105437076 | 0.21 |
AHNAK2 |
AHNAK nucleoprotein 2 |
7878 |
0.16 |
| chr2_74666737_74667617 | 0.19 |
RTKN |
rhotekin |
533 |
0.54 |
| chr5_131593477_131594185 | 0.19 |
PDLIM4 |
PDZ and LIM domain 4 |
431 |
0.81 |
| chr20_55204347_55205219 | 0.19 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
425 |
0.81 |
| chr7_100464814_100466140 | 0.18 |
TRIP6 |
thyroid hormone receptor interactor 6 |
717 |
0.47 |
| chr16_4985840_4986055 | 0.18 |
PPL |
periplakin |
1118 |
0.43 |
| chr1_156073604_156073917 | 0.16 |
LMNA |
lamin A/C |
10701 |
0.1 |
| chr17_7308835_7309309 | 0.16 |
NLGN2 |
neuroligin 2 |
879 |
0.24 |
| chr18_10454899_10455524 | 0.15 |
APCDD1 |
adenomatosis polyposis coli down-regulated 1 |
232 |
0.95 |
| chr3_194494197_194494348 | 0.15 |
FAM43A |
family with sequence similarity 43, member A |
87650 |
0.07 |
| chr17_70116228_70116580 | 0.14 |
SOX9 |
SRY (sex determining region Y)-box 9 |
757 |
0.79 |
| chr11_62312562_62314176 | 0.14 |
RP11-864I4.4 |
|
102 |
0.64 |
| chr22_50783104_50783895 | 0.14 |
PPP6R2 |
protein phosphatase 6, regulatory subunit 2 |
1739 |
0.21 |
| chr6_44424385_44424803 | 0.14 |
ENSG00000266619 |
. |
21216 |
0.17 |
| chr4_186560086_186560237 | 0.14 |
SORBS2 |
sorbin and SH3 domain containing 2 |
10599 |
0.18 |
| chr17_36861833_36863162 | 0.13 |
CTB-58E17.3 |
|
17 |
0.9 |
| chr3_24562978_24563646 | 0.13 |
ENSG00000265028 |
. |
386 |
0.87 |
| chr14_79745937_79746359 | 0.13 |
NRXN3 |
neurexin 3 |
101 |
0.98 |
| chr3_64430574_64431219 | 0.12 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
162 |
0.97 |
| chr2_189156294_189156511 | 0.12 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
6 |
0.98 |
| chr17_59476045_59476898 | 0.12 |
RP11-332H18.4 |
|
260 |
0.74 |
| chr2_218843405_218844030 | 0.12 |
TNS1 |
tensin 1 |
60 |
0.79 |
| chr6_34205466_34206794 | 0.12 |
HMGA1 |
high mobility group AT-hook 1 |
438 |
0.85 |
| chr2_85505218_85505417 | 0.12 |
ENSG00000221579 |
. |
20840 |
0.12 |
| chr16_3162631_3163041 | 0.12 |
ZNF205 |
zinc finger protein 205 |
40 |
0.8 |
| chr10_135148565_135149526 | 0.11 |
CALY |
calcyon neuron-specific vesicular protein |
1331 |
0.22 |
| chr11_118478413_118479735 | 0.11 |
PHLDB1 |
pleckstrin homology-like domain, family B, member 1 |
716 |
0.54 |
| chr9_130330127_130331202 | 0.11 |
FAM129B |
family with sequence similarity 129, member B |
703 |
0.67 |
| chr19_6740138_6741070 | 0.11 |
TRIP10 |
thyroid hormone receptor interactor 10 |
325 |
0.81 |
| chr3_49237110_49237583 | 0.11 |
CCDC36 |
coiled-coil domain containing 36 |
403 |
0.74 |
| chr11_131780761_131781452 | 0.10 |
NTM |
neurotrimin |
209 |
0.95 |
| chr6_129203821_129204789 | 0.10 |
LAMA2 |
laminin, alpha 2 |
37 |
0.99 |
| chrX_37430984_37431427 | 0.10 |
LANCL3 |
LanC lantibiotic synthetase component C-like 3 (bacterial) |
300 |
0.94 |
| chr1_35246823_35247104 | 0.10 |
GJB3 |
gap junction protein, beta 3, 31kDa |
173 |
0.91 |
| chr7_70159815_70160908 | 0.10 |
AUTS2 |
autism susceptibility candidate 2 |
33764 |
0.25 |
| chrX_110038687_110039773 | 0.10 |
CHRDL1 |
chordin-like 1 |
56 |
0.99 |
| chr7_555957_556150 | 0.10 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
2092 |
0.33 |
| chr17_76879212_76879961 | 0.10 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
9354 |
0.14 |
| chr7_95401616_95402462 | 0.10 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
123 |
0.98 |
| chr17_41622177_41622554 | 0.10 |
RP11-392O1.4 |
|
212 |
0.81 |
| chrX_68348217_68348929 | 0.10 |
PJA1 |
praja ring finger 1, E3 ubiquitin protein ligase |
33964 |
0.24 |
| chr20_51589535_51589784 | 0.10 |
TSHZ2 |
teashirt zinc finger homeobox 2 |
713 |
0.79 |
| chr8_145020311_145020609 | 0.10 |
PLEC |
plectin |
113 |
0.91 |
| chr5_139283809_139284341 | 0.10 |
NRG2 |
neuregulin 2 |
93 |
0.98 |
| chr15_61520624_61520983 | 0.10 |
RORA |
RAR-related orphan receptor A |
715 |
0.73 |
| chr15_80544489_80544852 | 0.10 |
FAH |
fumarylacetoacetate hydrolase (fumarylacetoacetase) |
96377 |
0.07 |
| chr1_3816010_3816785 | 0.10 |
C1orf174 |
chromosome 1 open reading frame 174 |
452 |
0.8 |
| chr2_1711742_1712354 | 0.09 |
PXDN |
peroxidasin homolog (Drosophila) |
15706 |
0.26 |
| chr1_34629900_34630098 | 0.09 |
CSMD2 |
CUB and Sushi multiple domains 2 |
876 |
0.55 |
| chr6_138427325_138427625 | 0.09 |
PERP |
PERP, TP53 apoptosis effector |
1173 |
0.59 |
| chr6_13924445_13925088 | 0.09 |
RNF182 |
ring finger protein 182 |
89 |
0.98 |
| chr1_199728481_199728632 | 0.09 |
ENSG00000200139 |
. |
128731 |
0.05 |
| chr6_146755289_146755690 | 0.09 |
ENSG00000222971 |
. |
93933 |
0.09 |
| chr14_101034010_101035263 | 0.09 |
BEGAIN |
brain-enriched guanylate kinase-associated |
192 |
0.93 |
| chr9_13568681_13569117 | 0.09 |
MPDZ |
multiple PDZ domain protein |
289310 |
0.01 |
| chr1_39598517_39598955 | 0.09 |
ENSG00000206654 |
. |
12021 |
0.17 |
| chr7_2842191_2842738 | 0.09 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
12428 |
0.25 |
| chr18_5629293_5629838 | 0.09 |
ENSG00000252432 |
. |
32484 |
0.15 |
| chr17_79317268_79317735 | 0.09 |
TMEM105 |
transmembrane protein 105 |
13027 |
0.13 |
| chr11_93063544_93063792 | 0.09 |
CCDC67 |
coiled-coil domain containing 67 |
143 |
0.97 |
| chr18_53253101_53253291 | 0.09 |
TCF4 |
transcription factor 4 |
1 |
0.99 |
| chr5_1110808_1112337 | 0.09 |
SLC12A7 |
solute carrier family 12 (potassium/chloride transporter), member 7 |
578 |
0.75 |
| chrX_153687680_153687951 | 0.09 |
PLXNA3 |
plexin A3 |
1194 |
0.23 |
| chr2_71127433_71128165 | 0.08 |
VAX2 |
ventral anterior homeobox 2 |
79 |
0.96 |
| chr17_1018223_1018374 | 0.08 |
ABR |
active BCR-related |
378 |
0.88 |
| chr1_27321840_27321991 | 0.08 |
TRNP1 |
TMF1-regulated nuclear protein 1 |
1105 |
0.44 |
| chr10_6780355_6780830 | 0.08 |
PRKCQ |
protein kinase C, theta |
158329 |
0.04 |
| chr16_576320_577633 | 0.08 |
CAPN15 |
calpain 15 |
741 |
0.43 |
| chr20_55205529_55205819 | 0.08 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
264 |
0.9 |
| chr3_50194015_50194506 | 0.08 |
RP11-493K19.3 |
|
742 |
0.47 |
| chr16_86542600_86543872 | 0.08 |
FOXF1 |
forkhead box F1 |
897 |
0.62 |
| chr2_235353840_235353991 | 0.08 |
ARL4C |
ADP-ribosylation factor-like 4C |
51329 |
0.19 |
| chrX_135230110_135230418 | 0.08 |
FHL1 |
four and a half LIM domains 1 |
6 |
0.98 |
| chrX_114796055_114796359 | 0.08 |
PLS3 |
plastin 3 |
625 |
0.52 |
| chrX_56258780_56259711 | 0.08 |
KLF8 |
Kruppel-like factor 8 |
235 |
0.96 |
| chr2_220492846_220493097 | 0.08 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
588 |
0.57 |
| chr7_1497923_1498578 | 0.08 |
MICALL2 |
MICAL-like 2 |
712 |
0.54 |
| chr5_155107689_155108219 | 0.08 |
ENSG00000200275 |
. |
164493 |
0.04 |
| chrX_48930794_48931951 | 0.08 |
PRAF2 |
PRA1 domain family, member 2 |
276 |
0.78 |
| chr7_45128395_45129126 | 0.08 |
NACAD |
NAC alpha domain containing |
247 |
0.87 |
| chr3_52566781_52567334 | 0.08 |
NT5DC2 |
5'-nucleotidase domain containing 2 |
490 |
0.67 |
| chr17_34998121_34998780 | 0.08 |
MRM1 |
mitochondrial rRNA methyltransferase 1 homolog (S. cerevisiae) |
40059 |
0.13 |
| chr9_132936080_132936268 | 0.08 |
NCS1 |
neuronal calcium sensor 1 |
1317 |
0.47 |
| chr15_41795749_41795900 | 0.08 |
ITPKA |
inositol-trisphosphate 3-kinase A |
9751 |
0.13 |
| chr11_63530349_63531105 | 0.08 |
ENSG00000264519 |
. |
34816 |
0.12 |
| chr22_50744615_50744936 | 0.08 |
PLXNB2 |
plexin B2 |
1242 |
0.27 |
| chr19_39528811_39528970 | 0.07 |
FBXO27 |
F-box protein 27 |
5465 |
0.12 |
| chrX_122317435_122317817 | 0.07 |
GRIA3 |
glutamate receptor, ionotropic, AMPA 3 |
380 |
0.93 |
| chr8_125739093_125740062 | 0.07 |
MTSS1 |
metastasis suppressor 1 |
280 |
0.94 |
| chr12_127939879_127940274 | 0.07 |
ENSG00000253089 |
. |
134176 |
0.05 |
| chr1_34631046_34631197 | 0.07 |
CSMD2 |
CUB and Sushi multiple domains 2 |
246 |
0.89 |
| chr17_34611752_34612896 | 0.07 |
CCL3L1 |
chemokine (C-C motif) ligand 3-like 1 |
13395 |
0.14 |
| chr13_113840712_113840863 | 0.07 |
RP11-98F14.11 |
|
21290 |
0.11 |
| chr8_68896954_68897148 | 0.07 |
PREX2 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
32698 |
0.23 |
| chr19_13129320_13129532 | 0.07 |
CTC-239J10.1 |
|
4101 |
0.11 |
| chr8_99439239_99439631 | 0.07 |
KCNS2 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 |
185 |
0.95 |
| chr9_140785598_140785749 | 0.07 |
RP11-188C12.3 |
|
1349 |
0.47 |
| chr16_31076512_31076791 | 0.07 |
AC135050.5 |
|
165 |
0.57 |
| chr19_10341031_10341948 | 0.07 |
ENSG00000264266 |
. |
400 |
0.34 |
| chr10_100027380_100027945 | 0.07 |
LOXL4 |
lysyl oxidase-like 4 |
345 |
0.9 |
| chr4_153274070_153274464 | 0.07 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
144 |
0.97 |
| chr11_66102741_66104078 | 0.07 |
RIN1 |
Ras and Rab interactor 1 |
468 |
0.53 |
| chr5_139126785_139127680 | 0.07 |
ENSG00000200756 |
. |
29557 |
0.16 |
| chr7_158608170_158608321 | 0.07 |
ESYT2 |
extended synaptotagmin-like protein 2 |
13918 |
0.24 |
| chr1_196577305_196577474 | 0.07 |
KCNT2 |
potassium channel, subfamily T, member 2 |
100 |
0.97 |
| chr4_21949961_21950371 | 0.07 |
KCNIP4 |
Kv channel interacting protein 4 |
202 |
0.77 |
| chr7_150759622_150760148 | 0.07 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
164 |
0.88 |
| chr6_28511380_28512171 | 0.07 |
GPX5 |
glutathione peroxidase 5 (epididymal androgen-related protein) |
17986 |
0.16 |
| chr6_113966553_113966868 | 0.07 |
ENSG00000266650 |
. |
42593 |
0.17 |
| chr3_79815928_79816150 | 0.07 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
926 |
0.74 |
| chr8_22224818_22225894 | 0.07 |
SLC39A14 |
solute carrier family 39 (zinc transporter), member 14 |
305 |
0.9 |
| chr3_173302379_173302636 | 0.07 |
NLGN1 |
neuroligin 1 |
162 |
0.98 |
| chr7_44185024_44185674 | 0.07 |
GCK |
glucokinase (hexokinase 4) |
394 |
0.74 |
| chr1_94091019_94091233 | 0.07 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
11472 |
0.22 |
| chr7_73868670_73869167 | 0.07 |
GTF2IRD1 |
GTF2I repeat domain containing 1 |
479 |
0.83 |
| chrX_107018688_107019917 | 0.07 |
TSC22D3 |
TSC22 domain family, member 3 |
43 |
0.97 |
| chr6_110679391_110679830 | 0.07 |
METTL24 |
methyltransferase like 24 |
135 |
0.97 |
| chr12_3308931_3310293 | 0.07 |
TSPAN9 |
tetraspanin 9 |
729 |
0.75 |
| chr6_154800458_154800845 | 0.07 |
CNKSR3 |
CNKSR family member 3 |
1026 |
0.69 |
| chr5_151065179_151065606 | 0.07 |
SPARC |
secreted protein, acidic, cysteine-rich (osteonectin) |
601 |
0.67 |
| chr8_94712271_94712761 | 0.07 |
FAM92A1 |
family with sequence similarity 92, member A1 |
36 |
0.63 |
| chr11_126872809_126873319 | 0.07 |
KIRREL3-AS3 |
KIRREL3 antisense RNA 3 |
259 |
0.89 |
| chr19_3097315_3097524 | 0.07 |
GNA11 |
guanine nucleotide binding protein (G protein), alpha 11 (Gq class) |
3011 |
0.15 |
| chr1_180881914_180882730 | 0.07 |
KIAA1614 |
KIAA1614 |
3 |
0.98 |
| chr2_230845561_230845747 | 0.07 |
ENSG00000206725 |
. |
7623 |
0.17 |
| chrX_51811538_51811884 | 0.07 |
MAGED4B |
melanoma antigen family D, 4B |
557 |
0.75 |
| chr1_16470072_16470223 | 0.07 |
RP11-276H7.2 |
|
11559 |
0.12 |
| chr14_50809347_50809713 | 0.07 |
CDKL1 |
cyclin-dependent kinase-like 1 (CDC2-related kinase) |
1683 |
0.33 |
| chr2_113997514_113997878 | 0.07 |
RP11-65I12.1 |
|
611 |
0.6 |
| chr11_128760172_128761028 | 0.07 |
KCNJ5 |
potassium inwardly-rectifying channel, subfamily J, member 5 |
651 |
0.68 |
| chr19_1355185_1356018 | 0.07 |
MUM1 |
melanoma associated antigen (mutated) 1 |
625 |
0.53 |
| chr12_3069053_3070004 | 0.07 |
TEAD4 |
TEA domain family member 4 |
445 |
0.77 |
| chr9_91793130_91793737 | 0.07 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
249 |
0.95 |
| chr5_167197787_167197938 | 0.07 |
TENM2 |
teneurin transmembrane protein 2 |
15854 |
0.23 |
| chr22_43115644_43116713 | 0.07 |
A4GALT |
alpha 1,4-galactosyltransferase |
24539 |
0.15 |
| chr19_51161641_51162497 | 0.07 |
C19orf81 |
chromosome 19 open reading frame 81 |
9024 |
0.12 |
| chr16_15529020_15529579 | 0.07 |
C16orf45 |
chromosome 16 open reading frame 45 |
967 |
0.56 |
| chr21_46875019_46875466 | 0.07 |
COL18A1 |
collagen, type XVIII, alpha 1 |
161 |
0.95 |
| chr3_52016673_52017699 | 0.07 |
ABHD14B |
abhydrolase domain containing 14B |
239 |
0.49 |
| chr19_45754620_45755932 | 0.06 |
MARK4 |
MAP/microtubule affinity-regulating kinase 4 |
726 |
0.57 |
| chr22_25348691_25349626 | 0.06 |
KIAA1671 |
KIAA1671 |
461 |
0.83 |
| chr17_42620836_42621052 | 0.06 |
FZD2 |
frizzled family receptor 2 |
13981 |
0.17 |
| chr17_48074174_48074657 | 0.06 |
DLX3 |
distal-less homeobox 3 |
1827 |
0.28 |
| chr14_31733126_31733307 | 0.06 |
ENSG00000199291 |
. |
5301 |
0.16 |
| chr1_215256670_215256888 | 0.06 |
KCNK2 |
potassium channel, subfamily K, member 2 |
81 |
0.99 |
| chr16_88850139_88851147 | 0.06 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
976 |
0.35 |
| chrX_135258076_135258227 | 0.06 |
FHL1 |
four and a half LIM domains 1 |
6066 |
0.24 |
| chr15_96865528_96865838 | 0.06 |
NR2F2-AS1 |
NR2F2 antisense RNA 1 |
191 |
0.93 |
| chr21_28214648_28215240 | 0.06 |
ADAMTS1 |
ADAM metallopeptidase with thrombospondin type 1 motif, 1 |
393 |
0.9 |
| chr18_48086527_48087582 | 0.06 |
MAPK4 |
mitogen-activated protein kinase 4 |
42 |
0.99 |
| chr10_102759781_102760726 | 0.06 |
LZTS2 |
leucine zipper, putative tumor suppressor 2 |
643 |
0.52 |
| chr14_101543718_101544133 | 0.06 |
ENSG00000207959 |
. |
10864 |
0.02 |
| chr2_218874842_218875310 | 0.06 |
TNS1 |
tensin 1 |
7358 |
0.19 |
| chr10_64564066_64565253 | 0.06 |
ADO |
2-aminoethanethiol (cysteamine) dioxygenase |
143 |
0.92 |
| chr17_47075525_47076053 | 0.06 |
IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
681 |
0.41 |
| chr3_125690962_125691349 | 0.06 |
ROPN1B |
rhophilin associated tail protein 1B |
3098 |
0.23 |
| chr5_79252602_79252869 | 0.06 |
RP11-168A11.1 |
|
20333 |
0.2 |
| chr22_29703700_29703885 | 0.06 |
GAS2L1 |
growth arrest-specific 2 like 1 |
744 |
0.49 |
| chr12_81471441_81472090 | 0.06 |
ACSS3 |
acyl-CoA synthetase short-chain family member 3 |
71 |
0.98 |
| chr17_43483002_43483407 | 0.06 |
ARHGAP27 |
Rho GTPase activating protein 27 |
96 |
0.95 |
| chr10_33622786_33623487 | 0.06 |
NRP1 |
neuropilin 1 |
174 |
0.97 |
| chr17_21279729_21280949 | 0.06 |
KCNJ12 |
potassium inwardly-rectifying channel, subfamily J, member 12 |
830 |
0.69 |
| chr16_29690426_29691035 | 0.06 |
QPRT |
quinolinate phosphoribosyltransferase |
227 |
0.89 |
| chr16_55542879_55544136 | 0.06 |
LPCAT2 |
lysophosphatidylcholine acyltransferase 2 |
597 |
0.77 |
| chr7_2678463_2678614 | 0.06 |
TTYH3 |
tweety family member 3 |
6628 |
0.2 |
| chr14_77964628_77965267 | 0.06 |
ISM2 |
isthmin 2 |
212 |
0.91 |
| chr2_119592973_119593692 | 0.06 |
EN1 |
engrailed homeobox 1 |
11922 |
0.28 |
| chr19_37743129_37743340 | 0.06 |
ZNF383 |
zinc finger protein 383 |
24782 |
0.13 |
| chr1_113265808_113265959 | 0.06 |
FAM19A3 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A3 |
2684 |
0.16 |
| chr6_44264722_44265466 | 0.06 |
TCTE1 |
t-complex-associated-testis-expressed 1 |
331 |
0.81 |
| chr12_116892640_116892791 | 0.06 |
ENSG00000264037 |
. |
26592 |
0.24 |
| chr5_141744007_141744207 | 0.06 |
AC005592.2 |
|
10999 |
0.25 |
| chr19_41104181_41104507 | 0.06 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
1203 |
0.38 |
| chr17_26732645_26733444 | 0.06 |
SLC46A1 |
solute carrier family 46 (folate transporter), member 1 |
184 |
0.8 |
| chr17_79989895_79990665 | 0.06 |
RAC3 |
ras-related C3 botulinum toxin substrate 3 (rho family, small GTP binding protein Rac3) |
220 |
0.81 |
| chr11_2230168_2230319 | 0.06 |
ENSG00000265258 |
. |
35950 |
0.09 |
| chr22_46299339_46299556 | 0.06 |
WNT7B |
wingless-type MMTV integration site family, member 7B |
69148 |
0.09 |
| chr20_62600161_62600928 | 0.06 |
ZNF512B |
zinc finger protein 512B |
674 |
0.45 |
| chr16_4422135_4423224 | 0.06 |
VASN |
vasorin |
830 |
0.49 |
| chr17_34059454_34059687 | 0.06 |
RASL10B |
RAS-like, family 10, member B |
902 |
0.43 |
| chr9_128729332_128729483 | 0.06 |
PBX3 |
pre-B-cell leukemia homeobox 3 |
101857 |
0.08 |
| chr16_85203170_85204140 | 0.06 |
CTC-786C10.1 |
|
1227 |
0.55 |
| chr10_112258642_112259418 | 0.06 |
DUSP5 |
dual specificity phosphatase 5 |
1434 |
0.41 |
| chr7_42533422_42533764 | 0.06 |
GLI3 |
GLI family zinc finger 3 |
256935 |
0.02 |
| chr6_43243068_43244292 | 0.06 |
SLC22A7 |
solute carrier family 22 (organic anion transporter), member 7 |
22056 |
0.11 |
| chr11_68451782_68452060 | 0.06 |
GAL |
galanin/GMAP prepropeptide |
22 |
0.98 |
| chr2_162931613_162931764 | 0.06 |
DPP4 |
dipeptidyl-peptidase 4 |
636 |
0.69 |
| chr12_50975264_50975415 | 0.06 |
ENSG00000207136 |
. |
51717 |
0.12 |
| chr16_68679865_68680079 | 0.06 |
RP11-615I2.2 |
|
99 |
0.91 |
| chr3_170469573_170469724 | 0.06 |
ENSG00000222411 |
. |
90313 |
0.08 |
| chr6_150740165_150740632 | 0.06 |
IYD |
iodotyrosine deiodinase |
49285 |
0.16 |
| chr21_38352575_38353327 | 0.06 |
HLCS |
holocarboxylase synthetase (biotin-(proprionyl-CoA-carboxylase (ATP-hydrolysing)) ligase) |
23 |
0.97 |
| chr7_51383611_51384133 | 0.06 |
COBL |
cordon-bleu WH2 repeat protein |
296 |
0.95 |
| chr3_85007676_85007999 | 0.06 |
CADM2 |
cell adhesion molecule 2 |
295 |
0.94 |
| chr2_213401284_213401530 | 0.06 |
ERBB4 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 4 |
1846 |
0.51 |
| chr9_126761184_126761335 | 0.06 |
LHX2 |
LIM homeobox 2 |
2690 |
0.23 |
| chr11_106888657_106888832 | 0.06 |
GUCY1A2 |
guanylate cyclase 1, soluble, alpha 2 |
133 |
0.98 |
| chr19_41725635_41725911 | 0.06 |
AXL |
AXL receptor tyrosine kinase |
633 |
0.58 |
| chr14_105434049_105434298 | 0.06 |
AHNAK2 |
AHNAK nucleoprotein 2 |
10521 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0060737 | prostate gland morphogenetic growth(GO:0060737) |
| 0.0 | 0.1 | GO:0045767 | obsolete regulation of anti-apoptosis(GO:0045767) |
| 0.0 | 0.1 | GO:0010511 | regulation of phosphatidylinositol biosynthetic process(GO:0010511) |
| 0.0 | 0.1 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.1 | GO:0036072 | intramembranous ossification(GO:0001957) direct ossification(GO:0036072) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0061299 | retina vasculature development in camera-type eye(GO:0061298) retina vasculature morphogenesis in camera-type eye(GO:0061299) |
| 0.0 | 0.0 | GO:0072182 | nephron tubule epithelial cell differentiation(GO:0072160) regulation of nephron tubule epithelial cell differentiation(GO:0072182) regulation of epithelial cell differentiation involved in kidney development(GO:2000696) |
| 0.0 | 0.1 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0072217 | negative regulation of metanephros development(GO:0072217) |
| 0.0 | 0.0 | GO:0060463 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.1 | GO:0033600 | negative regulation of mammary gland epithelial cell proliferation(GO:0033600) |
| 0.0 | 0.0 | GO:0010716 | regulation of extracellular matrix disassembly(GO:0010715) negative regulation of extracellular matrix disassembly(GO:0010716) regulation of extracellular matrix organization(GO:1903053) negative regulation of extracellular matrix organization(GO:1903054) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0021869 | forebrain ventricular zone progenitor cell division(GO:0021869) |
| 0.0 | 0.1 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.0 | 0.1 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.0 | 0.1 | GO:0010757 | negative regulation of plasminogen activation(GO:0010757) |
| 0.0 | 0.1 | GO:0022010 | central nervous system myelination(GO:0022010) axon ensheathment in central nervous system(GO:0032291) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.1 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.0 | 0.4 | GO:0007566 | embryo implantation(GO:0007566) |
| 0.0 | 0.1 | GO:0051594 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0003181 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.0 | GO:0030859 | polarized epithelial cell differentiation(GO:0030859) |
| 0.0 | 0.0 | GO:0032656 | regulation of interleukin-13 production(GO:0032656) |
| 0.0 | 0.1 | GO:1901890 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.0 | GO:0072070 | loop of Henle development(GO:0072070) |
| 0.0 | 0.2 | GO:0030252 | growth hormone secretion(GO:0030252) |
| 0.0 | 0.0 | GO:0060080 | inhibitory postsynaptic potential(GO:0060080) |
| 0.0 | 0.0 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0045197 | establishment or maintenance of epithelial cell apical/basal polarity(GO:0045197) |
| 0.0 | 0.1 | GO:0050435 | beta-amyloid metabolic process(GO:0050435) |
| 0.0 | 0.0 | GO:0045112 | integrin biosynthetic process(GO:0045112) |
| 0.0 | 0.0 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.0 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 0.0 | GO:0051590 | positive regulation of neurotransmitter transport(GO:0051590) |
| 0.0 | 0.0 | GO:0006591 | ornithine metabolic process(GO:0006591) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0045683 | negative regulation of epidermis development(GO:0045683) |
| 0.0 | 0.0 | GO:0072526 | pyridine-containing compound catabolic process(GO:0072526) |
| 0.0 | 0.1 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.0 | GO:0061004 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0051927 | obsolete negative regulation of calcium ion transport via voltage-gated calcium channel activity(GO:0051927) |
| 0.0 | 0.1 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.1 | GO:0042983 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0005606 | laminin-1 complex(GO:0005606) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0001527 | microfibril(GO:0001527) |
| 0.0 | 0.1 | GO:0005652 | nuclear lamina(GO:0005652) |
| 0.0 | 0.0 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.0 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.0 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001159 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0005021 | vascular endothelial growth factor-activated receptor activity(GO:0005021) |
| 0.0 | 0.1 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0009374 | biotin binding(GO:0009374) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.0 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.0 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.0 | GO:0042978 | ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.1 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.0 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.0 | 0.0 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.0 | GO:0050543 | icosatetraenoic acid binding(GO:0050543) |
| 0.0 | 0.0 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0070215 | obsolete MDM2 binding(GO:0070215) |
| 0.0 | 0.2 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0050431 | transforming growth factor beta binding(GO:0050431) |
| 0.0 | 0.0 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.0 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0043495 | protein anchor(GO:0043495) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.0 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0017147 | Wnt-protein binding(GO:0017147) |
| 0.0 | 0.1 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.1 | REACTOME SIGNALLING TO P38 VIA RIT AND RIN | Genes involved in Signalling to p38 via RIT and RIN |
| 0.0 | 0.1 | REACTOME INTEGRATION OF PROVIRUS | Genes involved in Integration of provirus |
| 0.0 | 0.0 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |