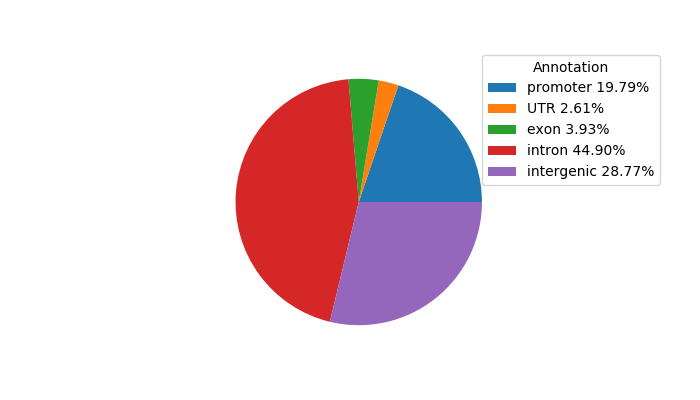
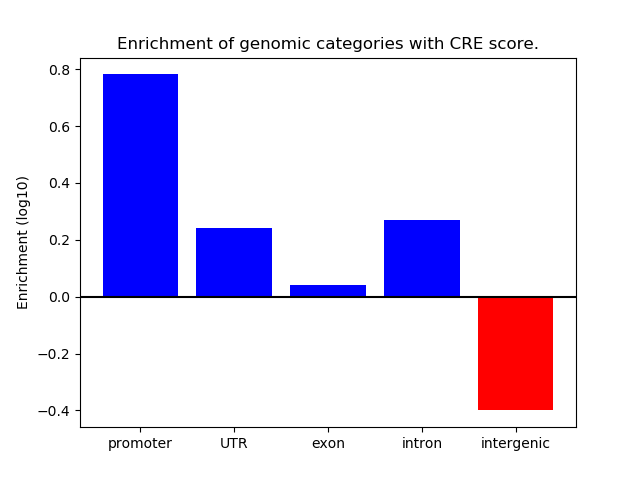
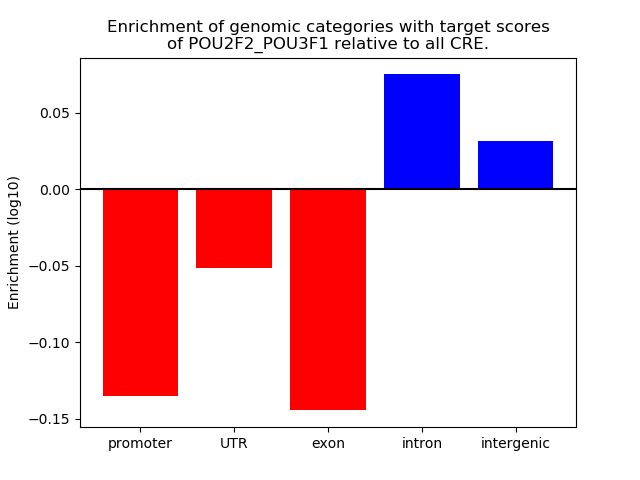
Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
Results for POU2F2_POU3F1
Z-value: 0.74
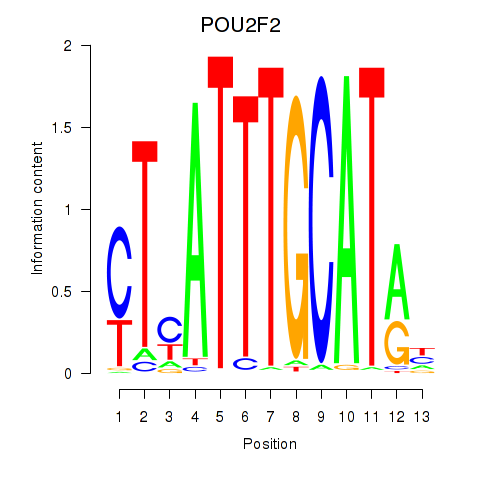
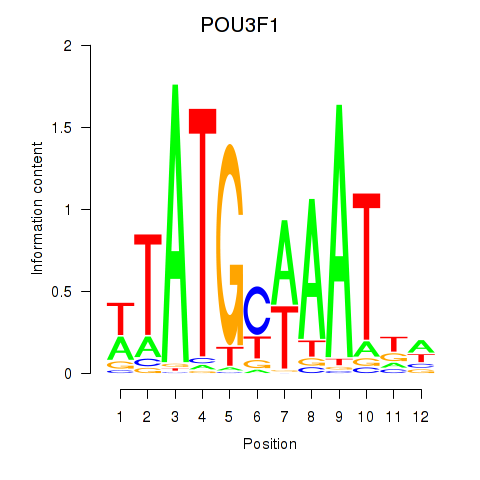
Transcription factors associated with POU2F2_POU3F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU2F2
|
ENSG00000028277.16 | POU class 2 homeobox 2 |
|
POU3F1
|
ENSG00000185668.5 | POU class 3 homeobox 1 |
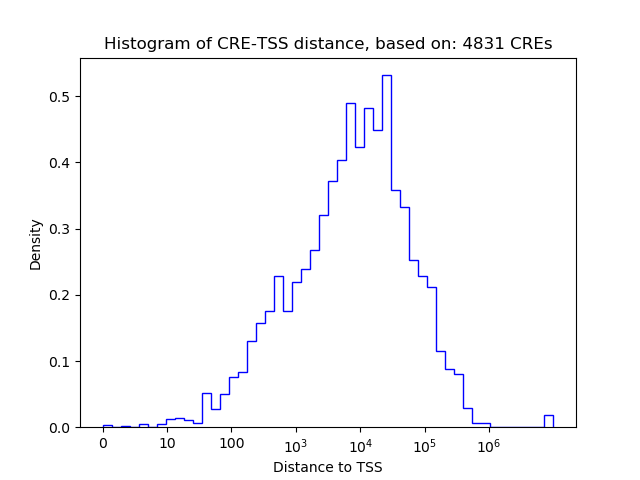
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr19_42617987_42618295 | POU2F2 | 2898 | 0.138953 | -0.72 | 3.0e-02 | Click! |
| chr19_42592755_42592969 | POU2F2 | 6695 | 0.106865 | -0.66 | 5.4e-02 | Click! |
| chr19_42618337_42618532 | POU2F2 | 2605 | 0.148628 | -0.52 | 1.5e-01 | Click! |
| chr19_42626536_42626691 | POU2F2 | 119 | 0.926435 | 0.49 | 1.8e-01 | Click! |
| chr19_42618636_42618787 | POU2F2 | 2328 | 0.160988 | -0.47 | 2.0e-01 | Click! |
| chr1_38504855_38505006 | POU3F1 | 7520 | 0.148058 | -0.56 | 1.1e-01 | Click! |
| chr1_38510021_38510235 | POU3F1 | 2322 | 0.237855 | -0.50 | 1.7e-01 | Click! |
| chr1_38512756_38512948 | POU3F1 | 402 | 0.815332 | -0.43 | 2.5e-01 | Click! |
| chr1_38512142_38512303 | POU3F1 | 228 | 0.913864 | -0.38 | 3.2e-01 | Click! |
| chr1_38513049_38513583 | POU3F1 | 866 | 0.543140 | -0.35 | 3.5e-01 | Click! |
Activity of the POU2F2_POU3F1 motif across conditions
Conditions sorted by the z-value of the POU2F2_POU3F1 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
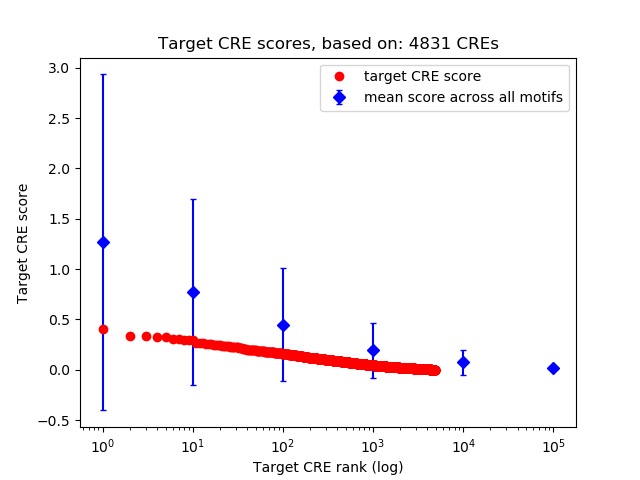
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr12_40551802_40551953 | 0.40 |
LRRK2 |
leucine-rich repeat kinase 2 |
38669 |
0.17 |
| chr20_47435196_47435347 | 0.34 |
PREX1 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 |
9149 |
0.27 |
| chr7_36763519_36763817 | 0.34 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
386 |
0.9 |
| chr2_198756913_198757256 | 0.33 |
PLCL1 |
phospholipase C-like 1 |
82102 |
0.1 |
| chr7_50351765_50352753 | 0.32 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
3941 |
0.34 |
| chr17_8857533_8857684 | 0.31 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
11416 |
0.24 |
| chrX_153190817_153191535 | 0.30 |
ARHGAP4 |
Rho GTPase activating protein 4 |
522 |
0.6 |
| chrX_78407653_78407829 | 0.30 |
GPR174 |
G protein-coupled receptor 174 |
18728 |
0.29 |
| chr19_41808988_41809179 | 0.30 |
CCDC97 |
coiled-coil domain containing 97 |
7011 |
0.11 |
| chr11_118305386_118306890 | 0.30 |
RP11-770J1.4 |
|
217 |
0.83 |
| chr11_6766362_6766695 | 0.26 |
GVINP1 |
GTPase, very large interferon inducible pseudogene 1 |
23417 |
0.11 |
| chr8_27237031_27237182 | 0.26 |
PTK2B |
protein tyrosine kinase 2 beta |
1062 |
0.59 |
| chr4_113209604_113209755 | 0.26 |
TIFA |
TRAF-interacting protein with forkhead-associated domain |
2620 |
0.23 |
| chr9_95822109_95822464 | 0.26 |
SUSD3 |
sushi domain containing 3 |
1225 |
0.45 |
| chr17_29641266_29641417 | 0.26 |
EVI2B |
ecotropic viral integration site 2B |
211 |
0.9 |
| chr13_99911056_99911338 | 0.25 |
GPR18 |
G protein-coupled receptor 18 |
515 |
0.8 |
| chr4_165040749_165040900 | 0.25 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
264317 |
0.02 |
| chr5_56484038_56484189 | 0.25 |
GPBP1 |
GC-rich promoter binding protein 1 |
12340 |
0.23 |
| chr3_71632242_71632973 | 0.25 |
FOXP1 |
forkhead box P1 |
297 |
0.91 |
| chr5_10632325_10632658 | 0.24 |
ANKRD33B-AS1 |
ANKRD33B antisense RNA 1 |
4154 |
0.28 |
| chr2_231586611_231586796 | 0.24 |
CAB39 |
calcium binding protein 39 |
8440 |
0.22 |
| chr5_130883505_130884015 | 0.24 |
RAPGEF6 |
Rap guanine nucleotide exchange factor (GEF) 6 |
15034 |
0.29 |
| chr21_15916676_15916915 | 0.23 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
1867 |
0.42 |
| chr7_33078848_33079779 | 0.23 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
1208 |
0.42 |
| chr1_239883177_239883989 | 0.23 |
CHRM3 |
cholinergic receptor, muscarinic 3 |
740 |
0.59 |
| chr11_34649048_34649199 | 0.23 |
EHF |
ets homologous factor |
3300 |
0.35 |
| chr6_13397968_13398119 | 0.23 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
10326 |
0.22 |
| chr7_137620232_137620510 | 0.23 |
CREB3L2 |
cAMP responsive element binding protein 3-like 2 |
1735 |
0.39 |
| chr1_6520196_6520500 | 0.22 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
5342 |
0.12 |
| chr18_22045724_22045875 | 0.22 |
HRH4 |
histamine receptor H4 |
5106 |
0.18 |
| chr14_99701823_99701974 | 0.22 |
AL109767.1 |
|
27387 |
0.19 |
| chr2_175456254_175456405 | 0.22 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
6012 |
0.19 |
| chr7_26192485_26193339 | 0.22 |
NFE2L3 |
nuclear factor, erythroid 2-like 3 |
1052 |
0.53 |
| chr9_274295_274630 | 0.22 |
DOCK8 |
dedicator of cytokinesis 8 |
1392 |
0.34 |
| chrX_129218649_129218800 | 0.22 |
ELF4 |
E74-like factor 4 (ets domain transcription factor) |
25612 |
0.18 |
| chr15_69074948_69075145 | 0.21 |
ENSG00000265195 |
. |
19218 |
0.22 |
| chr1_169679248_169679951 | 0.21 |
SELL |
selectin L |
1240 |
0.48 |
| chr9_34957337_34957488 | 0.20 |
KIAA1045 |
KIAA1045 |
72 |
0.97 |
| chr18_60965137_60965288 | 0.20 |
RP11-28F1.2 |
|
16103 |
0.17 |
| chr15_75080868_75081404 | 0.20 |
ENSG00000264386 |
. |
38 |
0.76 |
| chr22_27067492_27067685 | 0.20 |
CRYBA4 |
crystallin, beta A4 |
49660 |
0.13 |
| chr3_141129590_141129741 | 0.20 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
8005 |
0.24 |
| chr17_73341947_73342237 | 0.20 |
RP11-16C1.3 |
|
1606 |
0.24 |
| chr17_75878240_75878391 | 0.20 |
FLJ45079 |
|
344 |
0.91 |
| chr17_58602582_58603600 | 0.20 |
APPBP2 |
amyloid beta precursor protein (cytoplasmic tail) binding protein 2 |
393 |
0.59 |
| chr2_192014047_192014198 | 0.20 |
STAT4 |
signal transducer and activator of transcription 4 |
1575 |
0.44 |
| chr15_101786887_101787230 | 0.19 |
CHSY1 |
chondroitin sulfate synthase 1 |
5079 |
0.19 |
| chr19_44123855_44124353 | 0.19 |
ZNF428 |
zinc finger protein 428 |
78 |
0.93 |
| chr1_211780216_211780378 | 0.19 |
RP11-359E8.5 |
|
23230 |
0.15 |
| chr7_138729622_138729773 | 0.19 |
ZC3HAV1L |
zinc finger CCCH-type, antiviral 1-like |
8922 |
0.21 |
| chr2_233940706_233940970 | 0.19 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
15649 |
0.18 |
| chr7_31851936_31852087 | 0.19 |
ENSG00000223070 |
. |
18149 |
0.28 |
| chr6_35570444_35570718 | 0.19 |
ENSG00000212579 |
. |
49014 |
0.11 |
| chr17_8801498_8801803 | 0.19 |
PIK3R5 |
phosphoinositide-3-kinase, regulatory subunit 5 |
13174 |
0.21 |
| chr10_11218203_11218399 | 0.19 |
RP3-323N1.2 |
|
4962 |
0.24 |
| chr19_54873446_54873661 | 0.19 |
LAIR1 |
leukocyte-associated immunoglobulin-like receptor 1 |
997 |
0.38 |
| chr1_185663326_185663550 | 0.19 |
HMCN1 |
hemicentin 1 |
40245 |
0.17 |
| chr2_182176845_182176996 | 0.19 |
ENSG00000266705 |
. |
6541 |
0.33 |
| chr3_151027973_151028124 | 0.19 |
GPR87 |
G protein-coupled receptor 87 |
6692 |
0.18 |
| chr19_6801414_6802010 | 0.19 |
VAV1 |
vav 1 guanine nucleotide exchange factor |
17583 |
0.13 |
| chr20_52370448_52370599 | 0.18 |
ENSG00000238468 |
. |
85226 |
0.09 |
| chr10_14227081_14227239 | 0.18 |
RP11-397C18.2 |
|
110877 |
0.06 |
| chrX_48776392_48776920 | 0.18 |
PIM2 |
pim-2 oncogene |
355 |
0.76 |
| chr19_47378667_47378818 | 0.18 |
AP2S1 |
adaptor-related protein complex 2, sigma 1 subunit |
24493 |
0.14 |
| chr1_29253817_29254129 | 0.18 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
12882 |
0.18 |
| chr14_73885671_73885822 | 0.18 |
NUMB |
numb homolog (Drosophila) |
8967 |
0.19 |
| chr1_206729975_206731239 | 0.18 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
114 |
0.96 |
| chr3_3214293_3214763 | 0.18 |
CRBN |
cereblon |
6830 |
0.19 |
| chr14_35306917_35307068 | 0.18 |
ENSG00000251726 |
. |
7574 |
0.17 |
| chr8_66748492_66748643 | 0.18 |
PDE7A |
phosphodiesterase 7A |
2416 |
0.41 |
| chr3_197875188_197875339 | 0.18 |
AC073135.3 |
|
38186 |
0.14 |
| chr18_45018917_45019068 | 0.18 |
ENSG00000266582 |
. |
112125 |
0.07 |
| chr14_22747947_22748098 | 0.18 |
ENSG00000238634 |
. |
137135 |
0.04 |
| chr15_100050578_100050729 | 0.17 |
AC015660.1 |
HCG1993240; Serologically defined breast cancer antigen NY-BR-40; Uncharacterized protein |
12075 |
0.22 |
| chr11_9714574_9714725 | 0.17 |
ENSG00000201564 |
. |
12178 |
0.18 |
| chr2_230198282_230198433 | 0.17 |
PID1 |
phosphotyrosine interaction domain containing 1 |
62356 |
0.14 |
| chr19_18507804_18508682 | 0.17 |
LRRC25 |
leucine rich repeat containing 25 |
178 |
0.89 |
| chr19_16461941_16462107 | 0.17 |
EPS15L1 |
epidermal growth factor receptor pathway substrate 15-like 1 |
10740 |
0.14 |
| chr2_120687963_120688114 | 0.17 |
PTPN4 |
protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) |
686 |
0.74 |
| chrX_1640522_1640673 | 0.17 |
P2RY8 |
purinergic receptor P2Y, G-protein coupled, 8 |
15403 |
0.2 |
| chr2_198143570_198143721 | 0.17 |
ANKRD44-IT1 |
ANKRD44 intronic transcript 1 (non-protein coding) |
23598 |
0.16 |
| chr12_79960142_79960293 | 0.17 |
PAWR |
PRKC, apoptosis, WT1, regulator |
30097 |
0.18 |
| chr2_113945705_113945856 | 0.17 |
PSD4 |
pleckstrin and Sec7 domain containing 4 |
8077 |
0.14 |
| chrY_1590520_1590671 | 0.17 |
NA |
NA |
> 106 |
NA |
| chr1_236056952_236057103 | 0.17 |
LYST |
lysosomal trafficking regulator |
10155 |
0.17 |
| chr7_37448490_37448719 | 0.17 |
ENSG00000200113 |
. |
10627 |
0.2 |
| chr1_19789578_19789845 | 0.17 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
20868 |
0.15 |
| chr2_158977628_158977779 | 0.17 |
UPP2-IT1 |
UPP2 intronic transcript 1 (non-protein coding) |
6766 |
0.22 |
| chr11_121365458_121365609 | 0.17 |
RP11-730K11.1 |
|
41811 |
0.18 |
| chr13_46750556_46750707 | 0.17 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
5828 |
0.17 |
| chr3_172235814_172236037 | 0.16 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
5340 |
0.27 |
| chr2_198064690_198065196 | 0.16 |
ANKRD44 |
ankyrin repeat domain 44 |
2181 |
0.29 |
| chr10_59856470_59856621 | 0.16 |
ENSG00000238970 |
. |
141976 |
0.05 |
| chr7_36327271_36327770 | 0.16 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
9233 |
0.18 |
| chr11_121298288_121298439 | 0.16 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
24549 |
0.21 |
| chr7_25902012_25902163 | 0.16 |
ENSG00000199085 |
. |
87519 |
0.1 |
| chr4_35416147_35416298 | 0.16 |
ENSG00000206665 |
. |
81298 |
0.12 |
| chr8_90031831_90031982 | 0.16 |
RP11-586K2.1 |
|
319386 |
0.01 |
| chr1_167059294_167059481 | 0.16 |
GPA33 |
glycoprotein A33 (transmembrane) |
481 |
0.77 |
| chr7_2550958_2551215 | 0.16 |
LFNG |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
1077 |
0.47 |
| chr11_128595669_128596120 | 0.16 |
SENCR |
smooth muscle and endothelial cell enriched migration/differentiation-associated long non-coding RNA |
29976 |
0.16 |
| chr14_106471583_106471884 | 0.16 |
IGHV1-3 |
immunoglobulin heavy variable 1-3 |
10 |
0.91 |
| chr17_17603690_17604249 | 0.16 |
RAI1 |
retinoic acid induced 1 |
18160 |
0.17 |
| chr6_70219788_70219939 | 0.16 |
ENSG00000265362 |
. |
111402 |
0.07 |
| chr7_7960072_7960471 | 0.16 |
ENSG00000201747 |
. |
14121 |
0.18 |
| chr2_201985007_201985268 | 0.16 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
1923 |
0.24 |
| chr6_90066974_90067553 | 0.16 |
UBE2J1 |
ubiquitin-conjugating enzyme E2, J1 |
4696 |
0.23 |
| chr10_97154601_97154752 | 0.15 |
SORBS1 |
sorbin and SH3 domain containing 1 |
20776 |
0.25 |
| chr1_36270069_36270220 | 0.15 |
AGO4 |
argonaute RISC catalytic component 4 |
3629 |
0.22 |
| chr1_15342375_15342526 | 0.15 |
KAZN |
kazrin, periplakin interacting protein |
70035 |
0.12 |
| chrX_12989756_12990021 | 0.15 |
TMSB4X |
thymosin beta 4, X-linked |
3339 |
0.29 |
| chr6_16687583_16687734 | 0.15 |
RP1-151F17.1 |
|
73711 |
0.11 |
| chr3_146258052_146258203 | 0.15 |
PLSCR1 |
phospholipid scramblase 1 |
4181 |
0.23 |
| chr22_36805845_36806656 | 0.15 |
MYH9 |
myosin, heavy chain 9, non-muscle |
22187 |
0.14 |
| chr20_49073623_49073774 | 0.15 |
ENSG00000244376 |
. |
27678 |
0.17 |
| chr6_170604225_170605407 | 0.15 |
FAM120B |
family with sequence similarity 120B |
407 |
0.83 |
| chr12_57636595_57636960 | 0.15 |
NDUFA4L2 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 |
2302 |
0.16 |
| chr14_51280034_51280250 | 0.15 |
RP11-286O18.1 |
|
8456 |
0.15 |
| chr1_161523849_161524000 | 0.15 |
RP11-5K23.5 |
|
2944 |
0.16 |
| chr6_42513326_42513477 | 0.15 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
18399 |
0.19 |
| chr2_68995729_68995971 | 0.15 |
ARHGAP25 |
Rho GTPase activating protein 25 |
6083 |
0.26 |
| chr14_73931285_73931436 | 0.15 |
ENSG00000251393 |
. |
2231 |
0.24 |
| chr1_25887785_25888064 | 0.15 |
LDLRAP1 |
low density lipoprotein receptor adaptor protein 1 |
17853 |
0.19 |
| chr4_55809328_55809479 | 0.15 |
ENSG00000264332 |
. |
67696 |
0.11 |
| chr1_9711350_9712072 | 0.15 |
PIK3CD |
phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit delta |
79 |
0.97 |
| chr8_82066026_82066177 | 0.15 |
PAG1 |
phosphoprotein associated with glycosphingolipid microdomains 1 |
41798 |
0.19 |
| chr3_39006420_39006571 | 0.15 |
SCN11A |
sodium channel, voltage-gated, type XI, alpha subunit |
14443 |
0.19 |
| chr13_50810191_50810488 | 0.15 |
ENSG00000221198 |
. |
127034 |
0.05 |
| chr8_62604897_62605048 | 0.15 |
ASPH |
aspartate beta-hydroxylase |
2564 |
0.34 |
| chr7_133116374_133116525 | 0.15 |
EXOC4 |
exocyst complex component 4 |
144767 |
0.05 |
| chr11_40034818_40034969 | 0.15 |
ENSG00000221804 |
. |
151749 |
0.04 |
| chr18_9559122_9559273 | 0.15 |
RP11-881L2.1 |
|
28187 |
0.13 |
| chr1_25889724_25889930 | 0.14 |
LDLRAP1 |
low density lipoprotein receptor adaptor protein 1 |
19756 |
0.18 |
| chr11_47386705_47386856 | 0.14 |
MYBPC3 |
myosin binding protein C, cardiac |
12527 |
0.09 |
| chr6_43214536_43215130 | 0.14 |
TTBK1 |
tau tubulin kinase 1 |
894 |
0.48 |
| chr1_221927643_221927794 | 0.14 |
DUSP10 |
dual specificity phosphatase 10 |
12200 |
0.29 |
| chr20_33732566_33732717 | 0.14 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
318 |
0.85 |
| chr12_76658709_76658860 | 0.14 |
ENSG00000223273 |
. |
50747 |
0.15 |
| chr16_28235213_28235364 | 0.14 |
XPO6 |
exportin 6 |
12047 |
0.18 |
| chr4_2939568_2939719 | 0.14 |
NOP14-AS1 |
NOP14 antisense RNA 1 |
265 |
0.88 |
| chr9_97121768_97121919 | 0.14 |
HIATL1 |
hippocampus abundant transcript-like 1 |
14990 |
0.2 |
| chr14_61793464_61793615 | 0.14 |
PRKCH |
protein kinase C, eta |
92 |
0.97 |
| chr11_48041704_48041855 | 0.14 |
AC103828.1 |
|
4372 |
0.25 |
| chr16_3265391_3265542 | 0.14 |
OR1F1 |
olfactory receptor, family 1, subfamily F, member 1 |
11219 |
0.1 |
| chr16_50874988_50875494 | 0.14 |
ENSG00000199771 |
. |
28245 |
0.15 |
| chr14_35754560_35754711 | 0.14 |
ENSG00000265530 |
. |
6057 |
0.23 |
| chr20_55550076_55550227 | 0.14 |
ENSG00000251772 |
. |
24992 |
0.22 |
| chr15_52023462_52024365 | 0.14 |
LYSMD2 |
LysM, putative peptidoglycan-binding, domain containing 2 |
6405 |
0.15 |
| chr1_161605235_161605386 | 0.14 |
FCGR3B |
Fc fragment of IgG, low affinity IIIb, receptor (CD16b) |
3557 |
0.15 |
| chr6_130848326_130848477 | 0.14 |
ENSG00000202438 |
. |
46856 |
0.17 |
| chr7_38392270_38392581 | 0.14 |
AMPH |
amphiphysin |
110288 |
0.07 |
| chr1_235120289_235120440 | 0.14 |
ENSG00000239690 |
. |
80431 |
0.1 |
| chr3_172240144_172241280 | 0.14 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
553 |
0.83 |
| chr8_141607200_141607468 | 0.14 |
AGO2 |
argonaute RISC catalytic component 2 |
8653 |
0.24 |
| chrX_109351614_109351783 | 0.14 |
ENSG00000265584 |
. |
26352 |
0.2 |
| chr5_125785966_125786206 | 0.14 |
GRAMD3 |
GRAM domain containing 3 |
11951 |
0.22 |
| chr20_48448748_48448899 | 0.14 |
ENSG00000252123 |
. |
4294 |
0.19 |
| chr12_15102775_15102926 | 0.14 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
779 |
0.6 |
| chr17_40425804_40425955 | 0.14 |
AC003104.1 |
|
1178 |
0.32 |
| chr16_17510234_17510944 | 0.13 |
XYLT1 |
xylosyltransferase I |
54149 |
0.18 |
| chr1_206900182_206900333 | 0.13 |
ENSG00000199349 |
. |
21180 |
0.14 |
| chr18_13465441_13465744 | 0.13 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
578 |
0.59 |
| chr1_198896471_198896622 | 0.13 |
ENSG00000207759 |
. |
68264 |
0.12 |
| chr2_175357999_175358525 | 0.13 |
GPR155 |
G protein-coupled receptor 155 |
6440 |
0.21 |
| chr1_81935804_81935955 | 0.13 |
RP5-837I24.6 |
|
34905 |
0.21 |
| chr8_129289122_129289410 | 0.13 |
ENSG00000201782 |
. |
56516 |
0.16 |
| chr18_56307433_56307584 | 0.13 |
ALPK2 |
alpha-kinase 2 |
11319 |
0.15 |
| chr17_76714885_76715036 | 0.13 |
CYTH1 |
cytohesin 1 |
1853 |
0.36 |
| chr1_161154335_161154531 | 0.13 |
B4GALT3 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 3 |
7146 |
0.07 |
| chr2_216922950_216923179 | 0.13 |
PECR |
peroxisomal trans-2-enoyl-CoA reductase |
23475 |
0.15 |
| chr2_198006679_198006830 | 0.13 |
ANKRD44 |
ankyrin repeat domain 44 |
5874 |
0.27 |
| chr15_101792013_101793187 | 0.13 |
CHSY1 |
chondroitin sulfate synthase 1 |
463 |
0.8 |
| chr8_49416567_49416718 | 0.13 |
RP11-770E5.1 |
|
47485 |
0.19 |
| chr7_130643743_130643906 | 0.13 |
LINC-PINT |
long intergenic non-protein coding RNA, p53 induced transcript |
25044 |
0.22 |
| chr13_111108512_111108663 | 0.13 |
COL4A2-AS2 |
COL4A2 antisense RNA 2 |
7047 |
0.22 |
| chr10_73512307_73512458 | 0.13 |
C10orf54 |
chromosome 10 open reading frame 54 |
5005 |
0.22 |
| chr11_14666941_14667162 | 0.13 |
PDE3B |
phosphodiesterase 3B, cGMP-inhibited |
1674 |
0.36 |
| chr8_55064954_55065105 | 0.13 |
ENSG00000251835 |
. |
413 |
0.86 |
| chr4_90222893_90223044 | 0.13 |
GPRIN3 |
GPRIN family member 3 |
6193 |
0.32 |
| chr1_198651611_198651863 | 0.13 |
RP11-553K8.5 |
|
15547 |
0.24 |
| chr19_6890241_6890675 | 0.13 |
EMR1 |
egf-like module containing, mucin-like, hormone receptor-like 1 |
2868 |
0.21 |
| chrX_147462452_147462756 | 0.13 |
AC002368.4 |
|
119531 |
0.06 |
| chr3_45985242_45985393 | 0.13 |
CXCR6 |
chemokine (C-X-C motif) receptor 6 |
353 |
0.85 |
| chr8_21776103_21776686 | 0.13 |
XPO7 |
exportin 7 |
786 |
0.64 |
| chrX_118751905_118752056 | 0.13 |
NKRF |
NFKB repressing factor |
12122 |
0.15 |
| chr6_11986707_11986858 | 0.13 |
RP11-456H18.1 |
|
5463 |
0.27 |
| chr7_75085913_75086064 | 0.13 |
POM121C |
POM121 transmembrane nucleoporin C |
13995 |
0.12 |
| chr2_168252602_168252753 | 0.13 |
XIRP2 |
xin actin-binding repeat containing 2 |
208884 |
0.03 |
| chr3_53742016_53742167 | 0.12 |
CACNA1D |
calcium channel, voltage-dependent, L type, alpha 1D subunit |
41016 |
0.16 |
| chr12_98912505_98912656 | 0.12 |
TMPO-AS1 |
TMPO antisense RNA 1 |
2380 |
0.2 |
| chr14_52778923_52779116 | 0.12 |
PTGER2 |
prostaglandin E receptor 2 (subtype EP2), 53kDa |
2004 |
0.41 |
| chr11_23251355_23251506 | 0.12 |
ENSG00000264478 |
. |
241829 |
0.02 |
| chr3_39302441_39302592 | 0.12 |
CX3CR1 |
chemokine (C-X3-C motif) receptor 1 |
19009 |
0.19 |
| chr1_56111785_56112048 | 0.12 |
ENSG00000272051 |
. |
161271 |
0.04 |
| chr5_32583768_32584065 | 0.12 |
SUB1 |
SUB1 homolog (S. cerevisiae) |
1689 |
0.49 |
| chr14_99702038_99702570 | 0.12 |
AL109767.1 |
|
26981 |
0.19 |
| chr6_15882568_15882719 | 0.12 |
DTNBP1 |
dystrobrevin binding protein 1 |
219370 |
0.02 |
| chr2_32489606_32489792 | 0.12 |
NLRC4 |
NLR family, CARD domain containing 4 |
250 |
0.92 |
| chr12_125405765_125405916 | 0.12 |
UBC |
ubiquitin C |
3926 |
0.19 |
| chr1_181050920_181051071 | 0.12 |
IER5 |
immediate early response 5 |
6643 |
0.23 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.3 | GO:0008653 | lipopolysaccharide metabolic process(GO:0008653) |
| 0.0 | 0.1 | GO:0010040 | response to iron(II) ion(GO:0010040) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.0 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.1 | GO:0003056 | regulation of vascular smooth muscle contraction(GO:0003056) |
| 0.0 | 0.2 | GO:0050857 | positive regulation of antigen receptor-mediated signaling pathway(GO:0050857) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0061687 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 0.1 | GO:0051832 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.0 | 0.2 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.1 | GO:0032632 | interleukin-3 production(GO:0032632) |
| 0.0 | 0.1 | GO:0002606 | positive regulation of dendritic cell antigen processing and presentation(GO:0002606) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0002513 | tolerance induction to self antigen(GO:0002513) |
| 0.0 | 0.1 | GO:1903429 | regulation of neuron maturation(GO:0014041) regulation of cell maturation(GO:1903429) |
| 0.0 | 0.1 | GO:0002313 | mature B cell differentiation involved in immune response(GO:0002313) |
| 0.0 | 0.1 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0031936 | negative regulation of chromatin silencing(GO:0031936) |
| 0.0 | 0.0 | GO:0002335 | mature B cell differentiation(GO:0002335) |
| 0.0 | 0.1 | GO:0002693 | positive regulation of cellular extravasation(GO:0002693) |
| 0.0 | 0.0 | GO:0010831 | positive regulation of myotube differentiation(GO:0010831) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.0 | GO:0043374 | CD8-positive, alpha-beta T cell activation(GO:0036037) CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0002507 | tolerance induction(GO:0002507) |
| 0.0 | 0.1 | GO:0030853 | negative regulation of granulocyte differentiation(GO:0030853) |
| 0.0 | 0.0 | GO:0060049 | regulation of protein glycosylation(GO:0060049) |
| 0.0 | 0.1 | GO:0017121 | phospholipid scrambling(GO:0017121) |
| 0.0 | 0.0 | GO:0030011 | maintenance of cell polarity(GO:0030011) |
| 0.0 | 0.0 | GO:0009648 | photoperiodism(GO:0009648) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0001782 | B cell homeostasis(GO:0001782) |
| 0.0 | 0.2 | GO:0042347 | negative regulation of NF-kappaB import into nucleus(GO:0042347) |
| 0.0 | 0.0 | GO:0032060 | bleb assembly(GO:0032060) |
| 0.0 | 0.0 | GO:0070574 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.0 | GO:0002885 | positive regulation of hypersensitivity(GO:0002885) |
| 0.0 | 0.1 | GO:0060235 | lens induction in camera-type eye(GO:0060235) |
| 0.0 | 0.0 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.0 | GO:0048541 | Peyer's patch development(GO:0048541) |
| 0.0 | 0.1 | GO:0031061 | negative regulation of histone methylation(GO:0031061) |
| 0.0 | 0.1 | GO:0090205 | positive regulation of cholesterol metabolic process(GO:0090205) |
| 0.0 | 0.0 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.0 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0032509 | endosome transport via multivesicular body sorting pathway(GO:0032509) |
| 0.0 | 0.0 | GO:0072577 | endothelial cell apoptotic process(GO:0072577) epithelial cell apoptotic process(GO:1904019) regulation of epithelial cell apoptotic process(GO:1904035) regulation of endothelial cell apoptotic process(GO:2000351) |
| 0.0 | 0.0 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0016045 | detection of bacterium(GO:0016045) |
| 0.0 | 0.1 | GO:0042989 | sequestering of actin monomers(GO:0042989) |
| 0.0 | 0.0 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.1 | GO:0006244 | pyrimidine nucleotide catabolic process(GO:0006244) |
| 0.0 | 0.0 | GO:0045409 | negative regulation of tumor necrosis factor biosynthetic process(GO:0042536) negative regulation of interleukin-6 biosynthetic process(GO:0045409) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.1 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.0 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:0042119 | granulocyte activation(GO:0036230) neutrophil activation(GO:0042119) |
| 0.0 | 0.0 | GO:0046643 | regulation of gamma-delta T cell activation(GO:0046643) positive regulation of gamma-delta T cell activation(GO:0046645) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.0 | GO:0002679 | respiratory burst involved in defense response(GO:0002679) |
| 0.0 | 0.1 | GO:0060134 | prepulse inhibition(GO:0060134) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032045 | guanyl-nucleotide exchange factor complex(GO:0032045) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0098984 | asymmetric synapse(GO:0032279) neuron to neuron synapse(GO:0098984) |
| 0.0 | 0.3 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.0 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.0 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.0 | GO:0031264 | death-inducing signaling complex(GO:0031264) |
| 0.0 | 0.1 | GO:0030128 | AP-2 adaptor complex(GO:0030122) clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.1 | GO:0043190 | ATP-binding cassette (ABC) transporter complex(GO:0043190) |
| 0.0 | 0.0 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.0 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0001891 | phagocytic cup(GO:0001891) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.2 | GO:0047238 | glucuronosyl-N-acetylgalactosaminyl-proteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047238) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.2 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.7 | GO:0045028 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0003836 | beta-galactoside (CMP) alpha-2,3-sialyltransferase activity(GO:0003836) |
| 0.0 | 0.1 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.1 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.3 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.2 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.2 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0005164 | tumor necrosis factor receptor binding(GO:0005164) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.0 | GO:0016312 | inositol bisphosphate phosphatase activity(GO:0016312) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0004955 | prostaglandin receptor activity(GO:0004955) |
| 0.0 | 0.0 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 0.0 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0008409 | 5'-3' exonuclease activity(GO:0008409) |
| 0.0 | 0.0 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.2 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.0 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.1 | GO:0046870 | cadmium ion binding(GO:0046870) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.0 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.5 | SIG PIP3 SIGNALING IN B LYMPHOCYTES | Genes related to PIP3 signaling in B lymphocytes |
| 0.0 | 0.3 | PID TRAIL PATHWAY | TRAIL signaling pathway |
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.4 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.0 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.3 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.2 | REACTOME INFLAMMASOMES | Genes involved in Inflammasomes |
| 0.0 | 0.3 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |