Project
ENCODE: H3K4me3 ChIP-Seq of primary human cells
Navigation
Downloads
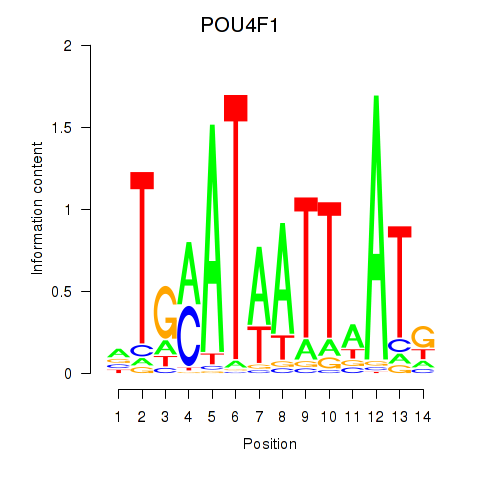
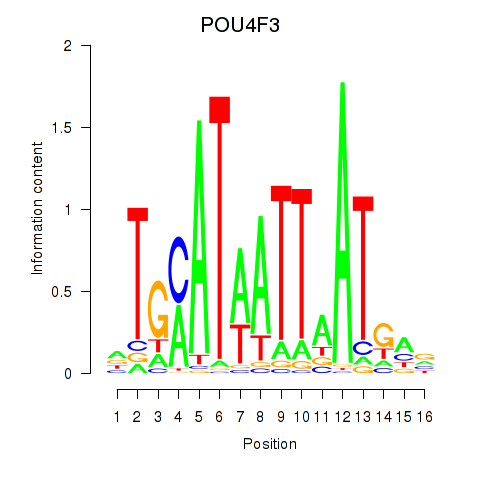
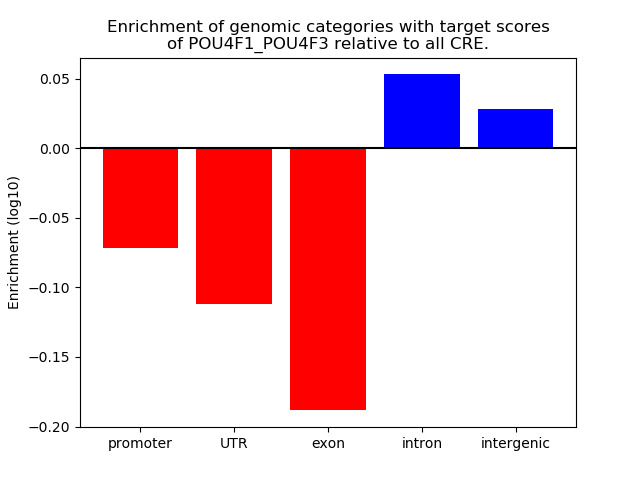
Results for POU4F1_POU4F3
Z-value: 1.20
Transcription factors associated with POU4F1_POU4F3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
POU4F1
|
ENSG00000152192.6 | POU class 4 homeobox 1 |
|
POU4F3
|
ENSG00000091010.4 | POU class 4 homeobox 3 |
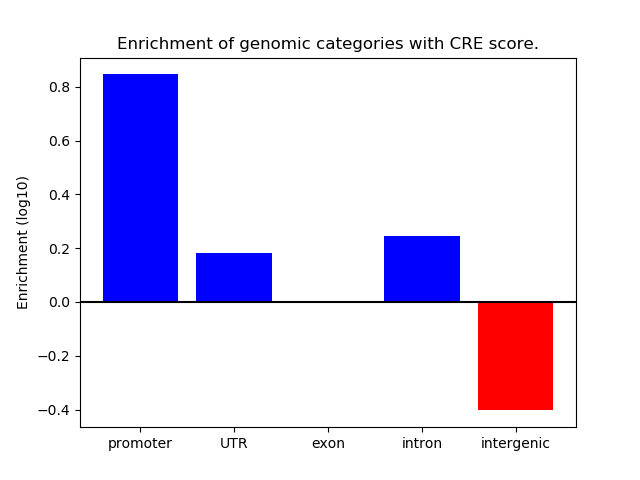
Correlations of motif activity and signal intensity at CREs associated with the motif's TFs:
This plot shows correlation between observed signal intensity of a CRE associated with the transcription factor across all samples and activity of the motif.
For each TF, only the top 5 correlated CREs are shown.
| CRE | Gene | Distance | Association probability | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|---|---|
| chr13_79182716_79183828 | POU4F1 | 5599 | 0.193498 | 0.55 | 1.2e-01 | Click! |
| chr13_79181751_79181902 | POU4F1 | 4153 | 0.209565 | 0.53 | 1.4e-01 | Click! |
| chr13_79182558_79182709 | POU4F1 | 4960 | 0.199043 | 0.47 | 2.0e-01 | Click! |
| chr13_79181975_79182128 | POU4F1 | 4378 | 0.206092 | 0.45 | 2.2e-01 | Click! |
| chr13_79176958_79177133 | POU4F1 | 628 | 0.709557 | -0.32 | 3.9e-01 | Click! |
| chr5_145719881_145720125 | POU4F3 | 1416 | 0.467441 | 0.79 | 1.1e-02 | Click! |
| chr5_145724296_145724632 | POU4F3 | 5877 | 0.231468 | 0.60 | 9.0e-02 | Click! |
| chr5_145725536_145725687 | POU4F3 | 7024 | 0.223641 | 0.38 | 3.2e-01 | Click! |
| chr5_145725300_145725451 | POU4F3 | 6788 | 0.225019 | -0.08 | 8.4e-01 | Click! |
Activity of the POU4F1_POU4F3 motif across conditions
Conditions sorted by the z-value of the POU4F1_POU4F3 motif activity
Move your cursor over a bar to see sample name and corresponding Z-value.
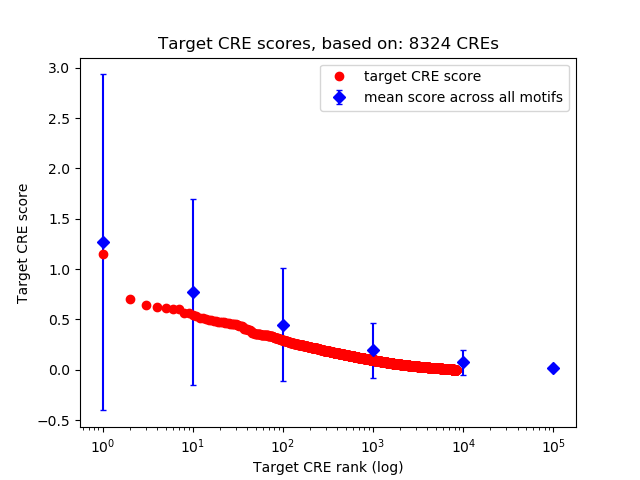
Top target CREs of the motif:
| Cis Regulatory Element (CRE) | Target Score | Top associated gene | Gene Info | Distance of CRE to TSS | CRE/Gene association probability |
|---|---|---|---|---|---|
| chr6_116833589_116833880 | 1.15 |
TRAPPC3L |
trafficking protein particle complex 3-like |
228 |
0.83 |
| chr12_91574306_91575101 | 0.70 |
DCN |
decorin |
50 |
0.99 |
| chr7_18549761_18549974 | 0.65 |
HDAC9 |
histone deacetylase 9 |
931 |
0.71 |
| chr9_22008020_22008450 | 0.62 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
717 |
0.61 |
| chr17_46646430_46646725 | 0.62 |
HOXB3 |
homeobox B3 |
4808 |
0.08 |
| chr18_811181_811332 | 0.61 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
983 |
0.49 |
| chr11_111849006_111849202 | 0.60 |
DIXDC1 |
DIX domain containing 1 |
1071 |
0.39 |
| chr4_108746841_108747185 | 0.57 |
SGMS2 |
sphingomyelin synthase 2 |
649 |
0.76 |
| chr2_18481003_18481326 | 0.56 |
ENSG00000212455 |
. |
259165 |
0.02 |
| chr1_162603392_162603656 | 0.55 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
1264 |
0.48 |
| chr2_36581700_36582677 | 0.53 |
CRIM1 |
cysteine rich transmembrane BMP regulator 1 (chordin-like) |
881 |
0.74 |
| chr6_100717098_100717249 | 0.51 |
RP1-121G13.2 |
|
157821 |
0.04 |
| chr11_86085513_86085741 | 0.51 |
CCDC81 |
coiled-coil domain containing 81 |
151 |
0.96 |
| chr9_89952337_89952906 | 0.51 |
ENSG00000212421 |
. |
77256 |
0.11 |
| chr1_88928130_88928341 | 0.50 |
ENSG00000239504 |
. |
15562 |
0.29 |
| chr11_68036812_68036963 | 0.50 |
C11orf24 |
chromosome 11 open reading frame 24 |
2528 |
0.3 |
| chr10_49812568_49813319 | 0.48 |
ARHGAP22 |
Rho GTPase activating protein 22 |
54 |
0.98 |
| chr6_47446841_47447070 | 0.48 |
CD2AP |
CD2-associated protein |
1430 |
0.42 |
| chr12_52258184_52258357 | 0.48 |
ANKRD33 |
ankyrin repeat domain 33 |
23474 |
0.16 |
| chr22_47241536_47241796 | 0.48 |
ENSG00000221672 |
. |
2137 |
0.4 |
| chr15_48010788_48011252 | 0.48 |
SEMA6D |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D |
334 |
0.93 |
| chr15_63050120_63050513 | 0.47 |
TLN2 |
talin 2 |
473 |
0.82 |
| chr9_97812256_97812766 | 0.47 |
ENSG00000207563 |
. |
34979 |
0.12 |
| chr13_80912266_80912417 | 0.46 |
SPRY2 |
sprouty homolog 2 (Drosophila) |
1453 |
0.53 |
| chr18_7572735_7572928 | 0.46 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
5014 |
0.33 |
| chr7_82070146_82070432 | 0.46 |
CACNA2D1 |
calcium channel, voltage-dependent, alpha 2/delta subunit 1 |
2742 |
0.43 |
| chr12_115119923_115120074 | 0.46 |
TBX3 |
T-box 3 |
1397 |
0.48 |
| chr16_27590300_27590451 | 0.46 |
KIAA0556 |
KIAA0556 |
5147 |
0.22 |
| chr5_141701461_141701673 | 0.46 |
SPRY4 |
sprouty homolog 4 (Drosophila) |
2180 |
0.34 |
| chr13_76213551_76213793 | 0.45 |
LMO7 |
LIM domain 7 |
3213 |
0.22 |
| chr6_152701966_152702349 | 0.45 |
SYNE1-AS1 |
SYNE1 antisense RNA 1 |
476 |
0.79 |
| chr3_114477857_114478359 | 0.44 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
10 |
0.99 |
| chr2_200446759_200446999 | 0.44 |
SATB2 |
SATB homeobox 2 |
110890 |
0.07 |
| chr3_79815928_79816150 | 0.43 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
926 |
0.74 |
| chr6_151597751_151597996 | 0.43 |
ENSG00000252431 |
. |
3874 |
0.18 |
| chr7_123389891_123390042 | 0.42 |
RP11-390E23.6 |
|
844 |
0.4 |
| chr5_102202312_102202463 | 0.41 |
PAM |
peptidylglycine alpha-amidating monooxygenase |
567 |
0.87 |
| chr12_71054468_71054762 | 0.41 |
PTPRB |
protein tyrosine phosphatase, receptor type, B |
23395 |
0.24 |
| chr1_95389567_95389941 | 0.41 |
CNN3 |
calponin 3, acidic |
1583 |
0.36 |
| chr8_97468793_97469144 | 0.40 |
ENSG00000202095 |
. |
25580 |
0.2 |
| chr4_183413334_183413747 | 0.40 |
TENM3 |
teneurin transmembrane protein 3 |
43388 |
0.17 |
| chr19_55678355_55678541 | 0.39 |
ENSG00000239137 |
. |
91 |
0.81 |
| chr6_72178134_72178480 | 0.39 |
ENSG00000207827 |
. |
64983 |
0.11 |
| chr4_157891142_157892336 | 0.38 |
PDGFC |
platelet derived growth factor C |
316 |
0.91 |
| chr1_200148726_200148877 | 0.37 |
ENSG00000221403 |
. |
34839 |
0.21 |
| chr3_13080776_13080942 | 0.36 |
IQSEC1 |
IQ motif and Sec7 domain 1 |
33758 |
0.22 |
| chr3_117454157_117454380 | 0.36 |
ENSG00000206889 |
. |
191216 |
0.03 |
| chrX_138909289_138909440 | 0.36 |
ATP11C |
ATPase, class VI, type 11C |
5083 |
0.25 |
| chr2_26624476_26625174 | 0.36 |
DRC1 |
dynein regulatory complex subunit 1 homolog (Chlamydomonas) |
41 |
0.98 |
| chr7_12811096_12811403 | 0.36 |
ENSG00000199470 |
. |
70735 |
0.11 |
| chr17_8532417_8532568 | 0.36 |
MYH10 |
myosin, heavy chain 10, non-muscle |
1543 |
0.44 |
| chr1_61692922_61693140 | 0.36 |
RP4-802A10.1 |
|
102626 |
0.08 |
| chr4_114897314_114897610 | 0.36 |
ARSJ |
arylsulfatase family, member J |
2690 |
0.37 |
| chr1_196620715_196621368 | 0.35 |
CFH |
complement factor H |
33 |
0.98 |
| chr2_242257443_242257594 | 0.35 |
HDLBP |
high density lipoprotein binding protein |
1042 |
0.41 |
| chr9_84947315_84947575 | 0.35 |
SPATA31D1 |
SPATA31 subfamily D, member 1 |
343758 |
0.01 |
| chrY_14922193_14922344 | 0.35 |
USP9Y |
ubiquitin specific peptidase 9, Y-linked |
36702 |
0.22 |
| chr12_91572375_91572546 | 0.35 |
DCN |
decorin |
131 |
0.98 |
| chr1_116089167_116089325 | 0.35 |
ENSG00000239984 |
. |
59846 |
0.11 |
| chr1_66737250_66737924 | 0.35 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
14257 |
0.29 |
| chr6_169653692_169653887 | 0.35 |
THBS2 |
thrombospondin 2 |
350 |
0.92 |
| chr11_100587578_100587729 | 0.35 |
CTD-2383M3.1 |
|
28967 |
0.17 |
| chr6_127840298_127840479 | 0.34 |
SOGA3 |
SOGA family member 3 |
52 |
0.98 |
| chr2_218193929_218194080 | 0.34 |
ENSG00000251982 |
. |
77725 |
0.11 |
| chr8_42072731_42073079 | 0.34 |
PLAT |
plasminogen activator, tissue |
7663 |
0.16 |
| chr6_152002769_152002982 | 0.34 |
ESR1 |
estrogen receptor 1 |
8756 |
0.26 |
| chr2_145787262_145787615 | 0.34 |
ENSG00000253036 |
. |
305200 |
0.01 |
| chr8_18870397_18870669 | 0.34 |
PSD3 |
pleckstrin and Sec7 domain containing 3 |
663 |
0.74 |
| chr13_74862980_74863131 | 0.34 |
ENSG00000206617 |
. |
296 |
0.95 |
| chr15_57512191_57512484 | 0.34 |
TCF12 |
transcription factor 12 |
673 |
0.79 |
| chr15_99788624_99789059 | 0.34 |
TTC23 |
tetratricopeptide repeat domain 23 |
895 |
0.51 |
| chr6_140373176_140373387 | 0.33 |
ENSG00000252107 |
. |
106550 |
0.08 |
| chr11_9587935_9588242 | 0.33 |
WEE1 |
WEE1 G2 checkpoint kinase |
7140 |
0.16 |
| chr2_202987834_202988283 | 0.33 |
AC079354.3 |
|
6277 |
0.17 |
| chr8_25317575_25317860 | 0.33 |
CDCA2 |
cell division cycle associated 2 |
1002 |
0.5 |
| chr2_226896266_226896823 | 0.33 |
NYAP2 |
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
622947 |
0.0 |
| chr10_63779003_63779237 | 0.33 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
29850 |
0.22 |
| chr7_66055880_66056152 | 0.33 |
KCTD7 |
potassium channel tetramerization domain containing 7 |
37852 |
0.16 |
| chr19_39186934_39187210 | 0.32 |
CTB-186G2.4 |
|
2826 |
0.16 |
| chr2_9348603_9349207 | 0.32 |
ASAP2 |
ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 |
2011 |
0.46 |
| chr15_52123506_52123657 | 0.32 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
1754 |
0.32 |
| chr2_62934247_62934470 | 0.32 |
EHBP1 |
EH domain binding protein 1 |
31 |
0.98 |
| chr9_96932049_96932529 | 0.32 |
ENSG00000199165 |
. |
5945 |
0.16 |
| chr18_9109419_9109570 | 0.32 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
5414 |
0.17 |
| chr1_78471558_78472034 | 0.32 |
DNAJB4 |
DnaJ (Hsp40) homolog, subfamily B, member 4 |
1258 |
0.34 |
| chr2_33516384_33516581 | 0.32 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
233 |
0.94 |
| chr18_53144293_53144444 | 0.31 |
ENSG00000264571 |
. |
2084 |
0.31 |
| chr15_37175433_37176047 | 0.31 |
ENSG00000212511 |
. |
30899 |
0.23 |
| chr21_18376217_18376955 | 0.31 |
ENSG00000239023 |
. |
285269 |
0.01 |
| chr6_109414275_109414426 | 0.31 |
SESN1 |
sestrin 1 |
1672 |
0.32 |
| chr13_33476928_33477100 | 0.31 |
KL |
klotho |
113193 |
0.06 |
| chr8_49831590_49831741 | 0.31 |
SNAI2 |
snail family zinc finger 2 |
2323 |
0.45 |
| chr5_10596142_10596297 | 0.31 |
ANKRD33B |
ankyrin repeat domain 33B |
31639 |
0.16 |
| chr1_8298719_8298899 | 0.30 |
ENSG00000200975 |
. |
32152 |
0.15 |
| chr17_79317268_79317735 | 0.30 |
TMEM105 |
transmembrane protein 105 |
13027 |
0.13 |
| chr18_53253489_53253863 | 0.30 |
TCF4 |
transcription factor 4 |
326 |
0.93 |
| chr15_70819174_70819403 | 0.30 |
UACA |
uveal autoantigen with coiled-coil domains and ankyrin repeats |
175332 |
0.03 |
| chr2_46928125_46928276 | 0.30 |
SOCS5 |
suppressor of cytokine signaling 5 |
1874 |
0.38 |
| chr2_219471971_219472122 | 0.30 |
PLCD4 |
phospholipase C, delta 4 |
442 |
0.69 |
| chr6_52925441_52925592 | 0.30 |
ICK |
intestinal cell (MAK-like) kinase |
1052 |
0.42 |
| chr3_185462581_185462876 | 0.29 |
ENSG00000265470 |
. |
22964 |
0.19 |
| chr2_216298275_216298426 | 0.29 |
FN1 |
fibronectin 1 |
2440 |
0.3 |
| chrX_17395799_17395950 | 0.29 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
2331 |
0.33 |
| chr5_33794654_33794915 | 0.29 |
ENSG00000201623 |
. |
93483 |
0.08 |
| chr3_60921693_60921844 | 0.29 |
ENSG00000212211 |
. |
79491 |
0.12 |
| chr12_60131988_60132146 | 0.29 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
30773 |
0.24 |
| chr7_151328064_151328292 | 0.29 |
PRKAG2 |
protein kinase, AMP-activated, gamma 2 non-catalytic subunit |
1261 |
0.55 |
| chr9_126270533_126270733 | 0.28 |
RP11-230L22.4 |
|
16470 |
0.24 |
| chr3_191217380_191217807 | 0.28 |
PYDC2 |
pyrin domain containing 2 |
38641 |
0.2 |
| chr4_77703399_77704071 | 0.28 |
RP11-359D14.3 |
|
19382 |
0.18 |
| chr4_53966969_53967196 | 0.28 |
RP11-752D24.2 |
|
155226 |
0.04 |
| chr3_189735394_189735583 | 0.28 |
ENSG00000265045 |
. |
96235 |
0.07 |
| chr15_77710162_77710461 | 0.28 |
PEAK1 |
pseudopodium-enriched atypical kinase 1 |
2131 |
0.32 |
| chr1_64240383_64241287 | 0.28 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
1121 |
0.63 |
| chr7_28726429_28726958 | 0.28 |
CREB5 |
cAMP responsive element binding protein 5 |
1095 |
0.68 |
| chr5_60598656_60599005 | 0.27 |
ZSWIM6 |
zinc finger, SWIM-type containing 6 |
29270 |
0.23 |
| chr2_170591300_170591489 | 0.27 |
KLHL23 |
kelch-like family member 23 |
668 |
0.64 |
| chr8_119086940_119087091 | 0.27 |
EXT1 |
exostosin glycosyltransferase 1 |
35638 |
0.24 |
| chr1_215447400_215447702 | 0.27 |
KCNK2 |
potassium channel, subfamily K, member 2 |
190691 |
0.03 |
| chr2_219435817_219435968 | 0.27 |
RQCD1 |
RCD1 required for cell differentiation1 homolog (S. pombe) |
2294 |
0.17 |
| chr11_46934289_46934467 | 0.27 |
LRP4 |
low density lipoprotein receptor-related protein 4 |
5795 |
0.19 |
| chr16_24697486_24697909 | 0.27 |
TNRC6A |
trinucleotide repeat containing 6A |
43319 |
0.18 |
| chr5_52707192_52707393 | 0.27 |
FST |
follistatin |
68947 |
0.13 |
| chr8_11382556_11382707 | 0.27 |
RP11-148O21.3 |
|
29085 |
0.11 |
| chr1_65730855_65731313 | 0.27 |
DNAJC6 |
DnaJ (Hsp40) homolog, subfamily C, member 6 |
647 |
0.77 |
| chr17_57230413_57230606 | 0.27 |
SKA2 |
spindle and kinetochore associated complex subunit 2 |
1347 |
0.23 |
| chr6_75913619_75913933 | 0.27 |
COL12A1 |
collagen, type XII, alpha 1 |
1268 |
0.52 |
| chr17_38708496_38708766 | 0.27 |
CCR7 |
chemokine (C-C motif) receptor 7 |
8634 |
0.17 |
| chr2_177418868_177419346 | 0.26 |
ENSG00000252027 |
. |
110297 |
0.06 |
| chr9_75169048_75169276 | 0.26 |
ENSG00000238402 |
. |
27010 |
0.21 |
| chr15_90543166_90543622 | 0.26 |
ZNF710 |
zinc finger protein 710 |
1230 |
0.4 |
| chr17_3572880_3573031 | 0.26 |
EMC6 |
ER membrane protein complex subunit 6 |
839 |
0.33 |
| chr3_9443823_9444009 | 0.26 |
SETD5-AS1 |
SETD5 antisense RNA 1 |
3653 |
0.18 |
| chr17_7156412_7156575 | 0.26 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
551 |
0.36 |
| chr9_16704056_16704545 | 0.26 |
BNC2 |
basonuclin 2 |
780 |
0.71 |
| chr20_17947911_17948977 | 0.26 |
SNX5 |
sorting nexin 5 |
657 |
0.53 |
| chr2_24676770_24677172 | 0.26 |
NCOA1 |
nuclear receptor coactivator 1 |
37830 |
0.18 |
| chr1_68149789_68149968 | 0.26 |
GADD45A |
growth arrest and DNA-damage-inducible, alpha |
866 |
0.65 |
| chr11_18368890_18369041 | 0.26 |
GTF2H1 |
general transcription factor IIH, polypeptide 1, 62kDa |
314 |
0.84 |
| chr4_170147421_170147572 | 0.26 |
RP11-327O17.2 |
|
24551 |
0.22 |
| chr6_113012091_113012242 | 0.26 |
ENSG00000252215 |
. |
158782 |
0.04 |
| chr8_38242197_38242597 | 0.25 |
LETM2 |
leucine zipper-EF-hand containing transmembrane protein 2 |
1328 |
0.3 |
| chr10_76716977_76717128 | 0.25 |
RP11-77G23.5 |
|
66957 |
0.1 |
| chr18_20558047_20558453 | 0.25 |
RBBP8 |
retinoblastoma binding protein 8 |
9445 |
0.15 |
| chr1_17766506_17766801 | 0.25 |
RCC2 |
regulator of chromosome condensation 2 |
433 |
0.8 |
| chr13_66190370_66190521 | 0.25 |
ENSG00000221685 |
. |
247860 |
0.02 |
| chr1_245329429_245329885 | 0.25 |
KIF26B |
kinesin family member 26B |
11370 |
0.2 |
| chr2_50371104_50371319 | 0.25 |
NRXN1 |
neurexin 1 |
124800 |
0.06 |
| chr8_15398472_15398721 | 0.25 |
TUSC3 |
tumor suppressor candidate 3 |
804 |
0.72 |
| chr10_119122644_119122795 | 0.25 |
ENSG00000222197 |
. |
4852 |
0.24 |
| chr8_119120675_119121215 | 0.25 |
EXT1 |
exostosin glycosyltransferase 1 |
1708 |
0.54 |
| chr17_14028855_14029146 | 0.25 |
ENSG00000252305 |
. |
51448 |
0.12 |
| chr8_118981794_118981945 | 0.25 |
EXT1 |
exostosin glycosyltransferase 1 |
140784 |
0.05 |
| chr3_150130220_150130998 | 0.25 |
TSC22D2 |
TSC22 domain family, member 2 |
1813 |
0.47 |
| chr15_38547811_38548278 | 0.25 |
SPRED1 |
sprouty-related, EVH1 domain containing 1 |
2662 |
0.42 |
| chr20_45114435_45114586 | 0.25 |
ZNF334 |
zinc finger protein 334 |
20499 |
0.19 |
| chr11_102039159_102039310 | 0.25 |
YAP1 |
Yes-associated protein 1 |
17578 |
0.19 |
| chr10_117930701_117930852 | 0.24 |
GFRA1 |
GDNF family receptor alpha 1 |
84531 |
0.1 |
| chr21_17792662_17793003 | 0.24 |
ENSG00000207638 |
. |
118577 |
0.06 |
| chr22_40673331_40673671 | 0.24 |
TNRC6B |
trinucleotide repeat containing 6B |
12494 |
0.23 |
| chr6_75316769_75316920 | 0.24 |
ENSG00000264884 |
. |
112715 |
0.07 |
| chr18_20139314_20139465 | 0.24 |
ENSG00000206827 |
. |
88156 |
0.09 |
| chr14_100260565_100260825 | 0.24 |
EML1 |
echinoderm microtubule associated protein like 1 |
923 |
0.67 |
| chr7_66120646_66120797 | 0.24 |
RP4-756H11.3 |
|
1148 |
0.52 |
| chr3_25193430_25193657 | 0.24 |
RARB |
retinoic acid receptor, beta |
22280 |
0.26 |
| chr4_74316638_74316988 | 0.24 |
AFP |
alpha-fetoprotein |
14849 |
0.2 |
| chr8_124281132_124281572 | 0.24 |
ZHX1-C8ORF76 |
ZHX1-C8ORF76 readthrough |
1725 |
0.21 |
| chr6_49294723_49295128 | 0.24 |
ENSG00000252457 |
. |
17596 |
0.27 |
| chr20_18446613_18446764 | 0.24 |
DZANK1 |
double zinc ribbon and ankyrin repeat domains 1 |
663 |
0.48 |
| chr6_27060216_27060434 | 0.24 |
ENSG00000222800 |
. |
17434 |
0.13 |
| chr5_54456050_54456451 | 0.24 |
GPX8 |
glutathione peroxidase 8 (putative) |
252 |
0.86 |
| chr18_60905231_60905454 | 0.24 |
ENSG00000238988 |
. |
43444 |
0.14 |
| chr11_82570104_82570255 | 0.24 |
PRCP |
prolylcarboxypeptidase (angiotensinase C) |
366 |
0.88 |
| chr6_45448601_45449137 | 0.24 |
RUNX2 |
runt-related transcription factor 2 |
58647 |
0.13 |
| chr7_44801800_44801951 | 0.24 |
ZMIZ2 |
zinc finger, MIZ-type containing 2 |
6071 |
0.17 |
| chr2_151091538_151091758 | 0.24 |
RND3 |
Rho family GTPase 3 |
250248 |
0.02 |
| chr17_46240634_46240785 | 0.24 |
ENSG00000221739 |
. |
6836 |
0.12 |
| chr7_80547175_80548098 | 0.24 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
863 |
0.76 |
| chr2_67620410_67620561 | 0.24 |
ETAA1 |
Ewing tumor-associated antigen 1 |
3966 |
0.38 |
| chr5_140940128_140940366 | 0.24 |
CTD-2024I7.13 |
|
2369 |
0.19 |
| chr9_81217104_81217255 | 0.23 |
PSAT1 |
phosphoserine aminotransferase 1 |
305120 |
0.01 |
| chr14_31464257_31464552 | 0.23 |
ENSG00000207952 |
. |
19544 |
0.17 |
| chr1_178836761_178836994 | 0.23 |
ANGPTL1 |
angiopoietin-like 1 |
1536 |
0.43 |
| chr10_79397825_79398297 | 0.23 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
66 |
0.98 |
| chr12_109195839_109196070 | 0.23 |
ENSG00000264043 |
. |
23907 |
0.13 |
| chr10_89917929_89918366 | 0.23 |
ENSG00000200891 |
. |
163695 |
0.04 |
| chr18_55889348_55889572 | 0.23 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
657 |
0.77 |
| chr15_88020923_88021173 | 0.23 |
ENSG00000207150 |
. |
3528 |
0.4 |
| chr14_95963717_95963868 | 0.23 |
RP11-1070N10.3 |
Uncharacterized protein |
18681 |
0.14 |
| chr11_89113858_89114276 | 0.23 |
NOX4 |
NADPH oxidase 4 |
109833 |
0.07 |
| chr1_110693882_110694033 | 0.23 |
SLC6A17 |
solute carrier family 6 (neutral amino acid transporter), member 17 |
849 |
0.49 |
| chr2_110366652_110366826 | 0.23 |
AC011753.5 |
|
3464 |
0.22 |
| chr2_70518346_70518567 | 0.23 |
SNRPG |
small nuclear ribonucleoprotein polypeptide G |
1452 |
0.31 |
| chr9_694286_694821 | 0.23 |
RP11-130C19.3 |
|
8998 |
0.22 |
| chr3_106059964_106060303 | 0.23 |
ENSG00000200610 |
. |
174611 |
0.04 |
| chr21_34918443_34918594 | 0.23 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
2721 |
0.18 |
| chr11_108777750_108777901 | 0.23 |
ENSG00000201243 |
. |
98419 |
0.08 |
| chr17_35851874_35852313 | 0.23 |
DUSP14 |
dual specificity phosphatase 14 |
523 |
0.79 |
| chr8_62621050_62621416 | 0.23 |
ASPH |
aspartate beta-hydroxylase |
5851 |
0.24 |
| chr20_33907930_33908081 | 0.22 |
FAM83C |
family with sequence similarity 83, member C |
27801 |
0.09 |
Network of associatons between targets according to the STRING database.

Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.2 | 0.5 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.1 | 0.4 | GO:0010766 | negative regulation of sodium ion transport(GO:0010766) |
| 0.1 | 0.5 | GO:0060437 | lung growth(GO:0060437) |
| 0.1 | 0.4 | GO:0050923 | regulation of negative chemotaxis(GO:0050923) |
| 0.1 | 0.3 | GO:0071692 | sequestering of extracellular ligand from receptor(GO:0035581) sequestering of TGFbeta in extracellular matrix(GO:0035583) protein localization to extracellular region(GO:0071692) maintenance of protein location in extracellular region(GO:0071694) extracellular regulation of signal transduction(GO:1900115) extracellular negative regulation of signal transduction(GO:1900116) |
| 0.1 | 0.1 | GO:0060648 | mammary gland bud morphogenesis(GO:0060648) |
| 0.1 | 0.3 | GO:0060083 | smooth muscle contraction involved in micturition(GO:0060083) |
| 0.1 | 0.5 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.1 | 0.4 | GO:0006930 | substrate-dependent cell migration, cell extension(GO:0006930) |
| 0.1 | 0.2 | GO:0060596 | mammary placode formation(GO:0060596) |
| 0.1 | 0.2 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.1 | 0.2 | GO:0002072 | optic cup morphogenesis involved in camera-type eye development(GO:0002072) |
| 0.1 | 0.2 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.1 | 0.1 | GO:0060594 | mammary gland specification(GO:0060594) |
| 0.1 | 0.1 | GO:0045636 | positive regulation of melanocyte differentiation(GO:0045636) positive regulation of pigment cell differentiation(GO:0050942) |
| 0.1 | 0.2 | GO:0060999 | positive regulation of dendritic spine development(GO:0060999) |
| 0.1 | 0.1 | GO:0006011 | UDP-glucose metabolic process(GO:0006011) |
| 0.1 | 0.2 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.2 | GO:0031394 | regulation of prostaglandin biosynthetic process(GO:0031392) positive regulation of prostaglandin biosynthetic process(GO:0031394) regulation of unsaturated fatty acid biosynthetic process(GO:2001279) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.2 | GO:0010800 | positive regulation of peptidyl-threonine phosphorylation(GO:0010800) |
| 0.0 | 0.1 | GO:0051305 | chromosome movement towards spindle pole(GO:0051305) |
| 0.0 | 0.0 | GO:0060242 | contact inhibition(GO:0060242) |
| 0.0 | 0.4 | GO:0035335 | peptidyl-tyrosine dephosphorylation(GO:0035335) |
| 0.0 | 0.5 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 0.2 | GO:0042023 | regulation of DNA endoreduplication(GO:0032875) negative regulation of DNA endoreduplication(GO:0032876) DNA endoreduplication(GO:0042023) |
| 0.0 | 0.1 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0001996 | positive regulation of heart rate by epinephrine-norepinephrine(GO:0001996) |
| 0.0 | 0.2 | GO:0043589 | skin morphogenesis(GO:0043589) |
| 0.0 | 0.2 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0060751 | epithelial cell proliferation involved in mammary gland duct elongation(GO:0060750) branch elongation involved in mammary gland duct branching(GO:0060751) |
| 0.0 | 0.3 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.4 | GO:0030219 | megakaryocyte differentiation(GO:0030219) |
| 0.0 | 0.1 | GO:0043497 | regulation of protein heterodimerization activity(GO:0043497) |
| 0.0 | 0.1 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0071502 | cellular response to temperature stimulus(GO:0071502) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:0042339 | keratan sulfate metabolic process(GO:0042339) |
| 0.0 | 0.1 | GO:0032328 | alanine transport(GO:0032328) |
| 0.0 | 0.1 | GO:0072033 | renal vesicle formation(GO:0072033) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0090195 | chemokine secretion(GO:0090195) regulation of chemokine secretion(GO:0090196) positive regulation of chemokine secretion(GO:0090197) |
| 0.0 | 0.1 | GO:0045722 | positive regulation of gluconeogenesis(GO:0045722) |
| 0.0 | 0.0 | GO:0006188 | IMP biosynthetic process(GO:0006188) IMP metabolic process(GO:0046040) |
| 0.0 | 0.1 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:2000008 | regulation of protein localization to cell surface(GO:2000008) |
| 0.0 | 0.1 | GO:0048845 | venous blood vessel morphogenesis(GO:0048845) |
| 0.0 | 0.1 | GO:0051451 | myoblast migration(GO:0051451) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.1 | GO:0050774 | negative regulation of dendrite morphogenesis(GO:0050774) |
| 0.0 | 0.1 | GO:0046882 | negative regulation of follicle-stimulating hormone secretion(GO:0046882) |
| 0.0 | 0.1 | GO:0045541 | negative regulation of cholesterol biosynthetic process(GO:0045541) negative regulation of cholesterol metabolic process(GO:0090206) |
| 0.0 | 0.1 | GO:0031657 | regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031657) positive regulation of cyclin-dependent protein serine/threonine kinase activity involved in G1/S transition of mitotic cell cycle(GO:0031659) |
| 0.0 | 0.0 | GO:0045989 | positive regulation of striated muscle contraction(GO:0045989) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.3 | GO:0031000 | response to caffeine(GO:0031000) |
| 0.0 | 0.1 | GO:0006048 | UDP-N-acetylglucosamine biosynthetic process(GO:0006048) |
| 0.0 | 0.1 | GO:0045628 | regulation of T-helper 2 cell differentiation(GO:0045628) |
| 0.0 | 0.1 | GO:0014012 | peripheral nervous system axon regeneration(GO:0014012) |
| 0.0 | 0.1 | GO:0006654 | phosphatidic acid biosynthetic process(GO:0006654) |
| 0.0 | 0.0 | GO:0034392 | negative regulation of smooth muscle cell apoptotic process(GO:0034392) |
| 0.0 | 0.1 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.1 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.0 | GO:0060687 | regulation of branching involved in prostate gland morphogenesis(GO:0060687) |
| 0.0 | 0.1 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.3 | GO:0043114 | regulation of vascular permeability(GO:0043114) |
| 0.0 | 0.1 | GO:0060997 | dendritic spine morphogenesis(GO:0060997) regulation of dendritic spine morphogenesis(GO:0061001) dendritic spine organization(GO:0097061) |
| 0.0 | 0.2 | GO:0045663 | positive regulation of myoblast differentiation(GO:0045663) |
| 0.0 | 0.1 | GO:0010107 | potassium ion import(GO:0010107) |
| 0.0 | 0.1 | GO:0072124 | glomerular mesangial cell proliferation(GO:0072110) regulation of glomerular mesangial cell proliferation(GO:0072124) |
| 0.0 | 0.2 | GO:0006907 | pinocytosis(GO:0006907) |
| 0.0 | 0.1 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:1902337 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) regulation of apoptotic process involved in morphogenesis(GO:1902337) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) regulation of apoptotic process involved in development(GO:1904748) |
| 0.0 | 0.3 | GO:0006957 | complement activation, alternative pathway(GO:0006957) |
| 0.0 | 0.1 | GO:0051497 | negative regulation of stress fiber assembly(GO:0051497) |
| 0.0 | 0.1 | GO:0002043 | blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:0002043) |
| 0.0 | 0.3 | GO:0048384 | retinoic acid receptor signaling pathway(GO:0048384) |
| 0.0 | 0.0 | GO:0010837 | regulation of keratinocyte proliferation(GO:0010837) |
| 0.0 | 0.0 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.0 | 0.0 | GO:0060998 | regulation of dendritic spine development(GO:0060998) |
| 0.0 | 0.0 | GO:0044557 | relaxation of smooth muscle(GO:0044557) relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.2 | GO:0034446 | substrate adhesion-dependent cell spreading(GO:0034446) |
| 0.0 | 0.2 | GO:0007616 | long-term memory(GO:0007616) |
| 0.0 | 0.1 | GO:0032376 | positive regulation of sterol transport(GO:0032373) positive regulation of cholesterol transport(GO:0032376) |
| 0.0 | 0.1 | GO:0060839 | endothelial cell fate commitment(GO:0060839) |
| 0.0 | 0.1 | GO:0051124 | synaptic growth at neuromuscular junction(GO:0051124) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0031033 | myosin filament organization(GO:0031033) |
| 0.0 | 0.3 | GO:0032012 | regulation of ARF protein signal transduction(GO:0032012) |
| 0.0 | 0.5 | GO:0048009 | insulin-like growth factor receptor signaling pathway(GO:0048009) |
| 0.0 | 0.2 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.5 | GO:0048010 | vascular endothelial growth factor receptor signaling pathway(GO:0048010) |
| 0.0 | 0.1 | GO:0021570 | rhombomere 4 development(GO:0021570) |
| 0.0 | 0.0 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0014056 | acetylcholine secretion, neurotransmission(GO:0014055) regulation of acetylcholine secretion, neurotransmission(GO:0014056) acetylcholine secretion(GO:0061526) |
| 0.0 | 0.1 | GO:0061384 | heart trabecula formation(GO:0060347) heart trabecula morphogenesis(GO:0061384) |
| 0.0 | 0.3 | GO:0007340 | acrosome reaction(GO:0007340) |
| 0.0 | 0.1 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0042753 | positive regulation of circadian rhythm(GO:0042753) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.1 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) tail-anchored membrane protein insertion into ER membrane(GO:0071816) |
| 0.0 | 0.1 | GO:0031641 | regulation of myelination(GO:0031641) |
| 0.0 | 0.0 | GO:0030146 | obsolete diuresis(GO:0030146) |
| 0.0 | 0.1 | GO:0010560 | positive regulation of glycoprotein biosynthetic process(GO:0010560) positive regulation of glycoprotein metabolic process(GO:1903020) |
| 0.0 | 0.4 | GO:0030199 | collagen fibril organization(GO:0030199) |
| 0.0 | 0.3 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0007026 | negative regulation of microtubule depolymerization(GO:0007026) |
| 0.0 | 0.1 | GO:0048013 | ephrin receptor signaling pathway(GO:0048013) |
| 0.0 | 0.1 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:2000144 | positive regulation of transcription initiation from RNA polymerase II promoter(GO:0060261) positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0048935 | peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.0 | GO:0021563 | glossopharyngeal nerve development(GO:0021563) glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:0019369 | arachidonic acid metabolic process(GO:0019369) |
| 0.0 | 0.1 | GO:0000729 | DNA double-strand break processing(GO:0000729) |
| 0.0 | 0.0 | GO:0051409 | response to nitrosative stress(GO:0051409) |
| 0.0 | 0.0 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0006002 | fructose 6-phosphate metabolic process(GO:0006002) |
| 0.0 | 0.1 | GO:0007008 | outer mitochondrial membrane organization(GO:0007008) protein import into mitochondrial outer membrane(GO:0045040) |
| 0.0 | 0.0 | GO:0031282 | regulation of guanylate cyclase activity(GO:0031282) |
| 0.0 | 0.1 | GO:0008634 | obsolete negative regulation of survival gene product expression(GO:0008634) |
| 0.0 | 0.1 | GO:0032486 | Rap protein signal transduction(GO:0032486) |
| 0.0 | 0.1 | GO:0071318 | cellular response to ATP(GO:0071318) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.1 | GO:0009169 | purine nucleoside monophosphate catabolic process(GO:0009128) ribonucleoside monophosphate catabolic process(GO:0009158) purine ribonucleoside monophosphate catabolic process(GO:0009169) |
| 0.0 | 0.0 | GO:0032288 | myelin assembly(GO:0032288) |
| 0.0 | 0.1 | GO:0046831 | regulation of RNA export from nucleus(GO:0046831) |
| 0.0 | 0.2 | GO:0032330 | regulation of chondrocyte differentiation(GO:0032330) |
| 0.0 | 0.0 | GO:0090036 | regulation of protein kinase C signaling(GO:0090036) positive regulation of protein kinase C signaling(GO:0090037) |
| 0.0 | 0.0 | GO:0021798 | forebrain dorsal/ventral pattern formation(GO:0021798) |
| 0.0 | 0.2 | GO:0060079 | excitatory postsynaptic potential(GO:0060079) |
| 0.0 | 0.0 | GO:0090136 | epithelial cell-cell adhesion(GO:0090136) |
| 0.0 | 0.0 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.1 | GO:0007567 | parturition(GO:0007567) |
| 0.0 | 0.1 | GO:0032693 | negative regulation of interleukin-10 production(GO:0032693) |
| 0.0 | 0.0 | GO:0060298 | positive regulation of cardiac muscle hypertrophy(GO:0010613) positive regulation of muscle hypertrophy(GO:0014742) positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.0 | GO:0051014 | actin filament severing(GO:0051014) |
| 0.0 | 0.0 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.0 | GO:0030309 | poly-N-acetyllactosamine metabolic process(GO:0030309) |
| 0.0 | 0.0 | GO:0051541 | elastin metabolic process(GO:0051541) |
| 0.0 | 0.2 | GO:0050999 | regulation of nitric-oxide synthase activity(GO:0050999) |
| 0.0 | 0.0 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
| 0.0 | 0.0 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.0 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:0019934 | cGMP-mediated signaling(GO:0019934) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.2 | GO:0030520 | intracellular estrogen receptor signaling pathway(GO:0030520) |
| 0.0 | 0.1 | GO:0032410 | negative regulation of transporter activity(GO:0032410) |
| 0.0 | 0.5 | GO:0007043 | cell-cell junction assembly(GO:0007043) |
| 0.0 | 0.0 | GO:0072385 | minus-end-directed organelle transport along microtubule(GO:0072385) |
| 0.0 | 0.0 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 0.0 | GO:0003148 | outflow tract septum morphogenesis(GO:0003148) |
| 0.0 | 0.0 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:1901983 | regulation of histone acetylation(GO:0035065) regulation of protein acetylation(GO:1901983) regulation of peptidyl-lysine acetylation(GO:2000756) |
| 0.0 | 0.1 | GO:0042249 | establishment of planar polarity of embryonic epithelium(GO:0042249) |
| 0.0 | 0.1 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.0 | GO:0003308 | negative regulation of Wnt signaling pathway involved in heart development(GO:0003308) |
| 0.0 | 0.3 | GO:0007098 | centrosome cycle(GO:0007098) |
| 0.0 | 0.0 | GO:0014721 | voluntary skeletal muscle contraction(GO:0003010) twitch skeletal muscle contraction(GO:0014721) |
| 0.0 | 0.0 | GO:0030488 | tRNA methylation(GO:0030488) |
| 0.0 | 0.0 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.0 | GO:0035020 | regulation of Rac protein signal transduction(GO:0035020) |
| 0.0 | 0.0 | GO:0050686 | negative regulation of mRNA processing(GO:0050686) negative regulation of mRNA metabolic process(GO:1903312) |
| 0.0 | 0.0 | GO:0006167 | AMP biosynthetic process(GO:0006167) |
| 0.0 | 0.1 | GO:0035414 | negative regulation of catenin import into nucleus(GO:0035414) |
| 0.0 | 0.1 | GO:0001893 | maternal placenta development(GO:0001893) |
| 0.0 | 0.1 | GO:0070071 | proton-transporting two-sector ATPase complex assembly(GO:0070071) |
| 0.0 | 0.2 | GO:0010811 | positive regulation of cell-substrate adhesion(GO:0010811) |
| 0.0 | 0.1 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0031146 | SCF-dependent proteasomal ubiquitin-dependent protein catabolic process(GO:0031146) |
| 0.0 | 0.0 | GO:0046826 | negative regulation of protein export from nucleus(GO:0046826) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.3 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.1 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.1 | 0.2 | GO:0032449 | CBM complex(GO:0032449) |
| 0.0 | 0.2 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.2 | GO:0031588 | nucleotide-activated protein kinase complex(GO:0031588) |
| 0.0 | 0.1 | GO:0036454 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.3 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.2 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0032059 | bleb(GO:0032059) |
| 0.0 | 0.1 | GO:0000306 | extrinsic component of vacuolar membrane(GO:0000306) extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.4 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.1 | GO:0005954 | calcium- and calmodulin-dependent protein kinase complex(GO:0005954) |
| 0.0 | 0.1 | GO:0098878 | ionotropic glutamate receptor complex(GO:0008328) neurotransmitter receptor complex(GO:0098878) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.0 | 0.1 | GO:0098645 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.0 | 0.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.3 | GO:0097517 | stress fiber(GO:0001725) contractile actin filament bundle(GO:0097517) |
| 0.0 | 0.1 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.1 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.2 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.0 | GO:0044291 | intercalated disc(GO:0014704) cell-cell contact zone(GO:0044291) |
| 0.0 | 0.1 | GO:0032156 | septin complex(GO:0031105) septin cytoskeleton(GO:0032156) |
| 0.0 | 0.1 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.1 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.0 | 0.1 | GO:0030315 | T-tubule(GO:0030315) |
| 0.0 | 0.1 | GO:0032588 | trans-Golgi network membrane(GO:0032588) |
| 0.0 | 0.1 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.4 | GO:0034704 | calcium channel complex(GO:0034704) |
| 0.0 | 0.1 | GO:0043034 | costamere(GO:0043034) |
| 0.0 | 0.0 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.1 | GO:0000796 | condensin complex(GO:0000796) |
| 0.0 | 0.0 | GO:0032432 | actin filament bundle(GO:0032432) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.9 | GO:0060076 | excitatory synapse(GO:0060076) |
| 0.0 | 0.0 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0043205 | fibril(GO:0043205) |
| 0.0 | 0.3 | GO:0000932 | cytoplasmic mRNA processing body(GO:0000932) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.0 | GO:0045254 | pyruvate dehydrogenase complex(GO:0045254) |
| 0.0 | 0.0 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.0 | 0.8 | GO:0001726 | ruffle(GO:0001726) |
| 0.0 | 0.9 | GO:0034705 | potassium channel complex(GO:0034705) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) core promoter proximal region sequence-specific DNA binding(GO:0000987) core promoter proximal region DNA binding(GO:0001159) |
| 0.2 | 0.7 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.2 | 0.5 | GO:0016842 | amidine-lyase activity(GO:0016842) |
| 0.1 | 0.5 | GO:0005010 | insulin-like growth factor-activated receptor activity(GO:0005010) |
| 0.1 | 0.3 | GO:0050508 | glucuronosyl-N-acetylglucosaminyl-proteoglycan 4-alpha-N-acetylglucosaminyltransferase activity(GO:0050508) |
| 0.1 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.1 | 0.3 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.2 | GO:0034056 | estrogen response element binding(GO:0034056) |
| 0.1 | 0.3 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.1 | 0.4 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 1.4 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.2 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.3 | GO:0043539 | protein serine/threonine kinase activator activity(GO:0043539) |
| 0.0 | 0.1 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.1 | GO:0005006 | epidermal growth factor-activated receptor activity(GO:0005006) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.2 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.0 | 0.1 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.4 | GO:0046970 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) histone deacetylase activity (H4-K16 specific)(GO:0034739) NAD-dependent histone deacetylase activity (H4-K16 specific)(GO:0046970) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.2 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.0 | 0.4 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.1 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0070548 | L-glutamine aminotransferase activity(GO:0070548) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.2 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 0.4 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.4 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.1 | GO:0031127 | galactoside 2-alpha-L-fucosyltransferase activity(GO:0008107) alpha-(1,2)-fucosyltransferase activity(GO:0031127) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.4 | GO:0008353 | RNA polymerase II carboxy-terminal domain kinase activity(GO:0008353) |
| 0.0 | 0.3 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.1 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.2 | GO:0003706 | obsolete ligand-regulated transcription factor activity(GO:0003706) |
| 0.0 | 0.1 | GO:0042153 | obsolete RPTP-like protein binding(GO:0042153) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0004948 | calcitonin receptor activity(GO:0004948) |
| 0.0 | 0.1 | GO:0016595 | glutamate binding(GO:0016595) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0016780 | phosphotransferase activity, for other substituted phosphate groups(GO:0016780) |
| 0.0 | 0.1 | GO:0042799 | histone methyltransferase activity (H4-K20 specific)(GO:0042799) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0000339 | RNA cap binding(GO:0000339) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.0 | GO:0005172 | vascular endothelial growth factor receptor binding(GO:0005172) |
| 0.0 | 0.1 | GO:0008266 | poly(U) RNA binding(GO:0008266) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) |
| 0.0 | 0.3 | GO:0005086 | ARF guanyl-nucleotide exchange factor activity(GO:0005086) |
| 0.0 | 0.1 | GO:0008484 | sulfuric ester hydrolase activity(GO:0008484) |
| 0.0 | 0.2 | GO:0030371 | translation repressor activity(GO:0030371) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.1 | GO:0004407 | histone deacetylase activity(GO:0004407) |
| 0.0 | 0.2 | GO:0035255 | ionotropic glutamate receptor binding(GO:0035255) |
| 0.0 | 0.1 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.1 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 0.1 | GO:0004791 | thioredoxin-disulfide reductase activity(GO:0004791) |
| 0.0 | 0.0 | GO:0016212 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.0 | 0.1 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.4 | GO:0005158 | insulin receptor binding(GO:0005158) |
| 0.0 | 0.2 | GO:0045296 | cadherin binding(GO:0045296) |
| 0.0 | 0.1 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0035035 | histone acetyltransferase binding(GO:0035035) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.1 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
| 0.0 | 0.1 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0003923 | GPI-anchor transamidase activity(GO:0003923) |
| 0.0 | 0.0 | GO:0031544 | procollagen-proline 3-dioxygenase activity(GO:0019797) peptidyl-proline 3-dioxygenase activity(GO:0031544) |
| 0.0 | 0.0 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0004579 | dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.0 | 0.1 | GO:0034416 | bisphosphoglycerate phosphatase activity(GO:0034416) |
| 0.0 | 0.1 | GO:0008242 | omega peptidase activity(GO:0008242) |
| 0.0 | 0.1 | GO:0005315 | inorganic phosphate transmembrane transporter activity(GO:0005315) |
| 0.0 | 0.1 | GO:0051879 | Hsp90 protein binding(GO:0051879) |
| 0.0 | 0.1 | GO:0008486 | diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) |
| 0.0 | 0.1 | GO:0001871 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0004622 | lysophospholipase activity(GO:0004622) |
| 0.0 | 0.1 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.0 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.7 | GO:0004714 | transmembrane receptor protein tyrosine kinase activity(GO:0004714) |
| 0.0 | 0.0 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.1 | GO:0005024 | transforming growth factor beta-activated receptor activity(GO:0005024) |
| 0.0 | 0.1 | GO:0090079 | translation regulator activity, nucleic acid binding(GO:0090079) |
| 0.0 | 0.0 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.0 | GO:0042805 | actinin binding(GO:0042805) |
| 0.0 | 0.1 | GO:0016565 | obsolete general transcriptional repressor activity(GO:0016565) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0008175 | tRNA methyltransferase activity(GO:0008175) |
| 0.0 | 0.1 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0031491 | nucleosome binding(GO:0031491) |
| 0.0 | 0.1 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0001727 | lipid kinase activity(GO:0001727) |
| 0.0 | 0.1 | GO:0005110 | frizzled-2 binding(GO:0005110) |
| 0.0 | 0.1 | GO:0070403 | NAD+ binding(GO:0070403) |
| 0.0 | 0.2 | GO:0005520 | insulin-like growth factor binding(GO:0005520) |
| 0.0 | 0.1 | GO:0015355 | secondary active monocarboxylate transmembrane transporter activity(GO:0015355) |
| 0.0 | 0.0 | GO:0016847 | 1-aminocyclopropane-1-carboxylate synthase activity(GO:0016847) |
| 0.0 | 0.1 | GO:0050780 | dopamine receptor binding(GO:0050780) |
| 0.0 | 0.0 | GO:0008192 | RNA guanylyltransferase activity(GO:0008192) |
| 0.0 | 0.0 | GO:0015193 | L-proline transmembrane transporter activity(GO:0015193) |
| 0.0 | 0.1 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.2 | GO:0030552 | cAMP binding(GO:0030552) |
| 0.0 | 0.0 | GO:0005148 | prolactin receptor binding(GO:0005148) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.4 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID S1P S1P3 PATHWAY | S1P3 pathway |
| 0.0 | 0.3 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.3 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.6 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.1 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 1.1 | PID AR PATHWAY | Coregulation of Androgen receptor activity |
| 0.0 | 0.0 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 0.4 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.0 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.1 | PID LYMPH ANGIOGENESIS PATHWAY | VEGFR3 signaling in lymphatic endothelium |
| 0.0 | 0.3 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.4 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
| 0.0 | 0.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.5 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 0.1 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 1.4 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.1 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.6 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.0 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.5 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.0 | 0.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.4 | REACTOME UNBLOCKING OF NMDA RECEPTOR GLUTAMATE BINDING AND ACTIVATION | Genes involved in Unblocking of NMDA receptor, glutamate binding and activation |
| 0.0 | 0.2 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.3 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME ROLE OF SECOND MESSENGERS IN NETRIN1 SIGNALING | Genes involved in Role of second messengers in netrin-1 signaling |
| 0.0 | 0.5 | REACTOME SMAD2 SMAD3 SMAD4 HETEROTRIMER REGULATES TRANSCRIPTION | Genes involved in SMAD2/SMAD3:SMAD4 heterotrimer regulates transcription |
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME CTLA4 INHIBITORY SIGNALING | Genes involved in CTLA4 inhibitory signaling |
| 0.0 | 0.2 | REACTOME REGULATION OF RHEB GTPASE ACTIVITY BY AMPK | Genes involved in Regulation of Rheb GTPase activity by AMPK |
| 0.0 | 0.8 | REACTOME NOTCH1 INTRACELLULAR DOMAIN REGULATES TRANSCRIPTION | Genes involved in NOTCH1 Intracellular Domain Regulates Transcription |
| 0.0 | 0.6 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.3 | REACTOME ACTIVATED POINT MUTANTS OF FGFR2 | Genes involved in Activated point mutants of FGFR2 |
| 0.0 | 0.4 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME REGULATION OF KIT SIGNALING | Genes involved in Regulation of KIT signaling |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.0 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.0 | REACTOME HEMOSTASIS | Genes involved in Hemostasis |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.1 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME RNA POL I TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase I Transcription Termination |
| 0.0 | 0.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.1 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.2 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.1 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME PRE NOTCH TRANSCRIPTION AND TRANSLATION | Genes involved in Pre-NOTCH Transcription and Translation |
| 0.0 | 0.2 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.2 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME AMINO ACID TRANSPORT ACROSS THE PLASMA MEMBRANE | Genes involved in Amino acid transport across the plasma membrane |
| 0.0 | 0.2 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.1 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.2 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.1 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |