Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
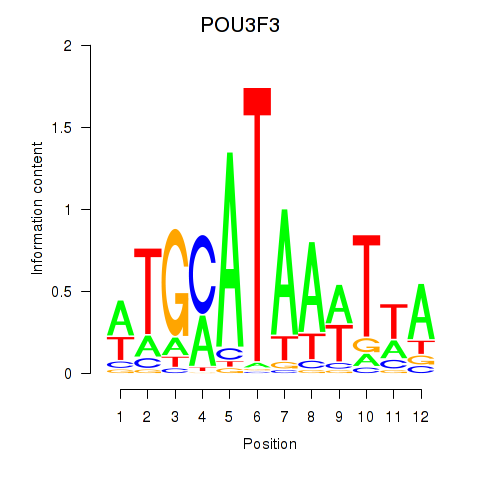
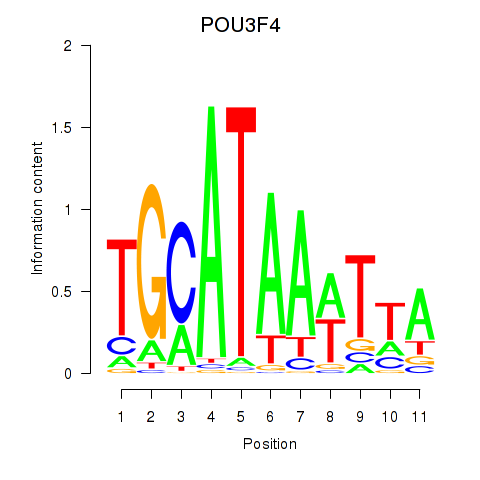
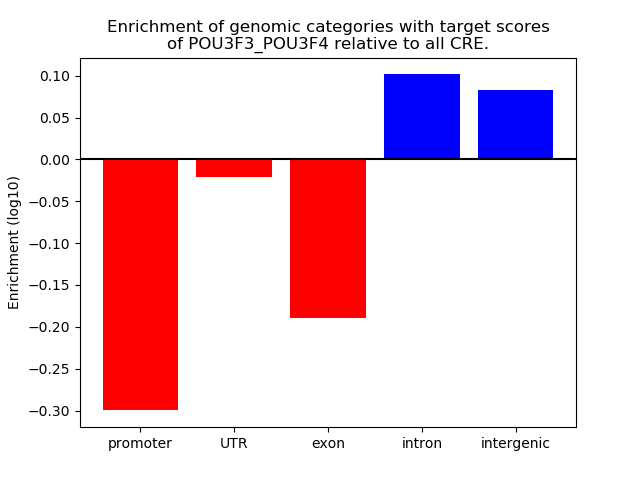
Results for POU3F3_POU3F4
Z-value: 1.75
Transcription factors associated with POU3F3_POU3F4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU3F3
|
ENSG00000198914.2 | POU class 3 homeobox 3 |
|
POU3F4
|
ENSG00000196767.4 | POU class 3 homeobox 4 |
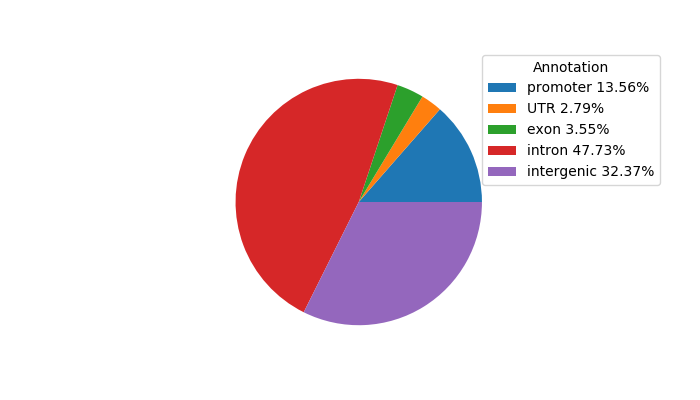
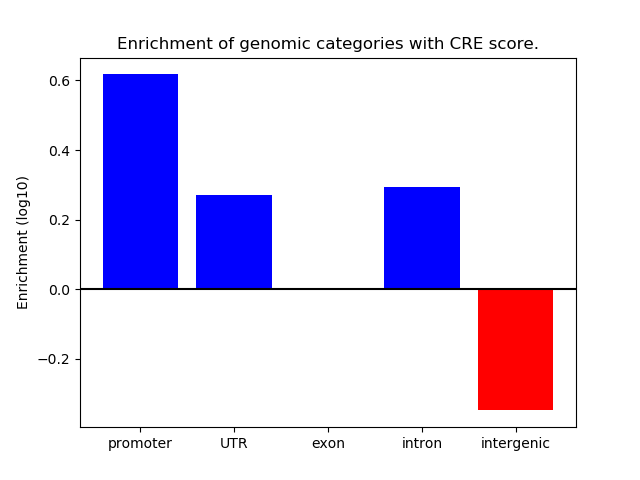
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
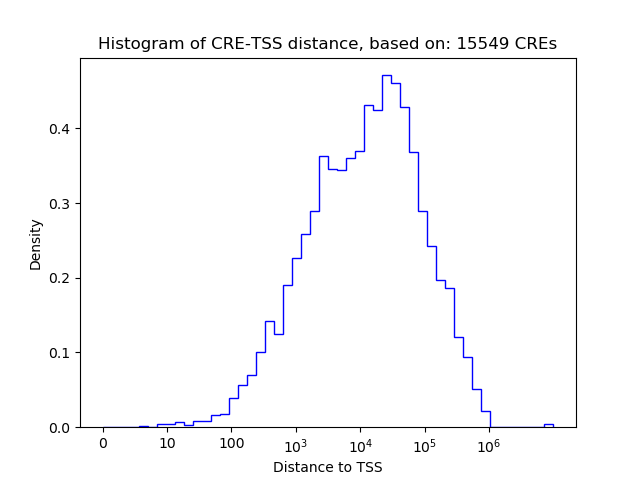
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr2_105471048_105471431 | POU3F3 | 730 | 0.471155 | -0.55 | 1.3e-01 | Click! |
| chr2_105471431_105471582 | POU3F3 | 463 | 0.684630 | -0.53 | 1.4e-01 | Click! |
| chr2_105473621_105473772 | POU3F3 | 1727 | 0.260320 | 0.52 | 1.5e-01 | Click! |
| chr2_105476021_105476172 | POU3F3 | 4127 | 0.156010 | -0.51 | 1.6e-01 | Click! |
| chr2_105474864_105475015 | POU3F3 | 2970 | 0.179506 | -0.33 | 3.8e-01 | Click! |
| chrX_82763168_82763319 | POU3F4 | 26 | 0.962329 | -0.51 | 1.6e-01 | Click! |
| chrX_82762361_82762512 | POU3F4 | 833 | 0.620699 | -0.41 | 2.8e-01 | Click! |
| chrX_82763349_82763500 | POU3F4 | 155 | 0.908200 | -0.36 | 3.4e-01 | Click! |
| chrX_82762744_82762895 | POU3F4 | 450 | 0.798138 | -0.29 | 4.5e-01 | Click! |
Activity of the POU3F3_POU3F4 motif across conditions
Conditions sorted by the z-value of the POU3F3_POU3F4 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
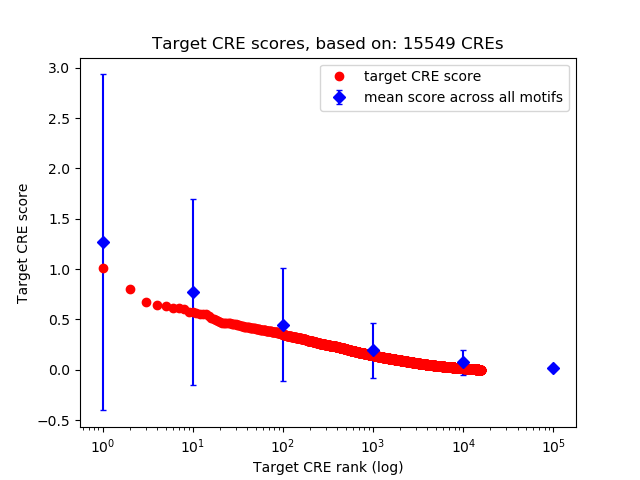
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr18_55863175_55863444 | 1.01 |
RP11-718I15.1 |
|
381 |
0.52 |
| chr18_32287572_32287755 | 0.80 |
DTNA |
dystrobrevin, alpha |
2585 |
0.38 |
| chr6_46375502_46375653 | 0.67 |
RCAN2 |
regulator of calcineurin 2 |
49136 |
0.16 |
| chr18_48718913_48719435 | 0.64 |
MEX3C |
mex-3 RNA binding family member C |
3955 |
0.31 |
| chr4_177254207_177254358 | 0.63 |
RP11-87F15.2 |
|
12673 |
0.21 |
| chr1_112603472_112603623 | 0.61 |
KCND3 |
potassium voltage-gated channel, Shal-related subfamily, member 3 |
71770 |
0.11 |
| chr7_51866473_51866624 | 0.61 |
ENSG00000242650 |
. |
82194 |
0.12 |
| chr12_85410552_85410703 | 0.60 |
TSPAN19 |
tetraspanin 19 |
747 |
0.74 |
| chr3_174059484_174059635 | 0.58 |
NAALADL2 |
N-acetylated alpha-linked acidic dipeptidase-like 2 |
99218 |
0.09 |
| chr19_52832897_52833072 | 0.57 |
ZNF610 |
zinc finger protein 610 |
6514 |
0.11 |
| chr10_119119895_119120046 | 0.56 |
ENSG00000222197 |
. |
2103 |
0.35 |
| chr13_70755828_70755979 | 0.55 |
KLHL1 |
kelch-like family member 1 |
73312 |
0.12 |
| chr2_191518131_191518282 | 0.55 |
NAB1 |
NGFI-A binding protein 1 (EGR1 binding protein 1) |
245 |
0.94 |
| chr10_52364321_52364493 | 0.55 |
ENSG00000238523 |
. |
13935 |
0.15 |
| chr12_39821314_39821465 | 0.54 |
KIF21A |
kinesin family member 21A |
15383 |
0.23 |
| chr4_83787091_83787242 | 0.52 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
918 |
0.55 |
| chr14_105479768_105479974 | 0.51 |
CDCA4 |
cell division cycle associated 4 |
7526 |
0.16 |
| chr7_155168058_155168209 | 0.50 |
AC008060.7 |
HCG2000720; Hypothetical gene supported by AK054822; Uncharacterized protein |
6638 |
0.18 |
| chr10_53873012_53873163 | 0.49 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
66550 |
0.14 |
| chr14_73885671_73885822 | 0.48 |
NUMB |
numb homolog (Drosophila) |
8967 |
0.19 |
| chr6_91424739_91424890 | 0.47 |
MAP3K7 |
mitogen-activated protein kinase kinase kinase 7 |
128050 |
0.06 |
| chr3_164285782_164285933 | 0.46 |
ENSG00000221755 |
. |
226728 |
0.02 |
| chr11_115065010_115065166 | 0.46 |
CADM1 |
cell adhesion molecule 1 |
46612 |
0.18 |
| chr6_41604382_41604533 | 0.46 |
MDFI |
MyoD family inhibitor |
163 |
0.94 |
| chr14_29242746_29242955 | 0.46 |
C14orf23 |
chromosome 14 open reading frame 23 |
849 |
0.55 |
| chr1_93221952_93222251 | 0.46 |
EVI5 |
ecotropic viral integration site 5 |
35850 |
0.13 |
| chr2_97452820_97452971 | 0.46 |
CNNM4 |
cyclin M4 |
1427 |
0.34 |
| chr1_186415843_186415994 | 0.46 |
PDC |
phosducin |
281 |
0.89 |
| chrX_26102260_26102411 | 0.46 |
MAGEB18 |
melanoma antigen family B, 18 |
54125 |
0.17 |
| chr4_166405048_166405199 | 0.45 |
CPE |
carboxypeptidase E |
65745 |
0.13 |
| chr2_202313137_202313288 | 0.45 |
TRAK2 |
trafficking protein, kinesin binding 2 |
3011 |
0.22 |
| chr1_93443435_93443586 | 0.45 |
ENSG00000239710 |
. |
2830 |
0.16 |
| chr10_118924530_118925015 | 0.44 |
ENSG00000266782 |
. |
2513 |
0.25 |
| chr1_101587818_101587969 | 0.44 |
ENSG00000252765 |
. |
10816 |
0.2 |
| chr10_127685162_127685313 | 0.44 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
1251 |
0.5 |
| chr7_87986606_87986757 | 0.43 |
STEAP4 |
STEAP family member 4 |
50475 |
0.15 |
| chr5_114866886_114867037 | 0.43 |
FEM1C |
fem-1 homolog c (C. elegans) |
13630 |
0.22 |
| chr3_173220593_173220744 | 0.43 |
NLGN1 |
neuroligin 1 |
81677 |
0.12 |
| chr18_53089837_53090372 | 0.43 |
TCF4 |
transcription factor 4 |
361 |
0.89 |
| chr2_181570330_181570481 | 0.42 |
ENSG00000264976 |
. |
88472 |
0.1 |
| chr4_126244718_126245045 | 0.42 |
FAT4 |
FAT atypical cadherin 4 |
7327 |
0.29 |
| chr10_63979683_63979834 | 0.42 |
RTKN2 |
rhotekin 2 |
16264 |
0.26 |
| chr8_59762363_59762514 | 0.42 |
ENSG00000201231 |
. |
35327 |
0.22 |
| chr17_68401721_68401883 | 0.42 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
236126 |
0.02 |
| chr1_21409721_21409872 | 0.42 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
5905 |
0.27 |
| chr2_43986075_43986259 | 0.42 |
ENSG00000252490 |
. |
13322 |
0.19 |
| chr1_51440088_51440239 | 0.41 |
CDKN2C |
cyclin-dependent kinase inhibitor 2C (p18, inhibits CDK4) |
4547 |
0.24 |
| chr4_95175515_95175666 | 0.41 |
SMARCAD1 |
SWI/SNF-related, matrix-associated actin-dependent regulator of chromatin, subfamily a, containing DEAD/H box 1 |
1145 |
0.65 |
| chr3_194592423_194592920 | 0.41 |
FAM43A |
family with sequence similarity 43, member A |
186049 |
0.02 |
| chr11_27335139_27335290 | 0.41 |
CCDC34 |
coiled-coil domain containing 34 |
49567 |
0.16 |
| chr12_133770634_133770785 | 0.40 |
ZNF268 |
zinc finger protein 268 |
2626 |
0.19 |
| chr4_22971290_22971441 | 0.40 |
GPR125 |
G protein-coupled receptor 125 |
453688 |
0.01 |
| chr2_175709747_175709898 | 0.40 |
CHN1 |
chimerin 1 |
1311 |
0.54 |
| chr7_97282168_97282319 | 0.40 |
ENSG00000199475 |
. |
53671 |
0.15 |
| chr13_74159314_74159465 | 0.40 |
ENSG00000206697 |
. |
358105 |
0.01 |
| chr3_29377379_29377667 | 0.40 |
ENSG00000216169 |
. |
33389 |
0.18 |
| chr4_161638888_161639039 | 0.40 |
ENSG00000221362 |
. |
286147 |
0.01 |
| chr8_70622919_70623070 | 0.40 |
RP11-102F4.2 |
|
2631 |
0.31 |
| chr6_139454805_139454996 | 0.40 |
HECA |
headcase homolog (Drosophila) |
1349 |
0.54 |
| chr20_34459115_34459914 | 0.39 |
ENSG00000201221 |
. |
16067 |
0.13 |
| chr3_60921693_60921844 | 0.39 |
ENSG00000212211 |
. |
79491 |
0.12 |
| chr3_81724848_81724999 | 0.39 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
67857 |
0.14 |
| chr10_3977780_3978372 | 0.39 |
KLF6 |
Kruppel-like factor 6 |
150603 |
0.04 |
| chr2_208339912_208340104 | 0.39 |
CREB1 |
cAMP responsive element binding protein 1 |
54453 |
0.13 |
| chr17_67056652_67056803 | 0.39 |
ABCA9 |
ATP-binding cassette, sub-family A (ABC1), member 9 |
320 |
0.9 |
| chr3_79030262_79030413 | 0.39 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
37357 |
0.2 |
| chr5_127036963_127037114 | 0.38 |
CTC-548H10.2 |
|
37317 |
0.19 |
| chr10_128613788_128613939 | 0.38 |
ENSG00000202487 |
. |
4487 |
0.28 |
| chrX_136337073_136337224 | 0.38 |
GPR101 |
G protein-coupled receptor 101 |
223315 |
0.02 |
| chr3_38247873_38248024 | 0.38 |
OXSR1 |
oxidative stress responsive 1 |
40952 |
0.12 |
| chr10_90753077_90753312 | 0.38 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
2047 |
0.24 |
| chr14_26016058_26016209 | 0.38 |
ENSG00000212270 |
. |
259785 |
0.02 |
| chr13_80614475_80614626 | 0.38 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
299244 |
0.01 |
| chr9_89574254_89574405 | 0.38 |
GAS1 |
growth arrest-specific 1 |
12225 |
0.31 |
| chr1_240669972_240670123 | 0.38 |
AL646016.1 |
Uncharacterized protein |
26543 |
0.18 |
| chr6_100909122_100909273 | 0.38 |
SIM1 |
single-minded family bHLH transcription factor 1 |
2356 |
0.39 |
| chr15_38549534_38549685 | 0.37 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
4227 |
0.36 |
| chr7_105432133_105432284 | 0.37 |
ATXN7L1 |
ataxin 7-like 1 |
84715 |
0.1 |
| chr1_189197213_189197364 | 0.37 |
ENSG00000252553 |
. |
438098 |
0.01 |
| chr5_104399872_104400023 | 0.37 |
ENSG00000252881 |
. |
283857 |
0.01 |
| chr10_70096270_70096993 | 0.37 |
PBLD |
phenazine biosynthesis-like protein domain containing |
3825 |
0.2 |
| chr3_156405026_156405177 | 0.37 |
TIPARP |
TCDD-inducible poly(ADP-ribose) polymerase |
6146 |
0.24 |
| chr18_29220350_29220501 | 0.37 |
ENSG00000252379 |
. |
28916 |
0.13 |
| chr8_131954085_131954236 | 0.37 |
RP11-737F9.1 |
|
6505 |
0.26 |
| chr2_216298275_216298426 | 0.37 |
FN1 |
fibronectin 1 |
2440 |
0.3 |
| chrY_23414698_23414849 | 0.37 |
CYorf17 |
chromosome Y open reading frame 17 |
133473 |
0.05 |
| chr10_6780355_6780830 | 0.37 |
PRKCQ |
protein kinase C, theta |
158329 |
0.04 |
| chr15_50151604_50151755 | 0.37 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
17213 |
0.21 |
| chr4_167130026_167130177 | 0.37 |
ENSG00000266254 |
. |
18816 |
0.28 |
| chr5_158301830_158301981 | 0.37 |
CTD-2363C16.1 |
|
108109 |
0.07 |
| chr14_81453709_81453897 | 0.36 |
CEP128 |
centrosomal protein 128kDa |
27955 |
0.21 |
| chr14_38888998_38889149 | 0.36 |
CLEC14A |
C-type lectin domain family 14, member A |
163499 |
0.04 |
| chr5_168003477_168003628 | 0.36 |
PANK3 |
pantothenate kinase 3 |
2803 |
0.26 |
| chr10_63402177_63402609 | 0.36 |
C10orf107 |
chromosome 10 open reading frame 107 |
20326 |
0.24 |
| chr4_123719562_123719754 | 0.36 |
FGF2 |
fibroblast growth factor 2 (basic) |
28205 |
0.16 |
| chr10_19314438_19314589 | 0.36 |
MALRD1 |
MAM and LDL receptor class A domain containing 1 |
178266 |
0.03 |
| chr13_99629621_99629906 | 0.36 |
DOCK9 |
dedicator of cytokinesis 9 |
481 |
0.85 |
| chr14_45559377_45559745 | 0.35 |
ENSG00000212615 |
. |
2115 |
0.2 |
| chr5_67577391_67577649 | 0.35 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
1385 |
0.58 |
| chr8_69986403_69986554 | 0.35 |
ENSG00000238808 |
. |
36331 |
0.24 |
| chr6_102835538_102835689 | 0.35 |
GRIK2 |
glutamate receptor, ionotropic, kainate 2 |
569368 |
0.0 |
| chr6_143256936_143257087 | 0.35 |
HIVEP2 |
human immunodeficiency virus type I enhancer binding protein 2 |
9327 |
0.26 |
| chr3_33061626_33061777 | 0.35 |
CCR4 |
chemokine (C-C motif) receptor 4 |
68635 |
0.09 |
| chr15_80361249_80361400 | 0.35 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
3582 |
0.28 |
| chr2_29340749_29340907 | 0.35 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
233 |
0.92 |
| chr1_164290160_164290619 | 0.35 |
ENSG00000199849 |
. |
30121 |
0.25 |
| chr16_63376236_63376387 | 0.35 |
RP11-96H17.3 |
|
275417 |
0.02 |
| chr6_13505550_13505701 | 0.35 |
GFOD1 |
glucose-fructose oxidoreductase domain containing 1 |
17731 |
0.14 |
| chr7_83098669_83098820 | 0.34 |
AC079799.2 |
|
114274 |
0.07 |
| chr7_136910239_136910390 | 0.34 |
hsa-mir-490 |
hsa-mir-490 |
43460 |
0.18 |
| chr15_35950174_35950564 | 0.34 |
DPH6 |
diphthamine biosynthesis 6 |
111976 |
0.07 |
| chrX_45625887_45626164 | 0.34 |
ENSG00000207725 |
. |
19495 |
0.22 |
| chr14_99712277_99712758 | 0.34 |
AL109767.1 |
|
16768 |
0.21 |
| chr15_59344636_59344787 | 0.34 |
RNF111 |
ring finger protein 111 |
20888 |
0.16 |
| chr10_55152331_55152482 | 0.33 |
ENSG00000252161 |
. |
65551 |
0.15 |
| chr18_29888572_29888839 | 0.33 |
RP11-344B2.2 |
|
21305 |
0.16 |
| chr4_117414615_117414766 | 0.33 |
MTRNR2L13 |
MT-RNR2-like 13 (pseudogene) |
194674 |
0.03 |
| chr21_38831812_38831963 | 0.33 |
DYRK1A |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
29200 |
0.2 |
| chr21_35304396_35305066 | 0.33 |
LINC00649 |
long intergenic non-protein coding RNA 649 |
1213 |
0.41 |
| chr4_39570973_39571124 | 0.33 |
UGDH |
UDP-glucose 6-dehydrogenase |
41117 |
0.11 |
| chr1_209558665_209558816 | 0.33 |
ENSG00000230937 |
. |
46738 |
0.19 |
| chr6_125364944_125365095 | 0.33 |
RNF217 |
ring finger protein 217 |
4339 |
0.31 |
| chr12_118472279_118472430 | 0.33 |
RFC5 |
replication factor C (activator 1) 5, 36.5kDa |
17820 |
0.18 |
| chr5_172286846_172286997 | 0.33 |
ERGIC1 |
endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 |
25507 |
0.16 |
| chr1_243687933_243688084 | 0.33 |
RP11-269F20.1 |
|
20826 |
0.27 |
| chr8_41277096_41277247 | 0.33 |
ENSG00000238936 |
. |
7003 |
0.23 |
| chrX_31453419_31453570 | 0.33 |
ENSG00000252903 |
. |
87185 |
0.1 |
| chr1_182991327_182991478 | 0.33 |
LAMC1 |
laminin, gamma 1 (formerly LAMB2) |
1193 |
0.49 |
| chr5_110560941_110561277 | 0.33 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
1325 |
0.51 |
| chr2_106372613_106372764 | 0.33 |
NCK2 |
NCK adaptor protein 2 |
10500 |
0.29 |
| chr1_38510289_38510696 | 0.32 |
POU3F1 |
POU class 3 homeobox 1 |
1958 |
0.27 |
| chr2_230148069_230148220 | 0.32 |
PID1 |
phosphotyrosine interaction domain containing 1 |
12143 |
0.27 |
| chr4_110217881_110218141 | 0.32 |
COL25A1 |
collagen, type XXV, alpha 1 |
5512 |
0.3 |
| chr7_157225535_157225734 | 0.32 |
AC006372.6 |
|
80354 |
0.09 |
| chr12_41084046_41084197 | 0.32 |
CNTN1 |
contactin 1 |
2123 |
0.46 |
| chr4_68593746_68593897 | 0.32 |
UBA6-AS1 |
UBA6 antisense RNA 1 (head to head) |
4041 |
0.2 |
| chr5_30785800_30785951 | 0.32 |
CDH6 |
cadherin 6, type 2, K-cadherin (fetal kidney) |
407982 |
0.01 |
| chr10_60580536_60580825 | 0.32 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
27385 |
0.23 |
| chr12_111003570_111003721 | 0.32 |
PPTC7 |
PTC7 protein phosphatase homolog (S. cerevisiae) |
17480 |
0.14 |
| chr14_38613607_38613758 | 0.32 |
CTD-2058B24.2 |
|
49804 |
0.14 |
| chr2_201248196_201249000 | 0.32 |
ENSG00000264798 |
. |
3300 |
0.23 |
| chr6_37483764_37484117 | 0.32 |
CCDC167 |
coiled-coil domain containing 167 |
16242 |
0.19 |
| chr10_75912778_75912929 | 0.32 |
ADK |
adenosine kinase |
1866 |
0.31 |
| chr2_213962198_213962349 | 0.32 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
51080 |
0.16 |
| chr1_8741272_8741496 | 0.32 |
RERE |
arginine-glutamic acid dipeptide (RE) repeats |
21894 |
0.23 |
| chr2_29850591_29850818 | 0.31 |
ENSG00000242699 |
. |
53485 |
0.16 |
| chr3_79625949_79626100 | 0.31 |
ENSG00000216116 |
. |
7810 |
0.3 |
| chr4_151600832_151601026 | 0.31 |
ENSG00000221235 |
. |
39911 |
0.19 |
| chr18_20558047_20558453 | 0.31 |
RBBP8 |
retinoblastoma binding protein 8 |
9445 |
0.15 |
| chr18_70644965_70645116 | 0.31 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
109856 |
0.07 |
| chr12_113797502_113797653 | 0.31 |
SLC8B1 |
solute carrier family 8 (sodium/lithium/calcium exchanger), member B1 |
279 |
0.85 |
| chr1_192547268_192547419 | 0.31 |
RGS1 |
regulator of G-protein signaling 1 |
2440 |
0.31 |
| chr6_14127798_14127949 | 0.31 |
CD83 |
CD83 molecule |
10001 |
0.25 |
| chr5_125703700_125704120 | 0.31 |
GRAMD3 |
GRAM domain containing 3 |
3095 |
0.37 |
| chr2_32940890_32941041 | 0.31 |
TTC27 |
tetratricopeptide repeat domain 27 |
37020 |
0.2 |
| chr21_35266767_35266918 | 0.31 |
ATP5O |
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
14557 |
0.14 |
| chr2_97593731_97594362 | 0.31 |
ENSG00000252845 |
. |
28506 |
0.12 |
| chr2_110376280_110376431 | 0.31 |
SOWAHC |
sosondowah ankyrin repeat domain family member C |
4444 |
0.21 |
| chr1_25060751_25060902 | 0.31 |
CLIC4 |
chloride intracellular channel 4 |
11022 |
0.2 |
| chr6_12280277_12280428 | 0.31 |
EDN1 |
endothelin 1 |
10244 |
0.3 |
| chr20_53950807_53950958 | 0.31 |
ENSG00000252089 |
. |
474531 |
0.01 |
| chr9_33446798_33447442 | 0.31 |
AQP3 |
aquaporin 3 (Gill blood group) |
489 |
0.76 |
| chr1_76788853_76789004 | 0.31 |
ST6GALNAC3 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
248524 |
0.02 |
| chr3_121373349_121373500 | 0.31 |
ENSG00000243544 |
. |
281 |
0.82 |
| chr18_13275881_13276032 | 0.31 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
2146 |
0.32 |
| chr4_110223153_110224430 | 0.31 |
COL25A1 |
collagen, type XXV, alpha 1 |
8 |
0.99 |
| chr12_61131831_61131982 | 0.31 |
ENSG00000251822 |
. |
374303 |
0.01 |
| chr7_79063706_79063857 | 0.31 |
ENSG00000234456 |
. |
18417 |
0.22 |
| chr2_40049399_40049550 | 0.31 |
SLC8A1-AS1 |
SLC8A1 antisense RNA 1 |
35881 |
0.19 |
| chrX_131763564_131763715 | 0.31 |
HS6ST2-AS1 |
HS6ST2 antisense RNA 1 |
38031 |
0.17 |
| chr6_106549276_106549427 | 0.30 |
RP1-134E15.3 |
|
1336 |
0.46 |
| chr3_24225373_24225524 | 0.30 |
THRB |
thyroid hormone receptor, beta |
18362 |
0.28 |
| chr5_165288349_165288500 | 0.30 |
ENSG00000252794 |
. |
251978 |
0.02 |
| chr15_24112084_24112235 | 0.30 |
ENSG00000206616 |
. |
89469 |
0.09 |
| chr22_46462327_46462478 | 0.30 |
RP6-109B7.4 |
|
3369 |
0.13 |
| chr16_75836248_75836399 | 0.30 |
ENSG00000199299 |
. |
58732 |
0.12 |
| chr1_154358065_154358416 | 0.30 |
IL6R |
interleukin 6 receptor |
19429 |
0.11 |
| chr4_75224942_75225093 | 0.30 |
EREG |
epiregulin |
5843 |
0.21 |
| chr13_79182716_79183828 | 0.30 |
POU4F1 |
POU class 4 homeobox 1 |
5599 |
0.19 |
| chr7_25902294_25902669 | 0.30 |
ENSG00000199085 |
. |
87125 |
0.1 |
| chr4_20261154_20261305 | 0.30 |
SLIT2 |
slit homolog 2 (Drosophila) |
4686 |
0.35 |
| chr13_82572321_82572472 | 0.29 |
ENSG00000222486 |
. |
732514 |
0.0 |
| chr16_4146823_4146974 | 0.29 |
ADCY9 |
adenylate cyclase 9 |
17129 |
0.21 |
| chr8_67524217_67524427 | 0.29 |
MYBL1 |
v-myb avian myeloblastosis viral oncogene homolog-like 1 |
820 |
0.61 |
| chr7_54625683_54625834 | 0.29 |
GS1-18A18.1 |
|
1095 |
0.57 |
| chr12_126874132_126874719 | 0.29 |
TMEM132B |
transmembrane protein 132B |
767383 |
0.0 |
| chr5_100224849_100225000 | 0.29 |
ST8SIA4 |
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
13994 |
0.28 |
| chr15_50977663_50977814 | 0.29 |
TRPM7 |
transient receptor potential cation channel, subfamily M, member 7 |
1230 |
0.45 |
| chr3_107850710_107850861 | 0.29 |
CD47 |
CD47 molecule |
40913 |
0.19 |
| chrX_141646229_141646380 | 0.29 |
ENSG00000266319 |
. |
48427 |
0.19 |
| chr9_12772195_12772422 | 0.29 |
LURAP1L |
leucine rich adaptor protein 1-like |
2712 |
0.28 |
| chr5_66445517_66445857 | 0.29 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
7425 |
0.28 |
| chr6_66722316_66722467 | 0.29 |
EYS |
eyes shut homolog (Drosophila) |
305273 |
0.01 |
| chr10_60495190_60495341 | 0.29 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
58030 |
0.16 |
| chr12_78337025_78337244 | 0.29 |
NAV3 |
neuron navigator 3 |
22922 |
0.28 |
| chr2_120041983_120042165 | 0.29 |
STEAP3-AS1 |
STEAP3 antisense RNA 1 |
35427 |
0.15 |
| chr7_28133742_28133893 | 0.29 |
JAZF1 |
JAZF zinc finger 1 |
22514 |
0.2 |
| chr12_55266839_55266990 | 0.29 |
MUCL1 |
mucin-like 1 |
18593 |
0.23 |
| chr4_140808579_140808865 | 0.29 |
MAML3 |
mastermind-like 3 (Drosophila) |
2484 |
0.4 |
| chr9_123956593_123956744 | 0.29 |
RAB14 |
RAB14, member RAS oncogene family |
7479 |
0.16 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0006883 | cellular sodium ion homeostasis(GO:0006883) |
| 0.1 | 0.4 | GO:0032714 | negative regulation of interleukin-5 production(GO:0032714) |
| 0.1 | 0.4 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.4 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.3 | GO:0010459 | negative regulation of heart rate(GO:0010459) |
| 0.1 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.2 | GO:0055098 | response to low-density lipoprotein particle(GO:0055098) |
| 0.1 | 0.2 | GO:0010613 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) |
| 0.1 | 0.2 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.2 | GO:0009822 | alkaloid catabolic process(GO:0009822) |
| 0.1 | 0.1 | GO:0019049 | evasion or tolerance of host defenses by virus(GO:0019049) avoidance of host defenses(GO:0044413) evasion or tolerance of host defenses(GO:0044415) avoidance of defenses of other organism involved in symbiotic interaction(GO:0051832) evasion or tolerance of defenses of other organism involved in symbiotic interaction(GO:0051834) |
| 0.1 | 0.2 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.1 | 0.4 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.1 | GO:0060197 | cloaca development(GO:0035844) cloacal septation(GO:0060197) |
| 0.1 | 0.3 | GO:0033084 | regulation of immature T cell proliferation in thymus(GO:0033084) |
| 0.1 | 0.3 | GO:0010667 | striated muscle cell apoptotic process(GO:0010658) cardiac muscle cell apoptotic process(GO:0010659) regulation of striated muscle cell apoptotic process(GO:0010662) negative regulation of striated muscle cell apoptotic process(GO:0010664) regulation of cardiac muscle cell apoptotic process(GO:0010665) negative regulation of cardiac muscle cell apoptotic process(GO:0010667) |
| 0.1 | 0.3 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.1 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.1 | 0.3 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.1 | 0.1 | GO:0032674 | interleukin-5 production(GO:0032634) regulation of interleukin-5 production(GO:0032674) |
| 0.1 | 0.2 | GO:0031223 | auditory behavior(GO:0031223) |
| 0.1 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.1 | 0.2 | GO:0031946 | regulation of glucocorticoid biosynthetic process(GO:0031946) |
| 0.1 | 0.3 | GO:0034331 | cell junction maintenance(GO:0034331) cell-cell junction maintenance(GO:0045217) |
| 0.1 | 0.1 | GO:0032713 | negative regulation of interleukin-4 production(GO:0032713) |
| 0.0 | 0.6 | GO:0014037 | Schwann cell differentiation(GO:0014037) |
| 0.0 | 0.1 | GO:0014889 | muscle atrophy(GO:0014889) |
| 0.0 | 0.2 | GO:0045898 | regulation of RNA polymerase II transcriptional preinitiation complex assembly(GO:0045898) |
| 0.0 | 0.1 | GO:0070254 | mucus secretion(GO:0070254) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.2 | GO:0090218 | positive regulation of lipid kinase activity(GO:0090218) |
| 0.0 | 0.2 | GO:0019852 | L-ascorbic acid metabolic process(GO:0019852) |
| 0.0 | 0.1 | GO:0010159 | specification of organ position(GO:0010159) |
| 0.0 | 0.1 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.1 | GO:0045589 | regulation of regulatory T cell differentiation(GO:0045589) |
| 0.0 | 0.1 | GO:0070141 | response to UV-A(GO:0070141) |
| 0.0 | 0.1 | GO:0015820 | branched-chain amino acid transport(GO:0015803) leucine transport(GO:0015820) |
| 0.0 | 0.1 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.1 | GO:0045002 | DNA double-strand break processing involved in repair via single-strand annealing(GO:0010792) double-strand break repair via single-strand annealing(GO:0045002) |
| 0.0 | 0.3 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.0 | GO:0001705 | ectoderm formation(GO:0001705) |
| 0.0 | 0.2 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.0 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0045066 | regulatory T cell differentiation(GO:0045066) |
| 0.0 | 0.2 | GO:0055091 | phospholipid homeostasis(GO:0055091) |
| 0.0 | 0.0 | GO:0060463 | lung lobe development(GO:0060462) lung lobe morphogenesis(GO:0060463) |
| 0.0 | 0.3 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.1 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.2 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.0 | 0.1 | GO:0006925 | inflammatory cell apoptotic process(GO:0006925) |
| 0.0 | 0.1 | GO:1904251 | regulation of bile acid biosynthetic process(GO:0070857) regulation of bile acid metabolic process(GO:1904251) |
| 0.0 | 0.1 | GO:0072048 | pattern specification involved in kidney development(GO:0061004) renal system pattern specification(GO:0072048) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.0 | GO:0046532 | regulation of photoreceptor cell differentiation(GO:0046532) |
| 0.0 | 0.1 | GO:0071436 | sodium ion export(GO:0071436) |
| 0.0 | 0.1 | GO:0003181 | atrioventricular valve development(GO:0003171) atrioventricular valve morphogenesis(GO:0003181) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0097576 | vacuole fusion(GO:0097576) |
| 0.0 | 0.2 | GO:0033327 | Leydig cell differentiation(GO:0033327) |
| 0.0 | 0.4 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.0 | GO:0045077 | negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.0 | GO:0048520 | positive regulation of behavior(GO:0048520) |
| 0.0 | 0.1 | GO:0006273 | lagging strand elongation(GO:0006273) |
| 0.0 | 0.3 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0042474 | middle ear morphogenesis(GO:0042474) |
| 0.0 | 0.0 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.1 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:0045007 | depurination(GO:0045007) |
| 0.0 | 0.0 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0010761 | fibroblast migration(GO:0010761) |
| 0.0 | 0.1 | GO:0003214 | cardiac left ventricle morphogenesis(GO:0003214) |
| 0.0 | 0.0 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0006828 | manganese ion transport(GO:0006828) |
| 0.0 | 0.0 | GO:0002887 | negative regulation of myeloid leukocyte mediated immunity(GO:0002887) negative regulation of leukocyte degranulation(GO:0043301) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.0 | GO:0033025 | mast cell homeostasis(GO:0033023) mast cell apoptotic process(GO:0033024) regulation of mast cell apoptotic process(GO:0033025) |
| 0.0 | 0.1 | GO:0045743 | positive regulation of fibroblast growth factor receptor signaling pathway(GO:0045743) |
| 0.0 | 0.6 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0045920 | negative regulation of exocytosis(GO:0045920) |
| 0.0 | 0.1 | GO:0021873 | forebrain neuroblast division(GO:0021873) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.2 | GO:0048207 | vesicle targeting, rough ER to cis-Golgi(GO:0048207) COPII vesicle coating(GO:0048208) COPII-coated vesicle budding(GO:0090114) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0033631 | cell-cell adhesion mediated by integrin(GO:0033631) |
| 0.0 | 0.1 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0055057 | neuronal stem cell division(GO:0036445) neuroblast division(GO:0055057) |
| 0.0 | 0.1 | GO:0015742 | alpha-ketoglutarate transport(GO:0015742) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.0 | GO:0051284 | negative regulation of ion transmembrane transporter activity(GO:0032413) negative regulation of release of sequestered calcium ion into cytosol(GO:0051280) positive regulation of sequestering of calcium ion(GO:0051284) negative regulation of calcium ion transmembrane transport(GO:1903170) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.1 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.1 | GO:0007132 | meiotic metaphase I(GO:0007132) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0070932 | histone H3 deacetylation(GO:0070932) histone H4 deacetylation(GO:0070933) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:0061154 | endothelial tube morphogenesis(GO:0061154) |
| 0.0 | 0.0 | GO:0060586 | multicellular organismal iron ion homeostasis(GO:0060586) |
| 0.0 | 0.1 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:0070172 | positive regulation of tooth mineralization(GO:0070172) |
| 0.0 | 0.1 | GO:0045780 | positive regulation of bone resorption(GO:0045780) positive regulation of bone remodeling(GO:0046852) |
| 0.0 | 0.1 | GO:0016115 | diterpenoid catabolic process(GO:0016103) terpenoid catabolic process(GO:0016115) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.1 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.0 | GO:0007207 | phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0060684 | epithelial-mesenchymal cell signaling(GO:0060684) |
| 0.0 | 0.0 | GO:0021615 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0046426 | negative regulation of JAK-STAT cascade(GO:0046426) negative regulation of STAT cascade(GO:1904893) |
| 0.0 | 0.0 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) positive regulation of cell junction assembly(GO:1901890) positive regulation of adherens junction organization(GO:1903393) |
| 0.0 | 0.1 | GO:0090292 | nuclear matrix organization(GO:0043578) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.0 | 0.1 | GO:0046337 | phosphatidylethanolamine biosynthetic process(GO:0006646) phosphatidylethanolamine metabolic process(GO:0046337) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.0 | GO:0061117 | negative regulation of cardiac muscle tissue growth(GO:0055022) negative regulation of cardiac muscle tissue development(GO:0055026) negative regulation of cardiac muscle cell proliferation(GO:0060044) negative regulation of heart growth(GO:0061117) |
| 0.0 | 0.1 | GO:0001945 | lymph vessel development(GO:0001945) |
| 0.0 | 0.0 | GO:0001743 | optic placode formation(GO:0001743) |
| 0.0 | 0.1 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.1 | GO:0070296 | sarcoplasmic reticulum calcium ion transport(GO:0070296) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0010508 | positive regulation of autophagy(GO:0010508) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.0 | GO:0070120 | ciliary neurotrophic factor-mediated signaling pathway(GO:0070120) |
| 0.0 | 0.1 | GO:0071800 | podosome assembly(GO:0071800) |
| 0.0 | 0.0 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.0 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.2 | GO:0006949 | syncytium formation(GO:0006949) |
| 0.0 | 0.2 | GO:0048008 | platelet-derived growth factor receptor signaling pathway(GO:0048008) |
| 0.0 | 0.1 | GO:0048841 | regulation of axon extension involved in axon guidance(GO:0048841) |
| 0.0 | 0.1 | GO:0050916 | sensory perception of sweet taste(GO:0050916) |
| 0.0 | 0.1 | GO:0051963 | regulation of synapse assembly(GO:0051963) |
| 0.0 | 0.0 | GO:0070977 | organ maturation(GO:0048799) bone maturation(GO:0070977) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.0 | GO:0072112 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell differentiation(GO:0072112) glomerular epithelial cell differentiation(GO:0072311) |
| 0.0 | 0.1 | GO:0055075 | potassium ion homeostasis(GO:0055075) |
| 0.0 | 0.0 | GO:0002911 | lymphocyte anergy(GO:0002249) regulation of T cell anergy(GO:0002667) positive regulation of T cell anergy(GO:0002669) T cell anergy(GO:0002870) regulation of lymphocyte anergy(GO:0002911) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.0 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.0 | GO:0090286 | cytoskeletal anchoring at nuclear membrane(GO:0090286) |
| 0.0 | 0.1 | GO:0021904 | dorsal/ventral neural tube patterning(GO:0021904) |
| 0.0 | 0.0 | GO:0061213 | positive regulation of mesonephros development(GO:0061213) positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.0 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.0 | GO:0006927 | obsolete transformed cell apoptotic process(GO:0006927) |
| 0.0 | 0.1 | GO:0060347 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.0 | GO:2000400 | positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.2 | GO:0000038 | very long-chain fatty acid metabolic process(GO:0000038) |
| 0.0 | 0.4 | GO:0007602 | phototransduction(GO:0007602) |
| 0.0 | 0.1 | GO:0009103 | lipopolysaccharide biosynthetic process(GO:0009103) |
| 0.0 | 0.1 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.1 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.0 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.2 | GO:0000737 | DNA catabolic process, endonucleolytic(GO:0000737) apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 0.0 | GO:0060117 | auditory receptor cell development(GO:0060117) |
| 0.0 | 0.0 | GO:0060535 | trachea morphogenesis(GO:0060439) trachea cartilage morphogenesis(GO:0060535) cartilage morphogenesis(GO:0060536) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.0 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0000381 | regulation of alternative mRNA splicing, via spliceosome(GO:0000381) |
| 0.0 | 0.0 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0021546 | rhombomere development(GO:0021546) |
| 0.0 | 0.1 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.0 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.0 | GO:0036315 | cellular response to sterol(GO:0036315) cellular response to cholesterol(GO:0071397) |
| 0.0 | 0.0 | GO:0031580 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.0 | GO:0060457 | negative regulation of digestive system process(GO:0060457) |
| 0.0 | 0.0 | GO:0090266 | positive regulation of spindle checkpoint(GO:0090232) regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) positive regulation of cell cycle checkpoint(GO:1901978) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.1 | GO:0042797 | 5S class rRNA transcription from RNA polymerase III type 1 promoter(GO:0042791) tRNA transcription from RNA polymerase III promoter(GO:0042797) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.2 | GO:0043488 | regulation of mRNA stability(GO:0043488) |
| 0.0 | 0.1 | GO:0043113 | receptor clustering(GO:0043113) |
| 0.0 | 0.1 | GO:0048240 | sperm capacitation(GO:0048240) |
| 0.0 | 0.0 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0032730 | regulation of interleukin-1 alpha production(GO:0032650) positive regulation of interleukin-1 alpha production(GO:0032730) |
| 0.0 | 0.0 | GO:0032825 | regulation of natural killer cell differentiation(GO:0032823) positive regulation of natural killer cell differentiation(GO:0032825) |
| 0.0 | 0.0 | GO:0034244 | negative regulation of DNA-templated transcription, elongation(GO:0032785) negative regulation of transcription elongation from RNA polymerase II promoter(GO:0034244) |
| 0.0 | 0.0 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.1 | GO:0001954 | positive regulation of cell-matrix adhesion(GO:0001954) |
| 0.0 | 0.0 | GO:0019532 | oxalate transport(GO:0019532) |
| 0.0 | 0.4 | GO:0007193 | adenylate cyclase-inhibiting G-protein coupled receptor signaling pathway(GO:0007193) |
| 0.0 | 0.0 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0030325 | adrenal gland development(GO:0030325) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.2 | GO:0043259 | laminin-10 complex(GO:0043259) |
| 0.1 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0005900 | oncostatin-M receptor complex(GO:0005900) |
| 0.0 | 0.3 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.2 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.2 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.1 | GO:0017059 | palmitoyltransferase complex(GO:0002178) serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.2 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.2 | GO:0031233 | intrinsic component of external side of plasma membrane(GO:0031233) |
| 0.0 | 0.1 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.0 | 0.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 0.1 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.0 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0033268 | node of Ranvier(GO:0033268) |
| 0.0 | 0.4 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.1 | GO:0071778 | obsolete WINAC complex(GO:0071778) |
| 0.0 | 0.1 | GO:0034993 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.0 | 0.1 | GO:0031265 | CD95 death-inducing signaling complex(GO:0031265) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.2 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0043256 | laminin complex(GO:0043256) |
| 0.0 | 0.1 | GO:0005682 | U5 snRNP(GO:0005682) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.0 | GO:0043240 | Fanconi anaemia nuclear complex(GO:0043240) |
| 0.0 | 0.1 | GO:0034045 | pre-autophagosomal structure membrane(GO:0034045) |
| 0.0 | 0.1 | GO:0090533 | cation-transporting ATPase complex(GO:0090533) |
| 0.0 | 0.1 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.0 | GO:0072669 | tRNA-splicing ligase complex(GO:0072669) |
| 0.0 | 0.2 | GO:0000299 | obsolete integral to membrane of membrane fraction(GO:0000299) |
| 0.0 | 0.0 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.1 | GO:0000127 | transcription factor TFIIIC complex(GO:0000127) |
| 0.0 | 0.2 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0035253 | ciliary rootlet(GO:0035253) |
| 0.0 | 0.0 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0031010 | ISWI-type complex(GO:0031010) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 0.3 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.2 | GO:0003953 | NAD+ nucleosidase activity(GO:0003953) |
| 0.1 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.1 | 0.5 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 0.1 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.2 | GO:0031705 | bombesin receptor binding(GO:0031705) endothelin B receptor binding(GO:0031708) |
| 0.1 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.1 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.1 | 0.3 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.1 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.5 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.4 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.2 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.3 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0055103 | ligase regulator activity(GO:0055103) ubiquitin-protein transferase regulator activity(GO:0055106) |
| 0.0 | 0.2 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.2 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0005384 | manganese ion transmembrane transporter activity(GO:0005384) |
| 0.0 | 0.1 | GO:0008900 | hydrogen:potassium-exchanging ATPase activity(GO:0008900) |
| 0.0 | 0.0 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.2 | GO:0000832 | inositol hexakisphosphate kinase activity(GO:0000828) inositol hexakisphosphate 5-kinase activity(GO:0000832) |
| 0.0 | 0.1 | GO:0031782 | melanocortin receptor binding(GO:0031779) type 3 melanocortin receptor binding(GO:0031781) type 4 melanocortin receptor binding(GO:0031782) |
| 0.0 | 0.1 | GO:0016454 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.3 | GO:0034593 | phosphatidylinositol bisphosphate phosphatase activity(GO:0034593) |
| 0.0 | 0.1 | GO:0015271 | outward rectifier potassium channel activity(GO:0015271) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.2 | GO:0004726 | non-membrane spanning protein tyrosine phosphatase activity(GO:0004726) |
| 0.0 | 0.3 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.2 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.0 | GO:0004104 | cholinesterase activity(GO:0004104) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.5 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.1 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.1 | GO:0004305 | ethanolamine kinase activity(GO:0004305) |
| 0.0 | 0.1 | GO:0031702 | type 1 angiotensin receptor binding(GO:0031702) |
| 0.0 | 0.2 | GO:0015385 | sodium:proton antiporter activity(GO:0015385) |
| 0.0 | 0.1 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.3 | GO:0043498 | obsolete cell surface binding(GO:0043498) |
| 0.0 | 0.1 | GO:0030284 | estrogen receptor activity(GO:0030284) |
| 0.0 | 0.1 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.1 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.0 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0035184 | histone threonine kinase activity(GO:0035184) |
| 0.0 | 0.2 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.1 | GO:0043295 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.2 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.0 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.1 | GO:0031821 | G-protein coupled serotonin receptor binding(GO:0031821) |
| 0.0 | 0.0 | GO:0004897 | ciliary neurotrophic factor receptor activity(GO:0004897) |
| 0.0 | 0.0 | GO:0044548 | S100 protein binding(GO:0044548) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.0 | GO:0030235 | nitric-oxide synthase regulator activity(GO:0030235) |
| 0.0 | 0.1 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0004470 | malic enzyme activity(GO:0004470) |
| 0.0 | 0.0 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.2 | GO:0032266 | phosphatidylinositol-3-phosphate binding(GO:0032266) |
| 0.0 | 0.3 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.0 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004594 | pantothenate kinase activity(GO:0004594) |
| 0.0 | 0.1 | GO:0070053 | thrombospondin receptor activity(GO:0070053) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0004083 | bisphosphoglycerate mutase activity(GO:0004082) bisphosphoglycerate 2-phosphatase activity(GO:0004083) phosphoglycerate mutase activity(GO:0004619) |
| 0.0 | 0.1 | GO:0015925 | galactosidase activity(GO:0015925) |
| 0.0 | 0.1 | GO:0000014 | single-stranded DNA endodeoxyribonuclease activity(GO:0000014) |
| 0.0 | 0.4 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 0.0 | GO:0032450 | maltose alpha-glucosidase activity(GO:0032450) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.2 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.1 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0016857 | racemase and epimerase activity, acting on carbohydrates and derivatives(GO:0016857) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.0 | GO:0034618 | arginine binding(GO:0034618) |
| 0.0 | 0.2 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.1 | GO:0004016 | adenylate cyclase activity(GO:0004016) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.0 | GO:0001601 | peptide YY receptor activity(GO:0001601) |
| 0.0 | 0.1 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.1 | GO:0033613 | activating transcription factor binding(GO:0033613) |
| 0.0 | 0.0 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) |
| 0.0 | 0.1 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.0 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.2 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.2 | GO:0045296 | cadherin binding(GO:0045296) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.7 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.3 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 0.0 | ST INTERLEUKIN 4 PATHWAY | Interleukin 4 (IL-4) Pathway |
| 0.0 | 0.9 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.9 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.5 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.8 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.0 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.2 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.1 | PID PI3KCI PATHWAY | Class I PI3K signaling events |
| 0.0 | 0.2 | NABA BASEMENT MEMBRANES | Genes encoding structural components of basement membranes |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.0 | PID IL2 1PATHWAY | IL2-mediated signaling events |
| 0.0 | 0.4 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.1 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.0 | PID TCPTP PATHWAY | Signaling events mediated by TCPTP |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.4 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 0.3 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.3 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.2 | REACTOME SIGNALLING BY NGF | Genes involved in Signalling by NGF |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME SIGNALING BY EGFR IN CANCER | Genes involved in Signaling by EGFR in Cancer |
| 0.0 | 0.3 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.0 | REACTOME REGULATION OF APOPTOSIS | Genes involved in Regulation of Apoptosis |
| 0.0 | 0.4 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.3 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.0 | 0.4 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.0 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE GOLGI MEMBRANE | Genes involved in Synthesis of PIPs at the Golgi membrane |
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.2 | REACTOME PEPTIDE HORMONE BIOSYNTHESIS | Genes involved in Peptide hormone biosynthesis |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.0 | REACTOME INHIBITION OF INSULIN SECRETION BY ADRENALINE NORADRENALINE | Genes involved in Inhibition of Insulin Secretion by Adrenaline/Noradrenaline |
| 0.0 | 0.0 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.2 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.1 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME SIGNALING BY ROBO RECEPTOR | Genes involved in Signaling by Robo receptor |
| 0.0 | 0.2 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.0 | REACTOME DOWNSTREAM TCR SIGNALING | Genes involved in Downstream TCR signaling |
| 0.0 | 0.0 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 0.1 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.0 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.7 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 0.3 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.2 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.1 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.3 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |